Abstract
We used fluorescence in situ hybridization (FISH) assays to identify t(14;18) translocations in archival paraffin-embedded tumor sections from non-Hodgkin lymphoma (NHL) cases enrolled in a population-based study. t(14;18) was identified in 54% of 152 cases, including 39% of diffuse large cell lymphomas (26 of 66 cases) and 84% of follicular lymphomas (36 of 43 cases). Eighty-seven percent of t(14;18)-positive cases and 57% of t(14;18)-negative cases expressed bcl-2. FISH assays detected twice as many t(14;18)-positive follicular lymphomas as PCR assays. Overall, study findings support the use of FISH assays to detect t(14;18) in archival tumor samples for epidemiologic studies of NHL subtypes.
Keywords: FISH, translocation, lymphoma, t(14:18), bcl-2
1. Introduction
Classification of lymphomas according to common somatic mutations has been suggested to reduce the etiologic heterogeneity of lymphoma outcomes for epidemiologic research[1–3]. Many lymphoma-associated chromosomal translocations juxtapose the immunoglobulin heavy chain (IGH) promoter on chromosome 14 with an oncogene whose subsequent overexpression contributes to lymphomagenesis [1,4]. For example, (14;18) translocations join the IGH promoter with the BCL2 gene at 18q21, resulting in constitutive expression of the anti-apoptotic bcl-2 protein.
A previous study used polymerase chain reaction (PCR) assays to detect t(14;18) in archival tumor samples from a subset of participants in the Factors Affecting Rural Men (FARM) study [5,6]. The population-based case-control study reported subtype-specific associations with various pesticides and other exposures, thus supporting the use of NHL subtypes based on t(14;18) translocations as outcomes in etiologic research.
We used FISH to identify t(14;18) translocations in the FARM study samples, and we also evaluated bcl-2 protein expression based on immunohistochemistry. In this report, we describe the frequency of t(14;18) translocations and bcl-2 expression (overall and according to NHL subtype) in archival paraffin-embedded lymphoma samples from the population-based FARM study, compare PCR and FISH assay results for t(14;18), and discuss the utility of FISH-based molecular subtyping for epidemiologic research.
2. Materials and methods
2.1. Study population
The FARM study, a population-based study of incident, pathologically confirmed hemolymphatic cancer cases including NHL in Iowa and Minnesota men, was conducted to evaluate associations with agricultural, other occupational, lifestyle, and medical factors. The study population and methods were previously described in detail [7,8]. Briefly, cases diagnosed between 1980 and 1983 among white men aged 30 or older were identified through hospital and pathology laboratory records (in Minnesota) and the State Health Registry of Iowa. The study was restricted to white men from areas other than metropolitan Minnesota in order to increase the proportion with agricultural exposures, the primary focus of the study.
Eighty-nine percent (694) of 780 ascertained NHL cases were enrolled, and the diagnosis of NHL was confirmed for 622 cases by a pathology review panel [7–9]. NHL cases were limited to solid tumors; therefore lymphocytic leukemias were not included in the NHL case subgroup. Controls were 1,245 white males aged 30 or older without a history of hemolymphatic cancer that were frequency-matched to cases by age (within five-year groups), state, and vital status. Between 1981 and 1984, detailed information on agricultural exposures, other occupational exposures, medical history, smoking, and other potential risk factors for NHL were obtained through in-person structured interviews of cases and controls, or the next-of-kin of deceased cases and controls [7,10–12].
2.2. t(14;18)-PCR study
In 1997, archival paraffin-embedded tumor blocks were requested and successfully retrieved for 248 FARM study NHL cases (40% of all NHL cases in the original study) [5,6]. Over half of the tumor blocks that were not retrieved had been destroyed or lost; the remainder were held at institutions that declined to participate in the t(14;18)-NHL study. Up to sixteen 5-micron sections and sixteen 10-micron sections were cut from each archival block. A single pathologist (GAD) reviewed the first, middle, and last sections to confirm the presence of tumor tissue and assign NHL subtypes according to the Revised European American Lymphoma classification scheme (REAL) [13]. Five-micron sections and slides were processed with diethyl procarbonate (DEPC) to prevent DNAse or RNAse from interfering with future assays [14].
t(14;18)-PCR assays were performed on DNA extracted from 10-micron sections cut from archival tumor blocks [5,6]. Each reaction used a consensus primer corresponding to the J segment of the IGH gene on chromosome 14 (5′-ACCTGAGGAGACGGTGAGC-3′) and a second primer corresponding to the BCL2 major breakpoint region (MBR) on chromosome 18 (MBR1: 5′-GAGAGTTGCTTTACGTG-GCCTG-3′). Negative samples were subjected to a second PCR reaction with the IGH consensus primer and a second BCL2 primer 360 base pairs upstream of the MBR primer (MBR2: 5′-CGCTTGACTCCTTTACGTGCTG-3′). Amplified DNA was isolated, denatured, and separated by gel electrophoresis, and transferred to a nylon membrane using a Southern blot procedure. Amplification was confirmed using radiolabeled probes that hybridized to DNA adjacent to each BCL2 primer site. A 175-base pair segment of the human hemoglobin beta (HBB) gene was amplified as an internal positive control to confirm that the DNA extracted from each sample was adequate for PCR amplification of t(14;18). Fifty-seven samples that were HBB-negative were classified as inadequate for PCR. Amplification products from 20 of the 68 t(14;18)-positive samples were sequenced to confirm that unique translocation breakpoints were amplified from each sample.
2.3. t(14;18)-FISH study
For the current study, commercially available FISH IGH and t(14;18) assays were run on 5-micron sections archived from FARM study tumor blocks (Vysis/Abbot Molecular, Des Plaines, IL). All sections were stored at 4°C prior to use. NHL subtypes were updated according to a modified version of the WHO classification scheme based on morphology only [15]. Cases that would be classified as chronic lymphocytic leukemias/small lymphocytic lymphomas (CLL/SLL) according to the WHO scheme are referred to as small lymphocytic lymphomas (SLL) since study cases were limited to solid tumors only. The majority of cases classified as diffuse large cell lymphomas (DLCL) were probably diffuse large B cell lymphomas (DLBCL); however, B-cell status was not confirmed. Cases classified as Burkitt lymphomas (BL) include both Burkitt and Burkitt-like lymphomas.
Assays were run according to the manufacturer’s instructions (Abbot Molecular/Vysis, Des Plaines, IL) with minor modifications (as described below). Translocations involving IGH on chromosome 14 were identified using the LSI® IGH Dual Color, Break Apart Rearrangement Probe which includes a 900 kilobase pair (kb) SpectrumGreen-labeled probe that hybridizes to the entire IGH variable (V) region, and a 250 kb SpectrumOrange-labeled probe that hybridizes to the 3′ flanking region (Fig. 1A) (Abbot Molecular/Vysis, Des Plaines, IL). In a translocation-negative cell, the expected signal pattern is two orange/green (yellow) fusion signals (Fig. 2A). In translocation-positive cells, separation of one pair of probes produces one orange and one green signal in addition to the fusion signal associated with the normal copy of IGH (Fig. 2B). t(14;18) translocations were identified using the LSI® IGH/BCL2 Dual Color, Dual Fusion Translocation Probe which includes a 1.5 mega base pair SpectrumGreen probe that spans the entire IGH locus plus 300 kb centromerically, and a 750 kb SpectrumOrange probe that covers BCL2 plus 250 kb beyond each end of the gene (Fig. 1C). The expected signal pattern in a normal nucleus is two orange and two green signals (Fig. 2C). In translocation-positive cells, juxtaposition of an orange and green probe produces one fusion signal in addition to the separate orange and green signals associated with the normal copies of IGH and BCL2 (Fig. 2D).
Fig. 1.
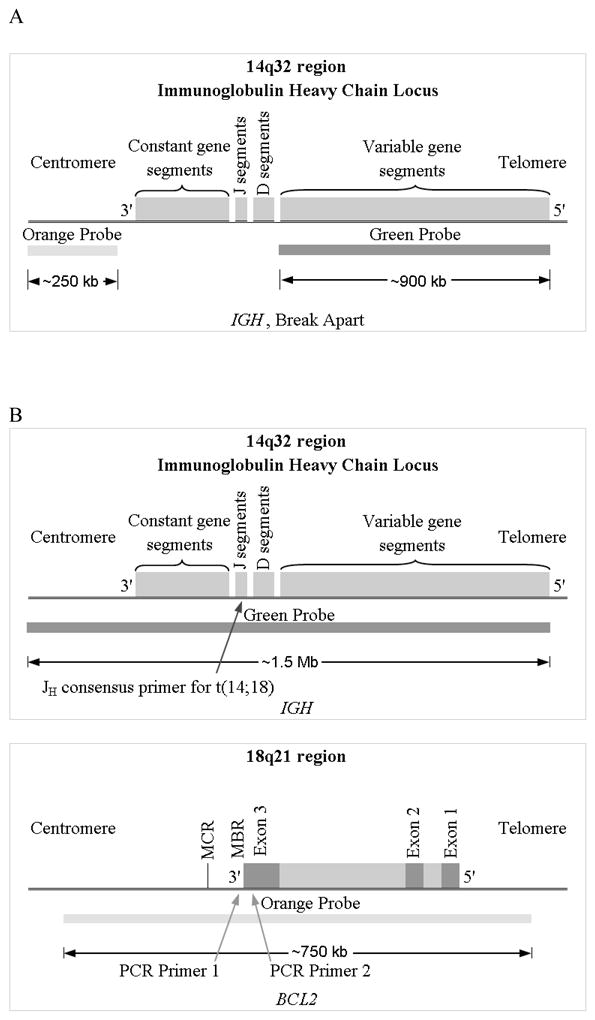
Gene regions corresponding to FISH probes used to detect chromosomal translocations. (A) IGH gene regions corresponding to LSI IGH Dual Color Break Apart probes used to detect IGH translocations (regardless of the partner gene). (B) Regions of IGH (upper) and BCL2 (lower) corresponding to LSI IGH/BCL2 Dual Color Dual Fusion probes to detect t(14;18). Arrows indicate the location of primers used in previous polymerase chain reaction (PCR) assays, including the Jh consensus primer on chromosome 14, the MBR1 primer for the BCL2 major breakpoint region (MBR) on chromosome 18, and the MBR2 primer 360 base pairs upstream of MBR1 (adapted from Abbot Molecular/Vysis, Des Plaines, IL).
Fig. 2.
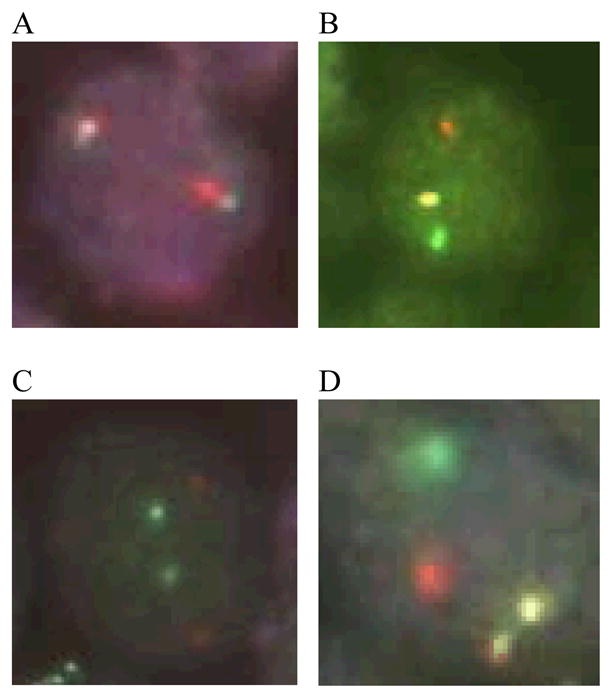
Photomicrographs showing signal patterns expected for FISH assays of normal cells and translocation positive cells. (A) Vysis LSI IGH Dual Color, Break Apart probe signal pattern expected for a translocation-negative cell (2 fusion signals) and (B) an IGH-positive cell (1 orange, 1 green, and 1 fusion signal). (C) Vysis LSI IGH/BCL2 Dual Color, Dual Fusion probe signal pattern expected for a t(14;18)-negative cell (2 orange, 2 green) and (D) a t(14;18)-positive cell (1 orange, 1 green, and 2 fusion signals).
We assayed all cases with at least two 5-micron sections available (n = 230) for any IGH translocation (IGH-FISH). To increase efficiency t(14;18)-FISH assays were performed only on samples that were of adequate quality to successfully classify them as positive or negative based on the IGH assay (n=166). Deparaffinization and pretreatment of archival 5-micron sections was performed using the Vysis® VP2000™ Processor (Abbott Molecular/Vysis, Des Plaines, IL). Next, a probe mixture containing a 7:1:2 ratio of buffer, fluorescent probe, and distilled water was applied to each slide. After a coverslip was placed over the tissue and sealed with rubber cement, the slide was placed in a HYBrite™ machine (Abbott Molecular/Vysis, Des Plaines, IL) where denaturation occurred at 75°C for 6 minutes, followed by hybridization at 37°C for 16 to 18 hours.
A rapid wash procedure was performed to remove unbound probe. Rubber cement was removed and slides were soaked for 60 minutes at ambient temperature in a 70 ml of solution of sodium chloride, sodium citrate and non-ionic detergent (2× SSC 0.1 NP40, pH 7–7.5). Next, coverslips were removed and slides were soaked for 2 minutes in 70 ml of 2× SSC 0.1 NP40 heated to 73°C. After slides were air-dried in darkness, 10μl of DAPI II counterstain was applied and a new coverslip was placed over the target area. Newly assayed slides were stored at −20°C until scoring.
2.4. FISH Scoring
FISH assayed sections were scored at room temperature using the 40× and 100× oil objectives on a Zeiss Axioskope 2 (Carl Zeiss, Thornwood, NY) equipped with a triple bandpass filter set (DAPI/Green/Orange V.2, Abbott Molecular/Vysis, Des Plaines, IL). Image capture was done using a color digital Axiocam 412–312 (Carl Zeiss, Thornwood, NY), and the software Openlab version 4.0.3 (Improvision, Inc, Lexington, MA).
Criteria for scoring were established before performing FISH assays. In other FISH studies, the cutoff commonly used is 5% (5 cells with translocation out of 100 cells) [16–21]. However, the samples in this study had fewer readable cells, thus the cutoff was more conservative. Cases were classified as translocation-positive if more than 30% of cells showed abnormal signal patterns. For each sample, a minimum of 25 non-overlapping lymphocytes with complete FISH signals were scored; samples with fewer than 25 readable cells were not classified. A complete FISH signal was defined as one that included the full set of orange, green or fusion signals expected in a positive or negative cell; therefore we did not score lymphocytes with truncated nuclei and incomplete signals, or overlapping cells with signals that could not be differentiated. In addition, the strength of both the orange and green FISH signals had to be adequate to distinguish separate signals from fusion signals. The appearance and quality of FISH signals varied depending on the source of the tissue, the adequacy and type of sample fixation, and the overall quality of the tissue section. Samples in which the quality of the sample or the strength of the FISH signals were inadequate to reliably identify positive cells (i.e. samples that did not include at least 25 cells that could be scored) were classified as unreadable for the assay. Cases that were unreadable for the IGH assay (n=64) were automatically classified as unreadable for t(14;18).
Cells other than lymphocytes, for example, epithelial or connective tissue cells in biopsies from extranodal sites, were not scored. Clonal evolution may result in both positive and negative neoplastic clones within a single tumor, and the distribution and clustering of neoplastic and non-neoplastic lymphocytes may vary depending on the tumor site and NHL subtype; therefore, each section was examined in its entirety for positive lymphocytes before being classified as translocation-negative. Non-neoplastic lymphocytes, which were present in all samples, served as internal negative controls.
t(14;18) assays were scored by two investigators (CC and JS). Concordance between the independent scores was 72% (121/169), including 9 cases that were classified as unreadable by both scorers. Discordant cases included sections that were classified as unreadable by only one scorer (26 cases). Discordant cases were subsequently reviewed and assigned a consensus score when possible, resulting in 152 successfully classified cases.
2.5. bcl-2 expression
bcl-2 immunostaining was performed on paraffin sections from 229 FARM study cases using a mouse monoclonal antibody (clone 124; Dako) corresponding to bcl-2 amino acids 41–54 (Dako Denmark A/S, Copenhagen, Denmark.). Cases were classified as bcl-2 positive if more than 20% of cells showed cytoplasmic staining. Twenty-one samples that were inadequate for scoring were classified as missing for bcl-2 expression. Scoring was performed by two investigators (WYH and JS). Independent scores were concordant for 92% of the cases assayed (191/208).
2.6. Data analysis
FISH data were entered into an Access database (Microsoft Corp., Redmond, WA). Univariate analyses were performed based on data from 227 cases with complete FISH, PCR, and immunostaining data using SAS® version 9.1. [22] The distribution of cases according to t(14;18) and bcl-2 expression was determined by WHO subtype, and t(14;18)-FISH assay results were compared with results of previous t(14;18)-PCR assays run on the same samples.
3. Results
3.1. Distribution of translocations detected by FISH
t(14;18)-FISH assays were successful on 152 out of 227 cases with complete FISH, bcl-2 and PCR data, including 82 (54%) that were t(14;18)-positive. Eighty-four percent of follicular lymphomas (FL) (36 of 43 FL cases successfully assayed) were t(14;18)-positive (Table 1). In comparison, only 44% of the FL cases successfully assayed by PCR were t(14;18)-positive (data not shown).
Table 1.
t(14;18)-positive lymphomas according to histologic subtype based on FISH assays of archival paraffin-embedded tumor samplesa.
| NHL Subtype | t(14;18)-positive | t(14;18)-negative | ||
|---|---|---|---|---|
| N | % | N | % | |
| Follicular | 36 | 84 | 7 | 16 |
| Diffuse Large Cell | 26 | 39 | 40 | 61 |
| Small lymphocytic | 5 | 33 | 10 | 67 |
| Burkitt/Burkitt-like | 6 | 50 | 6 | 50 |
| Mantle Cell | 3 | 100 | 0 | 0 |
| Marginal zone | 0 | 0 | 1 | 100 |
| Lymphoblastic | 0 | 0 | 1 | 100 |
| Lymphoplasmacytic | 0 | 0 | 1 | 100 |
| Unclassified | 6 | 60 | 4 | 40 |
| Total | 82 | 54 | 70 | 46 |
NHL subtypes were determined using a modified (World Health Organization) WHO classification scheme based on morphology only. Translocation status was determined using commercially available FISH assays for t(14;18) (Vysis, Abbott Molecular Inc., Des Plaines, IL).
3.2. bcl-2 expression
Over 75% of cases assayed for bcl-2 expression by immunostaining (157 of 206 readable cases) were bcl-2 positive, including 55 of 59 FL and 29 of 29 SLL (Table 2). bcl-2 expression was more likely to be positive in t(14;18)-positive cases (87% bcl-2 positive, 67 cases) than t(14;18)-negative cases (57% positive, 39 cases).
Table 2.
bcl-2 positive lymphomas according to histologic subtype based on IHC assays of archival paraffin-embedded tumor samples
| NHL Subtype | bcl-2-positive | bcl-2-negative | ||
|---|---|---|---|---|
| N | % | N | % | |
| Follicular | 55 | 93 | 4 | 7 |
| Diffuse Large Cell | 43 | 56 | 34 | 44 |
| Small lymphocytic | 29 | 100 | 0 | 0 |
| Burkitt/Burkitt-like | 7 | 50 | 7 | 50 |
| Mantle Cell | 4 | 100 | 0 | 0 |
| Marginal zone | 2 | 100 | 0 | 0 |
| Lymphoblastic | 0 | 0 | 1 | 100 |
| Lymphoplasmacytic | 1 | 100 | 0 | 0 |
| Unclassified | 16 | 84 | 3 | 16 |
| Total | 157 | 76 | 49 | 24 |
NHL subtypes were determined using a modified (World Health Organization) WHO classification scheme based on morphology only. bcl-2 expression was determined using a mouse monoclonal antibody (clone 124; Dako Denmark A/S, Copehagen, Denmark) corresponding to bcl-2 amino acids 41–54 run on paraffin-embedded tumor sections. 21 cases were not successfully assayed for bcl-2 expression.
3.3. t(14;18)-FISH versus t(14;18)-PCR
PCR and FISH assays for t(14;18) were concordant for 70% (92/132) of cases successfully evaluated by both assays, including 39 that were t(14;18)-positive and 53 that were t(14;18)-negative (Table 3). Among the discordant cases, 32 were PCR-negative/FISH-positive (18 FL, 4 DLCL, 94% bcl-2 positive), and 7 cases were PCR-positive/FISH-negative (1 FL, 3 DLCL, 86% bcl-2 positive). Overall, cases were more likely to be classified as t(14;18)-positive based on FISH (82 of 152 cases, 54%) than PCR (60 of 170 cases, 35%). The success rate (proportion of cases with interpretable results) of PCR assays (170 of 227 cases, 75%) was slightly higher than that of FISH assays (152 of 227 cases, 67%), but almost half of the samples that were inadequate for FISH (37 of 75) were also inadequate for PCR.
Table 3.
t(14;18)-positive and t(14 ;18)-negative cases based on FISH versus t(14;18)-PCR assays run on archival paraffin-embedded lymphoma samples
| t(14;18)-FISH | t(14;18)-PCR | |||
|---|---|---|---|---|
| Positive | Negative | Unreadableb | Total | |
| Positive | 40 | 32 | 10 | 82 |
| Negative | 7 | 53 | 10 | 70 |
| Unreadablea | 13 | 25 | 37 | 75 |
| Total | 60 | 110 | 57 | 227 |
t(14;18) status was determined using Vysis LSI IGH/BCL2 Dual Color, Dual Fusion FISH assays. PCR assays used a Jh consensus primer, an MBR1 primer corresponding to the BCL2 major breakpoint region (MBR), and a second primer 360 base pairs upstream of MBR1. Assays were run on archival paraffin-embedded tumor sections.
Unreadable FISH assays could not be scored because of inadequate sample or FISH signal strength.
Unreadable PCR assays were negative for the human hemoglobin beta (HBB) internal control and t(14;18).
4. Discussion
In general, FISH assay results were consistent with expectations based on previous studies, with t(14;18) identified in 84% of follicular lymphomas (FL) and 39% of diffuse large cell lymphomas (DLCL). Other FISH-based studies of FL in the United States (US), Canada, Europe, China and Malaysia have reported comparable proportions of t(14;18) positive cases (74–100%); however, Japanese studies have reported lower prevalences (56–64%) (Table 4a). US studies have also reported similar prevalences of t(14;18) among DLCL (20–32%), but results for study populations from other countries have varied widely (15–94%) (Table 4b). Some of the variation in t(14;18) lymphomas may reflect biological differences among populations [23], but differences in case selection and assay protocols may also contribute to variation among studies. Variation in the prevalence of t(14;18) DLCL also may reflect differences in diagnostic criteria among pathologists, who have been shown to be much less likely to agree on DLCL versus FL diagnoses [24].
Table 4.
Proportions of t(14;18)-positive lymphomas in selected studies of cases classified using FISH assays
| a. FOLLICULAR LYMPHOMA | |||||
|---|---|---|---|---|---|
| Study, year | Locationa | Case sampleb | Specimenc | Total cases | t(14;18) |
| N | % | ||||
| Current Study | US | Population- based | Paraffin | 43 | 84 |
| Gong et al., 2003 [34] | US | Hospital-based | FNA | 40 | 85 |
| Chiu et al., 2007d [31] | US | Population- based | Paraffin | 63 | 67 |
| Richmond et al., 2006 [35] | US | Hospital-based | FNA | 35 | 81 |
| Hirose et al., 2003 [36] | Japan | Convenience | Paraffin | 11 | 64 |
| Matsumoto et al., 2004 [37] | Japan | Hospital-based | Paraffin | 50 | 56 |
| Guo et al., 2005 [38] | China | Convenience | Frozen | 147 | 81 |
| Sekiguchi et al., 2005 [39] | Japan | Hospital-based | Paraffin | 47 | 60 |
| Peh et al., 2004 [40] | Malaysia | Hospital-based | Paraffin | 62 | 74 |
| Fan et al., 2003 [41] | Canada | Hospital-based | Paraffin | 24 | 83 |
| Godon et al., 2003 [19] | France | Convenience | Frozen | 63 | 100 |
| Bernicot et al., 2005 [42] | France | Hospital-based | Metaphase | 20 | 85 |
| Vaandrager et al., 2000e [43] | Netherlands | Convenience | Frozen | 40 | 88 |
| b. DIFFUSE LARGE B CELL LYMPHOMA | |||||
|---|---|---|---|---|---|
| Study | Locationa | Cases sampleb | Specimenc | Total Cases | t(14;18) |
| N | % | ||||
| Current Study | US | Population- based | Paraffin | 66 | 39 |
| Huang et al., 2002d [44] | US | Not specified | Paraffin | 35 | 20 |
| Gong et al., 2003 [34] | US | Hospital-based | FNA | 17 | 29 |
| Iqbal et al., 2004 [45] | US | Hospital-based | Paraffin | 141 | 24 |
| Chiu et al., 2007d [31] | US | Population- based | Paraffin | 65 | 32 |
| Cerretini et al., 2006 [46] | Argentina | Hospital-based | Metaphase | 30 | 30 |
| Hirose et al., 2003 [36] | Japan | Convenience | Paraffin | 61 | 15 |
| Godon et al., 2003 [19] | France | Convenience | Frozen | 17 | 94 |
| Bernicot et al., 2005 [42] | France | Hospital-based | Metaphase | 26 | 19 |
Country in which study was conducted: US, United States
Population-based: cases identified through state or national registries; Hospital-based: case series; Convenience: samples obtained from tissue/cytology files or banks
Paraffin: paraffin-embedded tumor sections; FNA: fine needle aspirate; Frozen: fresh-frozen tumor sample; Metaphase: metaphase spread from fresh tumor tissue.
FISH assays run on four representative 0.6-mm cores from each biopsy included on a tissue microarray.
FISH assays performed using yeast-artificial-chromosome (YAC)-DNA probes.
bcl-2 was expressed by the majority of cases in our population, but was more prevalent for t(14;18) positive (87%) than t(14;18) negative (57%) cases, consistent with expectations. In t(14;18) negative lymphomas bcl-2 expression may be increased by BCL2 amplification or hypomethylation, in response to EBV or CMV viral proteins, or through other mechanisms [25,26].
Overall, our results suggest that FISH was more sensitive than PCR for detecting t(14;18)-positive tumors, consistent with previous studies [16–19,27,28]. We expected that up to 25% of true t(14;18)-positive cases would have been false-negative based on PCR [29], which cannot detect translocation breakpoints that occur outside of the regions bounded by PCR primers. In addition, PCR may be less specific than FISH because of its potential to detect rare non-neoplastic t(14;18)-positive lymphocytes that may be present in biopsy samples [30]. This may explain discordant results for some or all the 7 study cases that were FISH-negative but PCR-positive for t(14;18). Sample contamination could also have caused false-positive PCR assays, but precautions were taken to prevent contamination, and sequencing confirmed unique t(14;18) breakpoints in a subset of samples [5].
Although we cannot confirm the sensitivity and specificity of FISH and PCR relative to a “gold standard” cytogenetic assay, we believe that the weight of the evidence supports FISH over PCR as the method of choice for detecting t(14;18) in archival samples for observational epidemiologic research. FISH also has advantages over karyotyping for observational research because it does not require fresh tissue, and can be performed on readily available archival samples, including paraffin-embedded sections over 25 years old, as in our study. In addition, running in situ FISH assays directly on tumor sections facilitates localization to neoplastic cells [29].
Disadvantages of FISH assays of preserved samples include the inability to score overlapping or truncated cells, which contributed to missing outcome data for approximately one-third of the cases assayed in our study. The cost of commercial FISH probes and the labor and training required to perform and score assays is also a concern, though costs and labor may be reduced through the use of tissue microarrays (TMAs) [31–33] and automated scoring [21].
In summary, the frequency of common translocations identified using FISH assays in a population-based sample of lymphoma cases was generally consistent with expectations. FISH appeared to be more sensitive to detect t(14;18)-positive tumors than PCR, though we were unable to formally evaluate either assay against a cytogenetic “gold standard.” Large-scale epidemiologic studies of lymphoma subtypes should consider the advantages of FISH over other assays and incorporate strategies for minimizing time and resources (e.g., by running assays on TMA sections). Large numbers of cases are needed to provide sufficient power to assess risk factors for translocation subtypes in population-based samples of lymphoma cases. However, the use of FISH assays to classify lymphomas according to t(14;18) may provide a relatively simple and efficient means of discovering risk factors and etiologic mechanisms for a common subset of lymphomas.
Acknowledgments
This study was supported in part by the UNC Lineberger Comprehensive Cancer Center (Population Sciences Research Award), the National Institute of Environmental Health Sciences (P30ES10126), the National Institutes of Health, National Cancer Institute (R03 CA71617-01), and the NIH Intramural Research Program (Division of Cancer Epidemiology and Genetics, National Cancer Institute). Special thanks to Sophia Wang and Charles Rabkin for their helpful comments, David W. Cowan from the Lineberger Comprehensive Cancer Center Immunohistochemistry/Hybridization Core Laboratory for guidance with FISH assays; Stephen Oglesbee and Boyd Yount from UNC-Chapel Hill; and Paige Tolbert from Emory University.
References
- 1.Medeiros LJ, Bagg A, Cossman J. Molecular genetic in the diagnosis and classification of lymphoid neoplasms. In: Jaffe ES, editor. Surgical Pathology of the Lymph Nodes and Related Organs. 2. Philadelphia: WB Saunders; 1994. pp. 58–97. [Google Scholar]
- 2.Potter M. Pathogenetic mechanisms in B-cell non-Hodgkin’s lymphomas in humans. Cancer Res. 1992;52:5522s–5528s. [PubMed] [Google Scholar]
- 3.Weisenburger DD. Pathological classification of non-Hodgkin’s lymphoma for epidemiological studies. Cancer Res. 1992;52:5456s–5462s. discussion 5462s–5464s. [PubMed] [Google Scholar]
- 4.Magrath I. Molecular basis of lymphomagenesis. Cancer Res. 1992;52:5529s–5540s. [PubMed] [Google Scholar]
- 5.Schroeder JC, Olshan AF, Baric R, Dent GA, Weinberg CR, Yount B, Cerhan JR, Lynch CF, Schuman LM, Tolbert PE, et al. Agricultural risk factors for t(14;18) subtypes of non-Hodgkin’s lymphoma. Epidemiology. 2001;12:701–9. doi: 10.1097/00001648-200111000-00020. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder JC, Olshan AF, Baric R, Dent GA, Weinberg CR, Yount B, Cerhan JR, Lynch CF, Schuman LM, Tolbert PE, et al. A case-control study of tobacco use and other non-occupational risk factors for t(14;18) subtypes of non-Hodgkin’s lymphoma (United States) Cancer Causes Control. 2002;13:159–68. doi: 10.1023/a:1014397920185. [DOI] [PubMed] [Google Scholar]
- 7.Blair A, Linos A, Stewart PA, Burmeister LF, Gibson R, Everett G, Schuman L, Cantor KP. Evaluation of risks for non-Hodgkin’s lymphoma by occupation and industry exposures from a case-control study. Am J Ind Med. 1993;23:301–12. doi: 10.1002/ajim.4700230207. [DOI] [PubMed] [Google Scholar]
- 8.Brown LM, Blair A, Gibson R, Everett GD, Cantor KP, Schuman LM, Burmeister LF, Van Lier SF, Dick F. Pesticide exposures and other agricultural risk factors for leukemia among men in Iowa and Minnesota. Cancer Res. 1990;50:6585–91. [PubMed] [Google Scholar]
- 9.Dick F, VanLier S, Banks P, Frizzera G, Witrak G, Gibson R, Everett G, Schuman L, Isacson P, O’Conor G, et al. Use of the working formulation for non-Hodgkin’s lymphoma in epidemiologic studies: agreement between reported diagnoses and a panel of experienced pathologists. J Natl Cancer Inst. 1987;78:1137–44. [PubMed] [Google Scholar]
- 10.Cantor KP, Blair A, Everett G, VanLier S, Burmeister L, Dick FR, Gibson RW, Schuman L. Hair dye use and risk of leukemia and lymphoma. Am J Public Health. 1988;78:570–1. doi: 10.2105/ajph.78.5.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linos A, Blair A, Gibson RW, Everett G, Van Lier S, Cantor KP, Schuman L, Burmeister L. Leukemia and non-Hodgkin’s lymphoma and residential proximity to industrial plants. Arch Environ Health. 1991;46:70–4. doi: 10.1080/00039896.1991.9937431. [DOI] [PubMed] [Google Scholar]
- 12.Pottern LM, Linet M, Blair A, Dick F, Burmeister LF, Gibson R, Schuman LM, Fraumeni JF., Jr Familial cancers associated with subtypes of leukemia and non-Hodgkin’s lymphoma. Leuk Res. 1991;15:305–14. doi: 10.1016/0145-2126(91)90005-e. [DOI] [PubMed] [Google Scholar]
- 13.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC, et al. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 1994;84:1361–92. [PubMed] [Google Scholar]
- 14.Schroeder JC. Agricultural and non-occupational risk factors for t(14;18) non-Hodgkin lymphoma. Chapel Hill: University of North Carolina; 1999. p. 157. [Google Scholar]
- 15.Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. JW Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARCPress; 2001. [Google Scholar]
- 16.Barrans SL, Evans PA, O’Connor SJ, Owen RG, Morgan GJ, Jack AS. The detection of t(14;18) in archival lymph nodes: development of a fluorescence in situ hybridization (FISH)-based method and evaluation by comparison with polymerase chain reaction. J Mol Diagn. 2003;5:168–75. doi: 10.1016/S1525-1578(10)60469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belaud-Rotureau MA, Parrens M, Carrere N, Turmo M, Ferrer J, de Mascarel A, Dubus P, Merlio JP. Interphase fluorescence in situ hybridization is more sensitive than BIOMED-2 polymerase chain reaction protocol in detecting IGH-BCL2 rearrangement in both fixed and frozen lymph node with follicular lymphoma. Hum Pathol. 2007;38:365–72. doi: 10.1016/j.humpath.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 18.Einerson RR, Kurtin PJ, Dayharsh GA, Kimlinger TK, Remstein ED. FISH is superior to PCR in detecting t(14;18)(q32;q21)-IgH/bcl-2 in follicular lymphoma using paraffin-embedded tissue samples. Am J Clin Pathol. 2005;124:421–9. doi: 10.1309/BLH8-MMK8-5UBQ-4K6R. [DOI] [PubMed] [Google Scholar]
- 19.Godon A, Moreau A, Talmant P, Baranger-Papot L, Genevieve F, Milpied N, Zandecki M, Avet-Loiseau H. Is t(14;18)(q32;q21) a constant finding in follicular lymphoma? An interphase FISH study on 63 patients. Leukemia. 2003;17:255–9. doi: 10.1038/sj.leu.2402739. [DOI] [PubMed] [Google Scholar]
- 20.Gu K, Chan WC, Hawley RC. Practical detection of t(14;18)(IgH/BCL2) in follicular lymphoma. Arch Pathol Lab Med. 2008;132:1355–61. doi: 10.5858/2008-132-1355-PDOBIF. [DOI] [PubMed] [Google Scholar]
- 21.Reichard KK, Hall BK, Corn A, Foucar MK, Hozier J. Automated analysis of fluorescence in situ hybridization on fixed, paraffin-embedded whole tissue sections in B-cell lymphoma. Mod Pathol. 2006;19:1027–33. doi: 10.1038/modpathol.3800630. [DOI] [PubMed] [Google Scholar]
- 22.SAS. Institute Inc. SAS®: Version 9.1. Cary (NC): SAS Institute, Inc; Copyright@2002–2003. [Google Scholar]
- 23.Segel MJ, Paltiel O, Zimran A, Gottschalk-Sabag S, Schibi G, Krichevski S, Ludkovski O, Ben Yehuda D. Geographic variance in the frequency of the t(14;18) translocation in follicular lymphoma: an Israeli series compared to the world. Blood Cells Mol Dis. 1998;24:62–72. doi: 10.1006/bcmd.1998.0171. [DOI] [PubMed] [Google Scholar]
- 24.Dick FR, VanLier SF, McKeen K, Everett GD, Blair A. Nonconcurrence in abstracted diagnoses of non-Hodgkin’s lymphoma. J Natl Cancer Inst. 1987;78:675–8. [PubMed] [Google Scholar]
- 25.Kusumoto S, Kobayashi Y, Sekiguchi N, Tanimoto K, Onishi Y, Yokota Y, Watanabe T, Maeshima AM, Ishida T, Inagaki H, et al. Diffuse large B-cell lymphoma with extra Bcl-2 gene signals detected by FISH analysis is associated with a “non-germinal center phenotype”. Am J Surg Pathol. 2005;29:1067–73. [PubMed] [Google Scholar]
- 26.Papakonstantinou G, Verbeke C, Hastka J, Bohrer M, Hehlmann R. bcl-2 expression in non-Hodgkin’s lymphomas is not associated with bcl-2 gene rearrangements. Br J Haematol. 2001;113:383–90. doi: 10.1046/j.1365-2141.2001.02727.x. [DOI] [PubMed] [Google Scholar]
- 27.Poetsch M, Weber-Matthiesen K, Plendl HJ, Grote W, Schlegelberger B. Detection of the t(14;18) chromosomal translocation by interphase cytogenetics with yeast-artificial-chromosome probes in follicular lymphoma and nonneoplastic lymphoproliferation. J Clin Oncol. 1996;14:963–9. doi: 10.1200/JCO.1996.14.3.963. [DOI] [PubMed] [Google Scholar]
- 28.Espinet B, Bellosillo B, Melero C, Vela MC, Pedro C, Salido M, Pijuan L, Florensa L, Besses C, Serrano S, et al. FISH is better than BIOMED-2 PCR to detect IgH/BCL2 translocation in follicular lymphoma at diagnosis using paraffin-embedded tissue sections. Leuk Res. 2008;32:737–42. doi: 10.1016/j.leukres.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Medeiros LJ, Carr J. Overview of the role of molecular methods in the diagnosis of malignant lymphomas. Arch Pathol Lab Med. 1999;123:1189–207. doi: 10.5858/1999-123-1189-OOTROM. [DOI] [PubMed] [Google Scholar]
- 30.Spagnolo DV, Ellis DW, Juneja S, Leong AS, Miliauskas J, Norris DL, Turner J. The role of molecular studies in lymphoma diagnosis: a review. Pathology. 2004;36:19–44. doi: 10.1080/00313020310001648404. [DOI] [PubMed] [Google Scholar]
- 31.Chiu BC, Dave BJ, Blair A, Gapstur SM, Chmiel JS, Fought AJ, Zahm SH, Weisenburger DD. Cigarette smoking, familial hematopoietic cancer, hair dye use, and risk of t(14;18)-defined subtypes of non-Hodgkin’s lymphoma. Am J Epidemiol. 2007;165:652–9. doi: 10.1093/aje/kwk044. [DOI] [PubMed] [Google Scholar]
- 32.Chiu BC, Dave BJ, Blair A, Gapstur SM, Zahm SH, Weisenburger DD. Agricultural pesticide use and risk of t(14;18)-defined subtypes of non-Hodgkin lymphoma. Blood. 2006;108:1363–9. doi: 10.1182/blood-2005-12-008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu BC, Dave BJ, Ward MH, Fought AJ, Hou L, Jain S, Gapstur S, Evens AM, Zahm SH, Blair A, et al. Dietary factors and risk of t(14;18)-defined subgroups of non-Hodgkin lymphoma. Cancer Causes Control. 2008;19:859–67. doi: 10.1007/s10552-008-9148-3. [DOI] [PubMed] [Google Scholar]
- 34.Gong Y, Caraway N, Gu J, Zaidi T, Fernandez R, Sun X, Huh YO, Katz RL. Evaluation of interphase fluorescence in situ hybridization for the t(14;18)(q32;q21) translocation in the diagnosis of follicular lymphoma on fine-needle aspirates: a comparison with flow cytometry immunophenotyping. Cancer. 2003;99:385–93. doi: 10.1002/cncr.11787. [DOI] [PubMed] [Google Scholar]
- 35.Richmond J, Bryant R, Trotman W, Beatty B, Lunde J. FISH detection of t(14;18) in follicular lymphoma on Papanicolaou-stained archival cytology slides. Cancer. 2006;108:198–204. doi: 10.1002/cncr.21917. [DOI] [PubMed] [Google Scholar]
- 36.Hirose Y, Masaki Y, Ozaki M. Fluorescence in situ hybridization detection of chromosome IGH/BCL2 translocations from paraffin-embedded tissue: evaluation in follicular lymphoma. Int J Hematol. 2003;78:154–9. doi: 10.1007/BF02983385. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto Y, Nomura K, Matsumoto S, Ueda K, Nakao M, Nishida K, Sakabe H, Yokota S, Horiike S, Nakamine H, et al. Detection of t(14;18) in follicular lymphoma by dual-color fluorescence in situ hybridization on paraffin-embedded tissue sections. Cancer Genet Cytogenet. 2004;150:22–6. doi: 10.1016/j.cancergencyto.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Guo Y, Karube K, Kawano R, Yamaguchi T, Suzumiya J, Huang GS, Ohshima K. Low-grade follicular lymphoma with t(14;18) presents a homogeneous disease entity otherwise the rest comprises minor groups of heterogeneous disease entities with Bcl2 amplification, Bcl6 translocation or other gene aberrances. Leukemia. 2005;19:1058–63. doi: 10.1038/sj.leu.2403738. [DOI] [PubMed] [Google Scholar]
- 39.Sekiguchi N, Kobayashi Y, Yokota Y, Kusumoto S, Tanimoto K, Watanabe T, Matsuno Y, Tobinai K. Follicular lymphoma subgrouping by fluorescence in situ hybridization analysis. Cancer Sci. 2005;96:77–82. doi: 10.1111/j.1349-7006.2005.00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peh SC, Shaminie J, Tai YC, Tan J, Gan SS. The pattern and frequency of t(14;18) translocation and immunophenotype in Asian follicular lymphoma. Histopathology. 2004;45:501–10. doi: 10.1111/j.1365-2559.2004.01979.x. [DOI] [PubMed] [Google Scholar]
- 41.Fan YS, Rizkalla K. Comprehensive cytogenetic analysis including multicolor spectral karyotyping and interphase fluorescence in situ hybridization in lymphoma diagnosis. a summary of 154 cases. Cancer Genet Cytogenet. 2003;143:73–9. doi: 10.1016/s0165-4608(02)00843-9. [DOI] [PubMed] [Google Scholar]
- 42.Bernicot I, Douet-Guilbert N, Le Bris MJ, Morice P, Abgrall JF, Berthou C, Morel F, De Braekeleer M. Characterization of IGH rearrangements in non-Hodgkin’s B-cell lymphomas by fluorescence in situ hybridization. Anticancer Res. 2005;25:3179–82. [PubMed] [Google Scholar]
- 43.Vaandrager JW, Schuuring E, Raap T, Philippo K, Kleiverda K, Kluin P. Interphase FISH detection of BCL2 rearrangement in follicular lymphoma using breakpoint-flanking probes. Genes Chromosomes Cancer. 2000;27:85–94. [PubMed] [Google Scholar]
- 44.Huang JZ, Sanger WG, Greiner TC, Staudt LM, Weisenburger DD, Pickering DL, Lynch JC, Armitage JO, Warnke RA, Alizadeh AA, et al. The t(14;18) defines a unique subset of diffuse large B-cell lymphoma with a germinal center B-cell gene expression profile. Blood. 2002;99:2285–90. doi: 10.1182/blood.v99.7.2285. [DOI] [PubMed] [Google Scholar]
- 45.Iqbal J, Sanger WG, Horsman DE, Rosenwald A, Pickering DL, Dave B, Dave S, Xiao L, Cao K, Zhu Q, et al. BCL2 translocation defines a unique tumor subset within the germinal center B-cell-like diffuse large B-cell lymphoma. Am J Pathol. 2004;165:159–66. doi: 10.1016/s0002-9440(10)63284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerretini R, Noriega MF, Narbaitz M, Slavutsky I. New chromosome abnormalities and lack of BCL-6 gene rearrangements in Argentinean diffuse large B-cell lymphomas. Eur J Haematol. 2006;76:284–93. doi: 10.1111/j.1600-0609.2005.00616.x. [DOI] [PubMed] [Google Scholar]


