Abstract
Cells form stress granule (SG), in response to unfavorable environments, to avoid apoptosis, but it is unclear whether and how SG formation and cellular apoptosis are coordinately regulated. In this study we detected the small GTPase, Ras homolog gene family member A (RhoA), and its downstream kinase, Rho-associated, coiled-coil containing protein kinase 1 (ROCK1), in SG, and found that their stress-induced activities were important for SG formation and subsequent global translational repression. Importantly, only activated RhoA and ROCK1 were sequestered into SG. Sequestration of activated ROCK1 into SG prevented ROCK1 from interacting with JNK-interacting protein 3 (JIP-3) and its activation of c-Jun N-terminal kinase (JNK), a pathway triggering apoptosis, thereby protecting cells from apoptosis. This study identifies a specific signaling pathway, mediated by RhoA and ROCK1, which determines cell fate by promoting SG formation or initiating apoptosis during stress.
Keywords: RhoA, ROCK1, stress granules, JIP-3, JNK, apoptosis, P19
1. Introduction
Stress granules (SGs) are subcellular structures that are formed in cells responding to various environmental stresses, such as high temperature, heavy metal poisoning or nutrient starvation [1-5]. It has been suggested that SG functions to protect housekeeping mRNAs and repress their translation by recruiting these mRNAs and certain mRNA-associated proteins. Through this mechanism, cells could also conserve energy and increase the availability of translation machinery for stress-related gene expression. The heat shock protein 70 transcript, for example, is excluded from SG and is actively translated under stress [3]. Recently, studies have shown that SG can protect cells from apoptosis, because actively disrupting SG would result in the release of apoptosis-inducing components from SG to trigger cell death [6, 7]. However, it is still unclear whether and how the decision to form SG or induce apoptosis, under stress, can be regulated by any specific signaling pathway.
RhoA, along with Rac1 and Cdc42, are the most studied Rho GTPases that regulate a wide variety of cellular processes, such as vesicle trafficking, cell-cycle progression and cytoskeleton rearrangement [8, 9]. The regulation of RhoA activity is mediated by the GDP/GTP activation cycle. Upon activation, GTP-bound RhoA signal is transmitted to many downstream effectors. Among these signal effectors, Rho-associated coiled-coil forming kinase (ROCK) is the most widely examined. The ROCK family, including ROCK1 and ROCK2, comprises of serine/threonine kinases that phosphorylate various downstream substrates. ROCK contains a Rho-binding domain (RBD) which, upon binding by RhoA, induces a conformational change at the carboxyl terminus and the activation of the kinase domain leading to actin filament stabilization and increased myosin ATPase activity [10, 11]. While it is generally believed that ROCK is important for cytoskeletal reorganization, it is unclear whether, under stress condition, ROCK plays a regulatory role in SG formation, which would require cytoskeleton and motor complex rearrangement [12-16]. In fact, ROCK1 can phosphorylate JIP-3 and activate JNK, which in turns would cause cytochrome C release and subsequent caspase-3 activation, thereby mediating UV irradiation-triggered apoptosis [14, 15, 17]. It is puzzling how ROCK1 can actively participate in events triggering stress response to determine cellular survival.
We have previously identified several critical SG components regulating SG dynamics, including a RNA-binding protein, growth factor receptor-bound protein 7 (Grb7) and dynein motor light chain (DLC2A) [16, 18]. In the current study, we found an additional regulatory signaling pathway, the RhoA/ROCK1 pathway that is important in cellular decision, to either form SG (survival) or trigger apoptosis (death). We found that under stress, RhoA and ROCK1 were activated, and both their activities were required for efficient SG formation. Further, both activated RhoA and ROCK1 were sequestered into SG. Forced disruption of SG formation resulted in the dissociation of RhoA and ROCK1 from other SG components. The released and activated ROCK1 would then interact with, and phosphorylate, JIP-3, thereby triggering apoptosis. This apoptotic pathway is inhibited in cells containing SG. This study provides the first example for a stress-initiated signaling pathway that determines cell fate by either promoting SG formation or triggering apoptosis.
2. Material and methods
2.1 Antibodies, reagents and plasmid constructions
The antibodies were from Santa Cruz Biotechnology (anti-Actin, anti-ROCK1, anti-ROCK2, anti-Cdc42, anti-Rac1, anti-RhoA, anti-JIP-3, anti-Dcp1a, anti-PABP, anti-HuR and anti-TIA-1), Upstate (anti-GST and anti-Phospho-Serine), Chemicon (anti-FMRP) and Cell Signaling (anti-JNK, anti-phspho-JNK). DAPI and AMP-PNP was from Roche. Annexin V apoptosis kit, EHNA, Y-27632, protease K, cycloheximide and puromycin are from Sigma Aldrich. C3 exotoxin was from BIOMOL International. RhoA activation kit and ROCK1 immunoblotting assay kit were from Cell Biolabs. 35S-Met/Cys was from MP Biomedical. The GFP-ROCK1 was made by inserting full-length ROCK1 cDNA into pAcGFP-C1 vector (Clontech). The GFP-ROCK1-K105A was made using a site-directed mutagenesis kit (Stratagene) with GFP-ROCK1 as template and a primer pair 5′-GGA AGG TTT ATG CTA TGG CGC TGC TCA GCA AAT TTG-3′ and 5′-CAA ATT TGC TGA GCA GCG CCA TAG CAT AAA CCT TCC-3′.
2.2 Western blotting, immunoprecipitation and immunohistochemistry
The above assays, especially for detecting SG-related components, are all as previously described [18, 19].
2.3 Heat shock treatment, arsenite treatment, protease resistance assay and metabolic labeling
Heat shock treatment was performed at 43°C for 30 minutes. Arsenite treatment was performed by treating cells in 0.5 mM sodium arsenite for 30 minutes. Protease resistance assay was performed as described [20] by using 100 μg total protein lysate with a 5-minute treatment of Protease K of indicated concentrations. Metabolic labeling was performed with 75% confluent P19 cells in 5-cm plates. 0.05 mCi 35S-Met/Cys was added and incubated for the time indicated. After washing with PBS, cells were harvested and lysate in PBS by sonication. 100 g total protein lysate was used for quantification by a liquid scintillation counter.
2.4 siRNA and transfection
siRNAs used in this study were from Qiagen. (RhoA, SI01401407 and SI01401428; ROCK1, SI01404487 and SI01404508; ROCK2, SI01404515 and SI01404522). siRNA transfection was performed by using Hiperfect transfection reagent (Qiagen).
2.5 RhoA and ROCK1 activity assay
These assays were conducted as manufacturer’s suggestion. In brief, for RhoA activation assay, the GST-Rhotekin was used to pull down active RhoA from control or heat shock-treated P19 cell lysate followed by Western blotting with anti-RhoA antibody. Regarding ROCK1 activation assay, immunoprecipitated complexes by anti-ROCK1 or anti-TIA-1 antibodies (depends on experimental design) were incubated with ROCK1 substrate protein, myosin phosphatase target subunit 1 (MYPT1). Activated ROCK1 is determined by the amount of phosphorylated MYPT1 detected by Western blotting with specific anti-phospho-MYPT1 antibody.
2.6 Apoptosis assay
The Annexin V apoptosis assay was performed as manufacturer’s protocol. In brief, control or heat shock-treated cells were suspended with buffer (10 mM HEPES/NaOH, pH 7.5 containing 0.14 M NaCl and 2.5 mM CaCl2). 2.5 μg/ml Annexin V FITC was added to suspended cells, incubated for 10 minutes at room temperature and monitored under Olympus IX70 inverted fluorescence microscope.
2.7 Cell scoring and statistical analysis
To quantify SG positive cells or SG number per cell, 100 cells were scored where SG was bigger than 1 □ m in the diameter [21]. The data throughout this study is presented as means ± standard deviations and analyzed with Student’s t-test where P < 0.05 was considered as significant.
3. Results
3.1 RhoA and its downstream kinase, ROCK1, are both present in SG
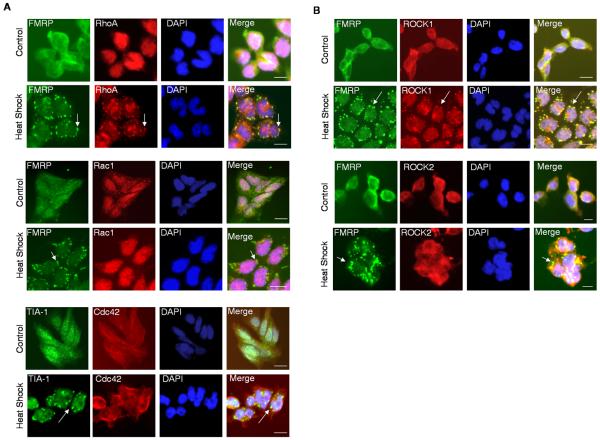
The most widely studied small GTPases, RhoA, Rac1 and Cdc42, are known to modulate cytoskeleton rearrangement in cells under stress. To first determine if these signaling molecules were physically associated with SG, we used a heat shock-triggered stress model of mouse embryonal carcinoma P19 cells [16, 18], to examine their cellular localization. We found that in the stressed P19 cytoplasm, the endogenous RhoA, but neither Rac1 nor Cdc42, was mostly localized in distinct foci overlapping with punctae of known SG components such as fragile X mental retardation protein (FMRP) and T-cell intracellular antigen 1 (TIA-1) (Fig 1A and Supplemental Figure S1 top). Two serine/threonine kinases, ROCK1 and ROCK2 that were known to transduce RhoA signal, were also tested in this model. It appeared that the immunoreactive signals of ROCK1, but not ROCK2, significantly overlapped with punctae positive for FMAP and TIA-1 (Fig 1B and Supplemental Figure S1 bottom). These overlapped signals were also detected in cells under another stress condition, 0.5 mM sodium arsenite treatment (Supplemental Figure S2). This would suggest that the distribution of RhoA and ROCK1 in SGs could be a common phenomenon.
Figure 1. RhoA and ROCK1 are recruited into SGs under heat shock stress.
(A) Immunohistochemistry of RhoA (top), Rac1 (middle) or Cdc42 (bottom) with SG marker, FRMP or TIA-1 in control or heat shock-treated cells as indicated. Clear granules were marked by arrows. (B) Immunohistochemistry of ROCK1 (top) or ROCK2 (bottom) in control or heat shock-treated cells as indicated. Bars, 25 μm. Clear granules were marked by arrows.
We found that a fraction of the FRMP-positive punctae did not overlap with ROCK1-positive punctae. To examine if ROCK1 could also be recruited into other cytoplasmic granules such as processing bodies (PBs), which might have contributed to the heterogeneity of immunohistochemistry results shown above, we examined the PB marker, decapping enzyme homolog 1a (Dcp1a). We found no significant colocalization of Dcp1a with either ROCK1 or ROCK2 (Supplemental Figure S3, top). Further, ROCK1, but not ROCK2, was colocalized with another SG marker protein poly-A binding protein (PABP) (Supplemental Figure S3, bottom), supporting that ROCK1 was indeed present in SGs but not PBs. The appearance of non-overlapping granules is consistent with other studies that also showed heterogeneous SGs [16, 18, 21].
3.2 RhoA and ROCK1 are important for SG formation
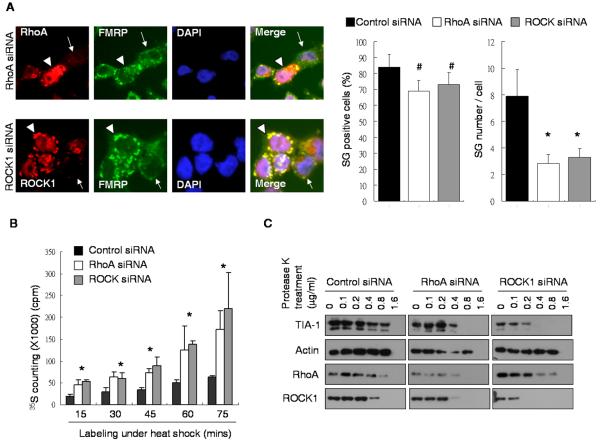
It is known that several types of stresses can activate RhoA and ROCK1 [17, 22, 23]; however, it is unclear whether RhoA/ROCK1, or their activation, could regulate SG formation in stressed cells. To determine whether RhoA and/or ROCK1 were required for SG formation, we conducted siRNA-specific knockdown of endogenous RhoA or ROCK1 in P19 cells and monitored the efficiency of SG formation under the heat shock condition (Fig 2A) or arsenite treatment (Supplemental Figure S4). As shown, RhoA- (top, pointed with an arrow) or ROCK1- (bottom, pointed with an arrow) silenced cells formed much fewer SG punctae (stained with FMRP), as compared to inefficiently silenced cells where RhoA and ROCK1 were retained (indicated by arrow heads) in the same culture under the same stress condition. We determined the efficiency of SG formation under RhoA- or ROCK-silencing by scoring both the SG punctae per efficiently silenced cell and the number of cells exhibiting apparent SG punctae among those silenced cells, in these two experiments. Quantification data (Fig 2A, right; Supplemental Figure S4, bottom) showed that knocking down RhoA or ROCK1 significantly reduced the efficiency of SG formation, as reflected by the significantly fewer SG punctae per silenced cell. However, the percentage of SG positive cells in both RhoA- and ROCK-1 knockdown cultures was not significantly different from that in the control culture (discussed later following Fig 3A).
Figure 2. Silencing RhoA or ROCK1 reduce SG formation.
(A) Immunohistochemistry of RhoA (top) or ROCK1 (bottom) with FMRP in heat shock-treated cells. Efficiently and inefficiently silenced cells were marked by arrows and arrow heads, respectively. Bars, 25 μm. Quantification of SG positive cells and average SG number per cell were shown on the right. (*P < 0.05; #P > 0.05) (B) Metabolic labeling of control (black bars), RhoA (white bars) or ROCK1 (gray bars) siRNAs transfected cells during different time points as indicated. (*P < 0.05) (C) Western blotting of TIA-1, Actin, RhoA and ROCK1 from control (left), RhoA (middle) or ROCK1 (right) siRNAs transfected cell lysate after the 5-minute Protease K treatment with indicated concentrations.
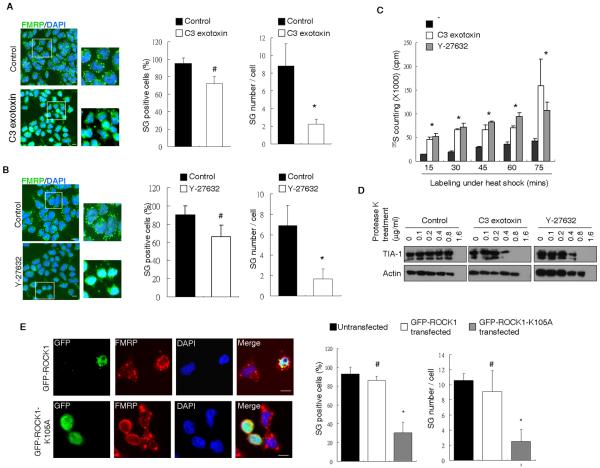
Figure 3. RhoA and ROCK1 activities are critical for SG formation.
(A) (B) Merged immunohistochemistry images from FRMP and DAPI staining of control- or (A) C3 exotoxin- or (B) Y-27632- treated cells. Indicated, enlarged areas were shown on the right. Bars, 25 μm. Quantification of of SG positive cells and average SG number per cell were shown on the right. (*P < 0.05; #P > 0.05) (C) Metabolic labeling of control (black bars), C3 exotoxin (white bars) or Y-27632 (gray bars) treated cells during different time points as indicated. (*P < 0.05) (D) Western blotting of TIA-1 and Actin from control (left), C3 exotoxin (middle) or Y-27632 (right) treated cell lysate after 5-minute Protease K treatment with indicated concentrations. (E) Immunohistochemistry of GFP and FMRP from GFP-ROCK1 or GFP-ROCK1-K105A transfected cells. Merged images were shown on the right. Bars, 25 μm.
To determine if ROCK2, which was not colocalized with SG markers, might be able to regulate SG formation or compensate for the silencing of ROCK1, we performed ROCK2 silencing (Supplemental Figure S5) and ROCK1/ROCK2 dual silencing (Supplemental Figure S6). It appeared that ROCK2 silencing did not affect SG formation and that ROCK1/ROCK2 dual silencing exerted a similar effect as that of ROCK1 silencing (Figure 2). This would suggest a specific role of ROCK1 in regulating SG dynamics.
To validate if the less effective SG formation (reflected by the fewer SG punctae per cell) under RhoA- or ROCK1-silencing would be consistent with the predicted translational effect, i.e. reduced translational repression, we measured the global translational rate by metabolic labeling with 35S-Cystenine/Methionine which would reflect the efficiency of de novo protein synthesis during stress. As shown (Fig 2B), the culture receiving specific siRNAs against RhoA- (white bars) or ROCK1 (gray bars) incorporated significantly higher counts of 35S, which indicated a higher translation rate, as compared to the control siRNA-transfected culture (black bars). The results supported that, upon knocking down RhoA or ROCK1, cells could no longer efficiently form SG and their translational repression was also suppressed.
It was proposed that SG was initiated by TIA-1 aggregation and the sensitivity of the TIA-1 aggregate to protease digestion would indicate SG integrity [20]. We then tested the effect of RhoA- and ROCK-silencing on the integrity of SG using the standard protease sensitivity assay, which monitored the remaining level of TIA-1 complex after different amounts of protease treatment for 5 minutes [20]. As shown (Fig 2C), in the control siRNA-transfected culture, TIA-1 complexes were sensitive to protease treatment at the concentration of approximately 0.8 μg/ml. However, in cultures receiving siRNAs against RhoA or ROCK1, TIA-1 complexes were more sensitive to protease treatment (sensitive to concentrations of 0.4 μg/ml and 0.2 μg/ml, respectively), indicating compromised SG integrity under RhoA or ROCK1 silencing. Together, these data showed that both RhoA and ROCK1 were important for stressed cells to efficiently form SG.
3.3 The activities of RhoA and ROCK1 are required for SG formation
The above data showed that RhoA and ROCK1 were physically associated with SG, and deleting either one component would compromise SG formation in stressed cells. It was of importance to determine whether the activities of RhoA or ROCK1 were required for SG formation. We administered a specific RhoA inhibitor, C3 exotoxin, to inactivate RhoA in P19 cells by lipotransfection [24]; the cells were then subjected to heat shock treatment. As shown in Fig 3A, SG formation, detected by immunohistochemistry (left) as well as the quantified data (middle and right), was less efficient in cells treated with the RhoA inhibitor. Similarly, administering a ROCK1-specific inhibitor, Y-27632, into P19 cultures also inhibited their SG formation (Fig 3B). We noticed that the effect of RhoA and ROCK1 on SG formation was significant only when it was determined by scoring the SG punctae numbers per efficiently silenced cell, but not by scoring SG-positive cells among all silenced cells, as shown in both Fig. 2A and Fig. 3A, 3B). This would indicate that RhoA and ROCK1 affected the formation of only a fraction of SG; other RhoA/ROCK1-independent pathways could contribute to SG formation as well. This is also consistent with the general view that SG punctae are heterogeneous.
The effects of RhoA and ROCK1 inhibitors were confirmed also by metabolic labeling (Fig 3C) and protease sensitivity assay (Fig 3D). As shown, C3 exotoxin and Y-27632 both were able to suppress the stress-induced translational repression, and the effect of these drugs was detected readily at the first time point monitored, 15-minute. The effect of these drugs was also reflected on the increased protease sensitivity of TIA-1 complex (sensitive to the concentrations of 0.4 μg/ml from drug-treated cells as compared to 0.8 μg/ml from the control cells).
To determine whether overexpression of the catalytically inactive ROCK1 could hamper SG formation, we introduced the GFP-fused wild type (GFP-ROCK1) or the enzymatically inactive (GFP-ROCK1-K105A) form of ROCK1 into P19 cells and monitored their SG formation. As shown in Fig 3E, both immunohistochemical images and quantification data indicated that overexpression of wild type ROCK1 had no significant effect on SG formation but expressing the inactive ROCK1 abolished SG formation. Together, the data showed that the activities of RhoA and ROCK1 were important for the efficiency of SG formation in stressed cells.
3.4 Active, but not inactive, RhoA and ROCK1 are sequestered into SG
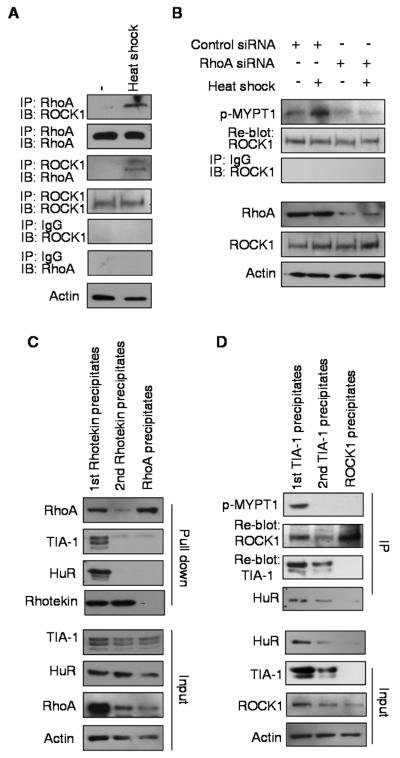
While both RhoA and ROCK1 could be detected in SG (Fig 1) and their activities were required for SG formation (Fig 3), we also noticed that certain RhoA and ROCK1 immuno-reactive signals could be detected outside SG punctae. A potentially important scenario is that there might be differential recruitment of active versus inactive forms of RhoA and ROCK1 into SG. To address this scenario, we first confirmed increased association of RhoA with ROCK1 following stress, in a reciprocal co-immunoprecipitation experiment (Fig 4A), and verified the stress-activated ROCK1 activity, which was indicated by phosphorylation of its specific substrate, p-MYPT (Fig 4B). Importantly, phosphorylation of the ROCK1 substrate was abolished by silencing RhoA (3rd and 4th columns of Fig 4B), which confirmed the RhoA-dependency of ROCK1 activation.
Figure 4. SGs differentially recruit activated RhoA and ROCK1.
(A) Western blotting of reciprocal co-immunoprecipitation of RhoA and ROCK1 from control or heat shock-treated cell lysate. IgG was used as precipitation control and Actin was used as input control. (B) Western blotting of phospho-MYPT1 with anti-ROCK1 antibody precipitated complex from control or RhoA siRNA transfected cells. IgG served as precipitation control. Input of RhoA, ROCK1 and Actin were shown on the bottom. (C) Western blotting of RhoA, TIA-1 and HuR from stressed-P19 cells as indicated from sequential pull down with GST-Rhotekin and anti-RhoA antibody. 1/8 input of each precipitation was shown on the bottom. (D) Western blotting of phospho-MYPT1, TIA-1 and HuR from sequential co-immunoprecipitation with anti-TIA-1 and anti-ROCK1 antibodies. 1/8 input of each precipitation was shown on the bottom.
We then examined whether active and inactive forms of RhoA were differentially distributed, i.e. within or outside SG by fractionating cellular lysates using sequential precipitation. As shown in Fig. 4C, lysate was first incubated with GST-fused Rhotekin, a specific activated RhoA-binding protein, allowing active RhoA to form complex with GST-Rhotekin. The mixture was then pulled down with glutathione-agarose beads, which effectively precipitated RhoA and SG components such as TIA-1 and HuR, in addition to GST-Rhotekin. The supernatant of the first precipitation was again incubated with GST-Rhotekin which would form complex with any remaining activated RhoA. The mixture was re-precipitated with glutathione-agarose beads, which pulled down only negligible amounts of RhoA and SG components; yet abundant Rhotekin was indeed present. Finally, the supernatant of the second precipitation was precipitated with RhoA antibody, which would precipitate any remaining RhoA that was presumably inactive and could not be co-precipitated with TIA-1 and HuR. This experiment showed that only activated RhoA was associated with SG, and the inactive RhoA was not in the SG fraction.
By using a similar strategy, we conducted another experiment to fractionate cell lysates with anti-TIA-1 antibody that precipitated SG-associated components, and examined ROCK1 activity as shown in Fig 4D. TIA-1 precipitate was incubated with the specific ROCK1 substrate MYPT1. It appeared that TIA-1 precipitate was able to phosphorylate MYPT1, as detected by p-MYPT1, and it indeed contained ROCK1, and SG components such as HuR, in addition to TIA-1. The supernatant of the first TIA-1 precipitate was then precipitated again with anti-TIA-1 antibody and the precipitate was incubated with MYPT-1, which could barely phosphorylate MYPT-1, indicating that most of TIA-1 associated, activated ROCK1 had been cleared out by the first TIA-1 precipitation. Importantly, the supernatant from the second TIA-1 precipitate still contained abundant ROCK1 (detected in anti-ROCK1 precipitate), which failed to phosphorylate MYPT-1. This would support that all activated ROCK1 had been precipitated by the first TIA-1 pull-down, which could phosphorylate MYPT-1. Those ROCK1 free from SG could not phosphorylate MYPT-1, and therefore were inactive.
To further rule out the possibility that active RhoA and ROCK1 could be in the SG-excluded complexes and precipitated with TIA-1 and HuR, we used SG blockers to inhibit SG formation as shown in Supplement Figure 2. The data showed that neither RhoA nor ROCK1 could be co-immunoprecipitated with TIA-1 when stressed cells were forced to abolish SG by treatment with SG blockers, cycloheximide or erythro-9-[3-(2-hydroxynonyl)] adenine (EHNA). This was not seen in experiments using two control drugs, puromycin or adenosine 5′-(β,γ-imido) triphosphate (AMP-PNP) [16] that could not disrupt SG.
3.5 SG inhibits apoptosis by sequestering activated ROCK1
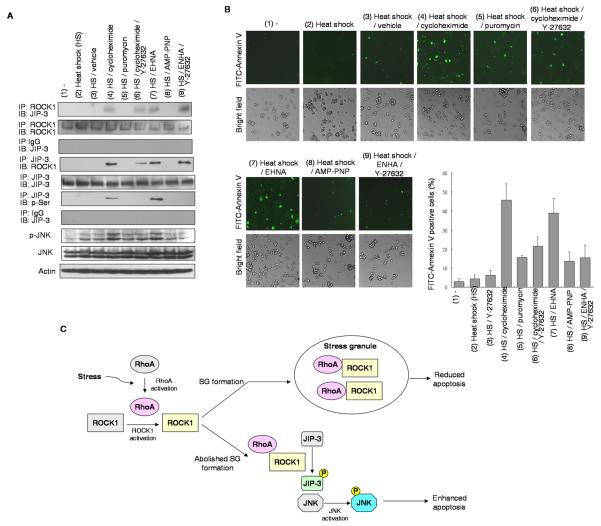
It is known that cells form SG to prevent apoptotic death; consistently, studies have also shown sequestration of apoptosis-related molecules into SG. Since ROCK1 is a known apoptosis contributor [17], it was intriguing that we detected certain ROCK1 immunoreactivity in SG and that only those ROCK1 molecules distributed to SG were enzymatically active. We then asked if the SG-sequestered, activated ROCK1 might be involved in apoptosis induction if it was not retained in SG. We first ruled out the association of SG with JIP-3, a SG-excluded protein and an indicator of JNK activation and apoptosis (Supplemental Figure S3), and monitored, by reciprocal co-immunoprecipitation (Fig 5A), the interaction between ROCK1 and JIP-3. Under normal or heat shock condition, ROCK1 and JIP-3 were not in the same complex (the 1st, 2nd and 3rd columns); however, disrupting SG during stress by treatment with cycloheximide (the 4th and 6th columns) or EHNA (the 7th and 9th columns), but not with puromycin (the 5th column) or AMP-PNP (the 8th column), stimulated complex formation between ROCK1 and JIP-3 (the 1st and 4th panels). The formation of ROCK1/JIP-3 complex in cycloheximide or EHNA-treated condition was not inhibited by ROCK1 inhibitor Y-27632, suggesting that ROCK1 activity was not required for its forming complex with JIP-3. As predicted, subsequent phosphorylation of JIP-3 by ROCK1 was observed (the 4th and 7th columns of the 6th panel) and it was inhibited by the ROCK1 inhibitor Y-27632 (the 6th and 9th columns of the 6th panel). Consistently, the p-JNK level was most robust when SG formation was disrupted under stress (the 8th panel), and it was inhibited by the ROCK1 inhibitor Y-27632. These data supported that JNK signaling pathway was activated when SG formation was forced to stop, and that blocking ROCK1 activity would block this effect.
Figure 5. Sequestration of ROCK1 into SGs inhibits apoptosis.
(A) Western blotting of reciprocal immunoprecipitation by anti-ROCK1 or anti-JIP-3 antibodies detected by anti-JIP-3, anti-ROCK1 or anti-phospho-Serine antibodies. Blotting of JNK, phospho-JNK and input control Actin were shown on the bottom. Heat shock and/or drug treatments were performed as indicated. (B) Immunohistochemistry from Annexin V FITC staining and bright field from heat shock and/or drug treatment as indicated. Quantification of FITC positive cells was shown on the bottom right. (C) Model for stress-activated RhoA and ROCK1 in determining SG formation and apoptosis.
Since activating JNK would lead to apoptosis, we then confirmed the fate of stressed cells when they were forced to stop SG formation. We examined the likelihood of apoptosis of the cultures under various conditions (as that shown in Fig 5A) by detection with FITC-Annexin V to monitor apoptotic cells. It appeared that a significantly increased cell population was FITC-Annexin V-positive when the culture was treated with both heat shock and cycloheximide or EHNA to block SG formation. This could be effectively rescued by adding the ROCK1 inhibitor, Y-27632, to the culture. Together, the results suggested that SG inhibited apoptosis by sequestering ROCK1. Forcing the activated ROCK1 out of SG would trigger apoptosis.
4. Discussion
In this study, by using the heat shock-induced cellular stress model, we show that stress activated RhoA and ROCK1 are important for SG formation in stressed cells. Active RhoA and ROCK1 are both sequestered into SG, which prevents their interaction with JIP-3, a critical component for JNK-mediated apoptosis. This is the first study identifying a signaling pathway that is able to trigger both SG formation and apoptosis (Fig 5C). The fate for cells to survive or die is determined, at least partially, by whether RhoA and ROCK1 can be successfully sequestered into SG. The data also support the important role for SG in inhibiting apoptosis by preventing JIP-3 phosphorylation and JNK activation. How the RhoA and ROCK1 are recruited into SG, and whether other RhoA/ROCK1 associated proteins are also in SG, remain to be examined.
The fact that SG selectively recruits active forms of RhoA and ROCK1 in stressed cells is interesting, which indicates a critical decision a cell has to make. It is known, as also shown in our data (Fig 4A), that one of the activating pathways of ROCK1 relies on its binding by RhoA. Therefore it is not a surprise that these two proteins are mobilized together under stressed condition. Since activation of these proteins requires conformational changes, it is possible that only activated RhoA or ROCK1 interacts with certain SG components, and are preferentially recruited into SG. It will be important to determine what SG components are required specifically for the recruitment of activated RhoA and ROCK1.
A possibility is that ROCK1 might act as a scaffold protein to regulate SG formation. However, since silencing ROCK1 had a similar effect as compared to treating cells with the ROCK inhibitor (Fig 2 and Fig 3) and the catalytically inactive ROCK1 significantly abolished SG formation (Fig 3E), it would suggest that ROCK1 enzyme activity, rather than its action as a scaffold action, is involved in the regulation of SG formation.
In addition to ROCK1, certain apoptosis related components can also be sequestered into SG in stressed cells to prevent apoptosis [6, 7]. As stress could simultaneously activate multiple, various apoptotic pathways, it would probably be most beneficial to a cell’s survival that various apoptosis-related components are all recruited into SG to prevent apoptosis. Because SG formation is initiated when cells encounter various stress sources for different kinds of cells, the compositions of SG are not likely to be identical among various cellular backgrounds. As such, it would be most critical to determine whether and how those SG-sequestered components, including ROCK1 in this study, are coordinated for sequestration into SG, or to be released to mediate apoptosis when SG formation is disrupted
Supplementary Material
Acknowledgement
This work is supported in part by NIH Grants DA11190, DA11806, DK54733, DK60521 and K02-DA13926.
Abbreviations
- RhoA
Ras homolog gene family member A
- ROCK
Rho-associated, coiled-coil containing protein kinase
- SG
stress granule
- JIP-3
JNK-interacting protein 3
- JNK
c-Jun N-terminal kinase
- EHNA
erythro-9-[3-(2-hydroxynonyl)] adenine AMP-PNP, adenosine 5′-(β,γ-imido) triphosphate
- FMRP
fragile X mental retardation protein
- TIA-1
T-cell intracellular antigen 1
- HuR
Hu antigen R
- Dcp1a
decapping enzyme homolog 1a
- PABP
poly-A binding protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kiebler MA, Bassell GJ. Neuron. 2006;51(6):685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- [2].Anderson P, Kedersha N. J Cell Biol. 2006;172(6):803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kedersha N, Anderson P. Biochem Soc Trans. 2002;30(Pt 6):963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- [4].Moore MJ. Science. 2005;309(5740):1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- [5].Anderson P, Kedersha N. Trends Biochem Sci. 2008;33(3):141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- [6].Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Nat Cell Biol. 2008;10(11):1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- [7].Eisinger-Mathason TS, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, Macara IG, Lannigan DA. Mol Cell. 2008;31(5):722–736. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Burridge K, Wennerberg K. Cell. 2004;116(2):167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- [9].Takai Y, Sasaki T, Matozaki T. Physiol Rev. 2001;81(1):153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- [10].Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. Embo J. 1996;15(8):1885–1893. [PMC free article] [PubMed] [Google Scholar]
- [11].Fujita A, Saito Y, Ishizaki T, Maekawa M, Fujisawa K, Ushikubi F, Narumiya S. Biochem J. 1997;328(Pt 3):769–775. doi: 10.1042/bj3280769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chernov KG, Barbet A, Hamon L, Ovchinnikov LP, Curmi PA, Pastre D. J Biol Chem. 2009 doi: 10.1074/jbc.M109.042879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Loschi M, Leishman CC, Berardone N, Boccaccio GL. J Cell Sci. 2009 doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dhanasekaran DN, Reddy EP. Oncogene. 2008;27(48):6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Igaki T. Apoptosis. 2009;14(8):1021–1028. doi: 10.1007/s10495-009-0361-7. [DOI] [PubMed] [Google Scholar]
- [16].Tsai NP, Tsui YC, Wei LN. Neuroscience. 2009;159(2):647–656. doi: 10.1016/j.neuroscience.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ongusaha PP, Qi HH, Raj L, Kim YB, Aaronson SA, Davis RJ, Shi Y, Liao JK, Lee SW. Sci Signal. 2008;1(47):ra14. doi: 10.1126/scisignal.1161938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tsai NP, Ho PC, Wei LN. Embo J. 2008;27(5):715–726. doi: 10.1038/emboj.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tsai NP, Bi J, Wei LN. Embo J. 2007;26(6):1522–1531. doi: 10.1038/sj.emboj.7601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Mol Biol Cell. 2004;15(12):5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ivanov PA, Chudinova EM, Nadezhdina ES. Exp Cell Res. 2003;290(2):227–233. doi: 10.1016/s0014-4827(03)00290-8. [DOI] [PubMed] [Google Scholar]
- [22].Sen U, Moshal KS, Singh M, Tyagi N, Tyagi SC. Mol Cell Biochem. 2007;302(12):133–143. doi: 10.1007/s11010-007-9435-4. [DOI] [PubMed] [Google Scholar]
- [23].Tzima E. Circ Res. 2006;98(2):176–185. doi: 10.1161/01.RES.0000200162.94463.d7. [DOI] [PubMed] [Google Scholar]
- [24].Borbiev T, Nurmukhambetova S, Liu F, Verin AD, Garcia JG. Anal Biochem. 2000;285(2):260–264. doi: 10.1006/abio.2000.4763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.