Abstract
Background
Estimation of HIV incidence rates is important for timing interventions, planning prevention studies, and monitoring the epidemic. This requires accurate estimation of the “recency period” (also known as the “window period”) between seroconversion and achievement of specific detectable levels of anti-HIV antibody titers, such as the standardized optical density (SOD) in the early phase of HIV-1 infection.
Methods
To obtain a better understanding of inter-patient variation of the recency period, prospective measurements of anti-viral antibody titers in the early phase of HIV-1 subtype C infection were quantified by Vironostika-LS. Time of seroconversion was estimated by Fiebig staging.
Results
The profiles of SOD values during the first year of infection commonly showed slow initial increase followed by a more rapid increase, though in some patients SOD values increased rapidly soon after seroconversion. Using an SOD cutoff of 1.0, the average duration of the recency period in subtype C infection in the local epidemic in Botswana was estimated to be 151 days (95% CI from 130 to 172 days) post-seroconversion. The recency period was significantly associated (p=0.007) with the level of viral replication during the first 2–3 months post-seroconversion. Reduction of SOD values after initiation of antiretroviral therapy (ART) was a dominant pattern in antiretroviral drug (ARV)-treated subjects.
Conclusions
Our data suggest that HIV incidence estimation based on sensitive/less sensitive EIA cross-sectional testing could be potentially improved by incorporation of viral load levels at the time of detection of a recent infection.
Keywords: Primary HIV-1 subtype C infection, detuned assay, viral load, SOD, Botswana, Vironostika
Introduction
Obtaining an accurate estimation of HIV incidence is critical for monitoring trends in the epidemic, assessing public health interventions, and developing prevention strategies. Reliable identification of individuals within the early phase of HIV infection opens an opportunity for behavioral and biomedical interventions directed toward reducing viral transmission, initiating early ART, and providing important information for designing new therapeutic and vaccine strategies through analysis of recently transmitted viruses and immune responses in primary HIV infection.
Longitudinal cohorts represent a classical way of studying HIV incidence by prospective follow-up of HIV-negative individuals with periodic testing for HIV. However, methods have been developed enabling the analysis of HIV incidence through cross-sectional sampling, which is significantly less expensive, faster, and logistically more feasible.1–11 The widespread use of cross-sectional methods for analyzing HIV incidence has resulted in a substantial increase of data on trends in the HIV/AIDS epidemic in different parts of the world, including studies in HIV-1 subtype C settings.12
However, as noted in a recent report of the Institute of Medicine,13 an important concern with the use of cross-sectional testing is the reliability and generalizability of results. Long-term infections present a challenge for HIV incidence testing because even a small number of such cases might inflate the incidence estimate,12 particularly if prevalence is high relative to incidence.14 Suppression of viral replication with or without ART is associated with lower titers of anti-HIV antibodies which may lead to misclassification of long-lasting infections as recent cases. Low levels of viral load have been found to account for misclassification,15 and have been found in samples tested discordantly by alternative lab methods.16 Although it is known that patients treated with ART have decreased anti-HIV titers,15, 17 the frequency and the time line for such decrease are still unclear.
Different methods for estimating HIV incidence—such as detuned Vironostika, BED, and avidity assay—can perform similarly well,18–20 and might complement each other (e.g., detuned Vironostika and avidity assay) in refining false-positive samples.16 However, a recent report from in Cote d'Ivoire demonstrated a wide range of estimated HIV incidence (1.2% to 11.2%) by different lab methods 21. It is likely that any particular diagnostic method might require adjustment to a local epidemic before it can be reliably used for accurate estimation of HIV incidence.
The rationale for this study was based on the assumption that with frequent longitudinal measurements, even a small number of cases within the early phase of HIV infection can provide a reliable estimate of the duration of the recency period, and can serve as a prototype for seroconversion panels for calibration of the recency period, which in turn can provide a more accurate estimate of HIV incidence in the local epidemic. In this study we characterized SOD evolution during the first year of infection in a cohort of subjects identified with primary HIV-1 subtype C infection, calibrated the recency period, and investigated potential associations between the recency period and viral RNA load.
Methods
Study subjects and testing
A total of 70 subjects including 8 acute and 62 recent cases from a primary HIV-1 subtype C infection study cohort in Botswana22 who were enrolled from March 2004 to December 2007 and who completed at least four study visits were included in the current analysis. There were 17 male (2 acute) and 53 female (6 acute) participants. Ages ranged from 19 to 53 years old. The age distributions of acutely and recently infected participants were similar, as were those of males and female. All subjects were Botswana nationals. All infections were caused by HIV-1 subtype C.23 The study was approved by Institutional Review Boards in Botswana and the US. Written informed consent was obtained from each participant.
Acutely or recently infected subjects were identified in a prospective study cohort of postnatal HIV-negative women or through VCT-based referrals.22 Participants in the prospective study were screened for HIV infection bimonthly. Referred subjects were screened for HIV infection cross-sectionally. The RNA+Ab− status of acutely infected subjects was defined by a positive HIV-1 RT-PCR test combined with a negative HIV-1 serology in double EIA. Initial screening for HIV-1/2 antibodies was performed with rapid EIA (Determine HIV-1/2, Abbott and Uni-Gold HIV Kit, Trinity Biotech) and/or regular EIA (Murex HIV 1.2.0, Abbott and Ortho HIV-1/2 Ab Capture, Ortho Diagnostics). The “window period” of the third-generation immunoassays used in the study was estimated to be about two weeks.24 Recent HIV-1 infections were identified by applying a two-step testing algorithm, the Serologic Testing Algorithm for Recent HIV Seroconversion (STARHS)2 using the Vironostika HIV-1 Plus O Microelisa System (bioMérieux) according to the protocol described elsewhere.3, 4 A total of 615 samples from 51 subjects with multiple measurements of SOD were analyzed.
Acutely infected subjects had weekly visits for the first two months, bi-weekly visits for the next two months, and monthly visits for the following eight months. Recently infected subjects had monthly visits. After the first year, study visits for both acutely and recently infected subjects became quarterly. Quantification of viral RNA in plasma was performed by COBAS AmpliPrep/COBAS Amplicor HIV-1 Monitor Test, ver. 1.5, according to the manufacturer’s instructions. The level of detection ranged from 50 copies/ml for the ultrasensitive method and 400 copies/ml for the standard method to 750,000 copies/ml. Specimens exceeding the upper level of detection were re-tested by 10-fold dilutions. Quantification of the total proviral DNA was performed by the method described elsewhere25 by targeting HIV-1 gag p24 and using the TaqMan Universal PCR master mix on the Applied Biosystems 7500 Realtime PCR system. Quantification of CD4+ cells was performed by flow cytometry using the four-color FACSCalibur. Individuals whose CD4+ T cell count dropped below 200 cells/mm3 or who developed opportunistic infection had access to ART (Combivir (AZT/3TC) 300/150 mg BID plus NVP 200 mg BID if female, or EFV 600 mg QD if male) free of charge, in accordance with Botswana National Program guidelines.
Adjustment of time 0 by Fiebig stage
Subjects with acute infection were RNA+Ab− and the estimated time of their seroconversion was computed as a mid-point between their last Ab− and Ab+ tests (average time difference = 19.5 days). To estimate the seroconversion time point for the recently infected subjects, the stage of HIV infection was categorized by applying the Fiebig algorithm.26 The earliest available plasma specimens were analyzed by HIV Blot 2.2 Western Blot Assay (Genelabs Diagnostics S.A.). The distribution of 62 recent cases by Fiebig staging26 included 10 (16.1%) cases in stage IV; 18 (29.0%) cases in stage V; 4 (6.5%) cases on the edge of stages V and VI (extremely faint p31 band evident for a transition from stage V to stage VI); and 30 (48%) cases in stage VI. According to the original paper26 and application of Fiebig staging to HIV-1 subtype C samples,27 the assumption was made that the beginning of Fiebig stage III coincides with the time of detectable seroconversion (time 0), and the mean duration of Fiebig stage III is 3 days, of stage IV is 6 days, and of stage V is 70 days. The mean duration of stage VI is not reliably estimable. Based on these estimates, the time from seroconversion until detection was assumed to average 6 days for subjects in stage IV (3 days of phase III and 3 days to the mid-point of phase IV), 44 days for subjects in stage V (9 days of phases III and IV and 35 days to the midpoint of phase V), and 79 days for stage V/VI (9 days of phases III and IV, and 70 days of phase V). Since the adjustment for stage VI was not considered reliable because of the uncertainty associated with the estimated recency period, subjects identified within Fiebig stage VI were excluded from analyses related to identification of the recency period.
Statistical analysis
Data were summarized with means ± standard deviations or means ± 95% confidence intervals. For subjects whose SOD values had not crossed 1, the time to SOD crossing 1 was assumed to be right-censored at the time of the last follow-up. Kaplan-Meier curves were used to describe the proportion of subjects whose SOD values remained below 1 during the 400 days after seroconversion, separately for two groups defined by RNA values, with the Gehan-Wilcoxon test used for comparisons. Mean time to SOD crossing a specified level, in the presence of censoring observations, was obtained by calculating the area under the Kaplan-Meier curve, assuming an exponential tail in situations when the Kaplan-Meier estimator did not reach zero. All reported p-values are 2-sided.
Results
Profile of SOD increase in primary HIV-1 subtype C infection
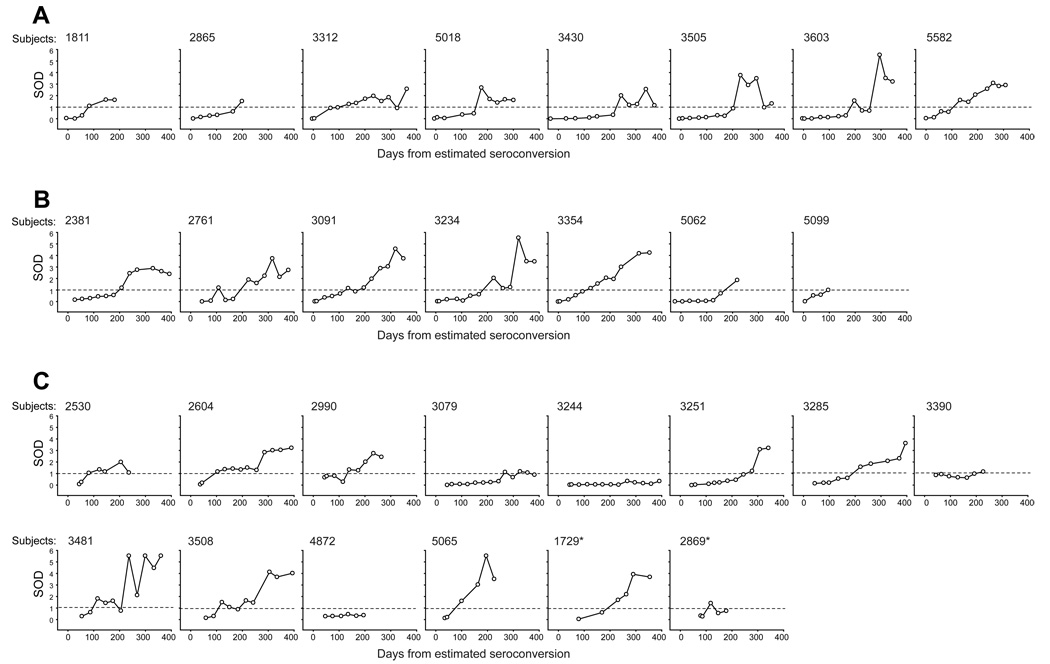
As seen in Figure 1, antiviral antibody titers quantified by detuned EIA were uniformly close to 0 during the early post-seroconversion period (when available), but thereafter demonstrated substantial heterogeneity in rates of growth in acutely (Fig. 1A) and recently (Fiebig stage IV: Fig. 1B, and Fiebig stages V and V/VI: Fig. 1C) infected subjects. As a result, subjects’ values reached thresholds (such as 1.0) at substantially different time points. The profiles of SOD values demonstrated slow initial increase soon after seroconversion and were followed by a more rapid increase in most cases. However, in two (6.9%) of 29 subjects identified in Fiebig stage V/VI or earlier, SOD values did not reach the threshold of 1.0 during the follow-up period (695 days for subject 3244; 195 days for subject 4872). The profiles of SOD evolution in 22 subjects identified within Fiebig stage VI (Fig. 1D) were similar to profiles in subjects with earlier Fiebig stages.
Figure 1.
Pre-ART SOD evolution in primary HIV-1 subtype C infection. Time 0 corresponds to the estimated time of seroconversion. Threshold at the SOD level of 1.0 is shown by dashed line. Post-ART SOD values are not shown. A. Acutely infected subjects, n=8. B. Subjects with recent infection: time of seroconversion (time 0) was adjusted by Fiebig stage IV (indeterminate WB), n=7. C. Subjects with recent infection: time of seroconversion (time 0) was adjusted by Fiebig stage V (WB+, p31-; n=12) and V/VI (WB+, faint p31 at the edge of detection; n=2; shown with asterisks). D. Subjects with recent infection: time of seroconversion (time 0) was adjusted by Fiebig stage VI (completely developed WB). Subjects 3244 and 4872 did not cross the threshold SOD level of 1.0 during the time of observation.
To characterize the profile of SOD dynamics in primary HIV-1 subtype C infection, the SOD values were analyzed within 50-day intervals post-seroconversion separately in groups of acutely and recently infected subjects. Similar profiles of SOD kinetics were evident within groups up to 300 days post-seroconversion followed by heterogeneity afterwards (data not shown). This similarity supports the value of using Fiebig staging to adjust for average seroconversion time.
Recency period is a time of reaching SOD threshold
To analyze the kinetics of SOD values, we computed the estimated time of crossing specified SOD levels from 0.1 to 1.0 by linearly interpolating between the SOD values observed just before and after reaching the specified SOD value. The heterogeneity in reaching specified SOD levels between acutely infected subjects is shown in Table 1. A generally gradual increase of the duration from the estimated time of seroconversion to the estimated time of reaching the specified SOD levels from 0.1 to 1.0 was accompanied by wide 95% CIs reflecting substantial variance between subjects. For the group of acutely infected subjects, the SOD value of 1.0 was reached in an average of 155 days (95% CI from 119 to 191 days) post-seroconversion.
Table 1.
Estimated duration from seroconversion to a specified SOD threshold from 0.2 to 1.0, days.
| Acutely infected subjects |
SOD threshold | ||||
|---|---|---|---|---|---|
| 0.2 | 0.4 | 0.6 | 0.8 | 1.0 | |
| 1811 | 47.3 | 59.3 | 66.0 | 72.8 | 79.5 |
| 2865 | 57.0 | 118.8 | 159.6 | 170.5 | 178.3 |
| 3312 | 16.2 | 30.3 | 44.3 | 58.4 | 99.2 |
| 5018 | 67.9 | 123.9 | 149.4 | 152.0 | 154.6 |
| 3340 | 153.7 | 213.3 | 217.0 | 220.7 | 224.4 |
| 3505 | 118.7 | 177.6 | 187.7 | 197.8 | 203.5 |
| 3603 | 135.0 | 167.3 | 172.5 | 177.6 | 182.7 |
| 5582 | 46.3 | 57.2 | 68.1 | 106.8 | 115.6 |
| Mean: | 80.3 | 118.5 | 133.1 | 144.6 | 154.7 |
| 95% CI, from | 46.2 | 73.1 | 88.4 | 103.6 | 118.8 |
| 95% CI, to | 114.3 | 163.8 | 177.8 | 185.5 | 190.7 |
A similar analysis of the estimated time of reaching different SOD levels was performed for groups of recently infected subjects identified within Fiebig stages IV and V–V/VI (data not shown). The mean time to reach the SOD threshold of 1.0 in the group of recently infected subjects, n=20, was similar to the acute infection group: 149.7 (95% CI from 124 to 176). For the entire cohort, the mean time to reach the SOD threshold of 1.0 was 151 days (95% CI from 130 to 172 days). The SOD values for subject 3244 increased at a very slow rate and remained low during an extended period; as a result, his/her time to crossing each specified SOD level was markedly higher than those of the remaining 28 subjects and heavily influenced the results. We therefore excluded his/her data in estimating mean time to crossing each specified SOD level, and note that this exclusion is likely to result in an underestimation of the recency periods.
Virologic and immunologic parameters in subjects 3244 and 4872
To investigate why subjects may have continuously low SOD values, we analyzed viral RNA dynamics in plasma, cell-associated proviral DNA, and CD4+ T cell counts in subjects 3244 and 4872 for 695 days and 195 days, respectively (Fig. 4). Both subjects had low levels of viral RNA and proviral DNA, and relatively high levels of CD4+ T cells with remarkable fluctuations. A transient spike of viral RNA in subject 4872 at day 108 did not result in changes in other studied parameters. While no single underlying reason could be identified, it seems that efficient control of the viral replication and preservation of CD4+ T cells (or their fast recovery) might contribute to the observed delay of the anti-HIV antibody titers increase, and in turn, present a potential problem for the incidence analysis based on cross-sectional sampling.
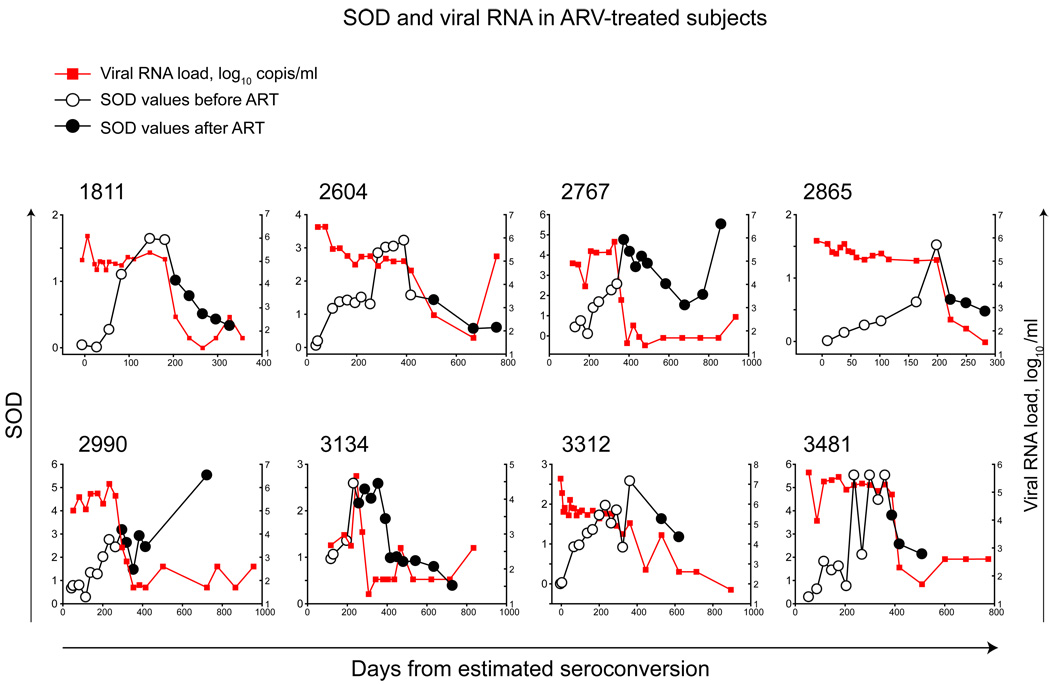
Figure 4.
SOD in ARV-treated subjects. Time 0 corresponds to the estimated time of seroconversion. Note: scales for time and SOD differ between subjects. Pre-ART measurements of SOD are indicated by open circles. Post-ART measurements are indicated by filled circles. Curves of viral RNA load are indicated by red squares and red lines.
Association of early viral replication with recency period
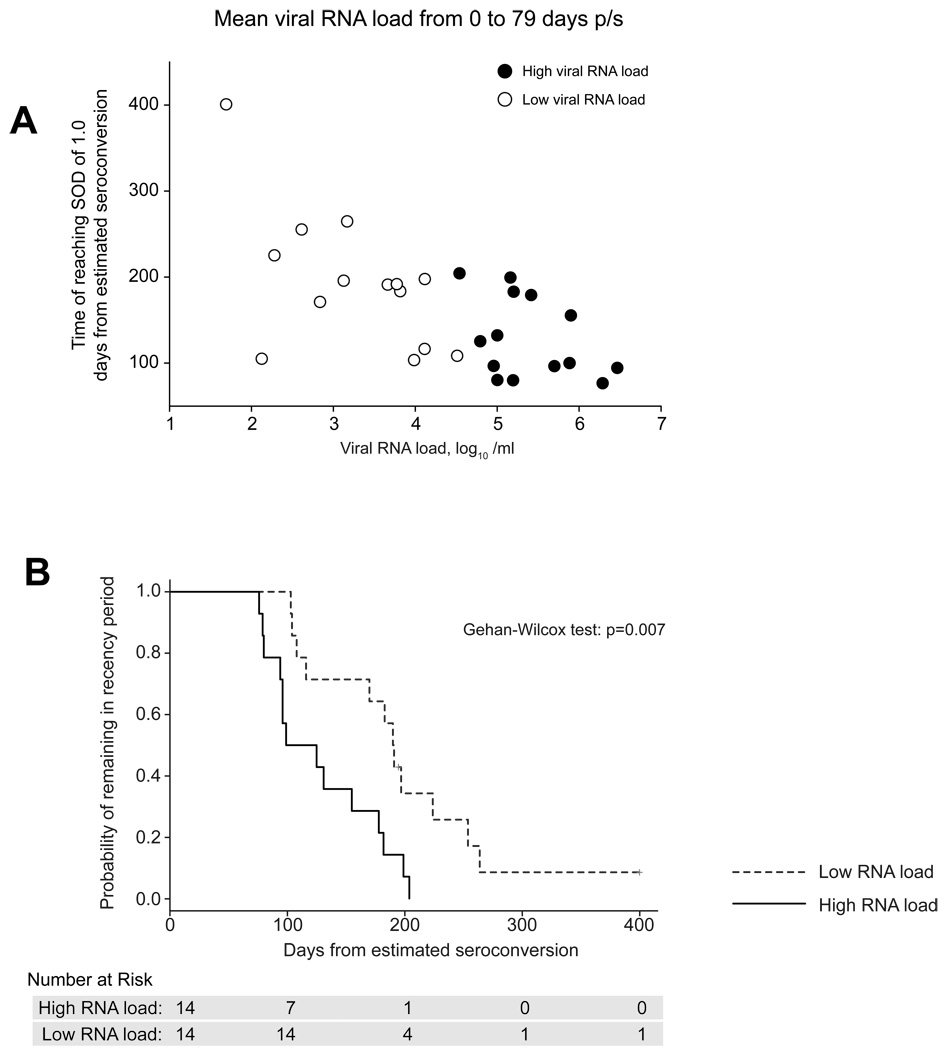
We assessed whether viral RNA load within the early post-seroconversion period of HIV-1C is associated with recency period. We computed each subject’s average viral load in the first 79 days following seroconversion (which corresponds to the end of Fiebig stage V) and their estimated time post seroconversion of reaching an SOD threshold of 1.0. The resulting scatterplot (Fig. 3A) suggests that subjects with higher early viral load levels have shorter recency periods (R2=0.419, p<0.001).
Figure 3.
Viral load by reaching SOD threshold and probability of recency period by viral load. A. A scatterplot of individual viral load levels from 0 to 79 days post estimated seroconversion and corresponding times to crossing the SOD threshold of 1.0. Median of 4.53 log10 copies/ml was used to divide subjects into groups of high (filled circles) and low (open circles) viral load. B. Kaplan-Meier estimates of the cumulative probability of remaining in recency period, by early HIV-1 viral load level. Subjects were considered to be in the high (or low) RNA group if their average RNA value from seroconversion to the end of Fiebig stage V was greater (or smaller) than the median value across all subjects. Time of seroconversion was calculated as a midpoint between the last seronegative and the first seropositive test.
Second, we classified each subject as having a high (above median) or low (below median) earlyviral load and used time-to-event methods to assess the association between viral load group (high versus low) and time until reaching an SOD threshold of 1.0. We performed 3 analyses, based on different ways of defining ‘high’ and ‘low’ viral load, and how the date of seroconversion was defined. In all analyses viral load groups were compared using the Generalized Wilcoxon test to account for censored data (subjects whose SODs never crossed 1). In the first analysis (Fig. 3B), days from seroconversion were calculated based on midpoint adjustment. That is, the time from seroconversion for a subject found by Western Blot to be in a specific Fiebig stage was taken to be the midpoint of this Feibig interval. The subject’s early viral load was then defined as the average of her/his RNA values from estimated seroconversion to the end of Fiebig stage V, and classified as high (or low) RNA group if his/her average RNA value during this period was greater (or smaller) than the median value across subjects. By these definitions, recency period was significantly shorter for subjects in the high RNA group than for those in the low RNA group (p=0.007). In the second analysis, a simulation was performed with randomly chosen time for each subject. As in the first analysis, RNA group was significantly associated with the recency period (the median p-value obtained from 2,000 such analyses was 0.01; data not shown). In the third analysis, viral load values for each subject were randomly chosen among all available RNA measurements from seroconversion to the end of Fiebig Stage V, as opposed to the mean viral load value in this period in the first and second analyses. As in the first two analyses, RNA group was significantly associated with time to SOD crossing 1 (p=0.003; data not shown).
Overall, the observed patterns suggest that early levels of viral RNA load are associated with the recency period. It is likely that the level of viral replication reflected by the level of viral RNA load drives the recency period, which suggests that viral load might be usefully incorporated into methods for estimating HIV incidence rates.
Reduction of SOD values in ARV-treated subjects
Longitudinal SOD measurements were available in 8 of 13 subjects who initiated ART (Fig. 6; filled circles). The reduction of SOD values after starting ART was a dominant pattern, despite high heterogeneity among subjects in the time and level of the drop, or subsequent increase of SOD values (e.g., subjects 2767 and 2990). It is important to determine whether initiation of ART is followed by reduction of SOD values below the threshold, and if so, what the fraction of those subjects is in subtype C infection. In our study, four of eight studied subjects dropped their SOD values below the threshold of 1.0, which may be due, at least in part, to a short period of follow-up after initiation of ART in subjects 3312 and 3481. However, subjects 2767 and 2990 demonstrated a transient drop that did not reach the threshold, followed by an increase in SOD values. Surprisingly, the expected increase of viral load was not observed and SOD increase was accompanied by suppressed viral RNA load below detectable levels and by stable proviral DNA load in both subjects.
Discussion
The goal of the study was to characterize SOD evolution during the first year of infection in a cohort of subjects identified with primary HIV-1 subtype C infection, and to estimate the recency period, expressed as a duration from the estimated time of seroconversion to the estimated time of crossing the SOD threshold of 1.0 as measured by detuned assay with the Vironostika EIA kit. Western blot kinetics in sequential samples were found to be helpful in identifying recently infected subjects and estimating time of seroconversion. The SOD in primary HIV-1 infection showed gradual increase over time in most analyzed cases. It has been suggested that decline in any two consecutive SOD values indicates an old infection.28 Our data (see Fig. 1) suggest that fluctuations in SOD values are relatively common within the early phase of HIV-1 subtype C infection. The duration of the recency period with SOD cut-off of 1.0 was estimated to be 151 days (95% CI from 130 to 172 days) post-seroconversion.
We found that the recency period was significantly associated with early post-seroconversion viral load. Subjects with higher RNA load reached the SOD threshold earlier than subjects with low viral load. In most ARV-treated subjects the drop of viral load was followed by reduction of SOD values. While viral load is a major marker of viral replication, the recency period mirrors the increase of virus-specific antibody titers. It seems plausible that the level of viral replication after seroconversion in HIV-1 subtype C infection determines the rate of antiviral antibodies elicited. The diverse levels of viral RNA in different settings may contribute to the differential performance of detuned EIA tests in non-B subtype epidemics. However, the higher levels of early viral load reported in subtype E-infected individuals (as compared with subtype B’ infection) in Thailand29 are in contrast with the longer recency period of HIV-1 subtype E,28 suggesting a need for dedicated studies able to address relationships between viral load and recency period.
We used Fiebig staging to determine the average time of seroconversion for individuals detected with recent infection, enabling the estimation of the average recency period until an SOD threshold. The notable similarity between average recency times for these subjects and for those with acute infections, where seroconversion time was determined directly, supports this use of Fiebig staging in future studies. The finding that viral load is significantly associated with recency duration suggests that estimation of HIV incidence using cross-sectional samples of standard and detuned EIA, or the BED assay, might be improved if information about viral load and/or Western blot could be incorporated into the analysis.
We note that the average recency periods obtained in this study, when used in conjunction with cross-sectional (sensitive/less sensitive) antibody testing to estimate HIV incidence rates, depend critically on the test used. Thus, the estimate of 151 days, which was obtained based on Vironostika EIA, could not be used directly in incidence estimation if another test were used to determine whether subjects were in recency period or not. However, the key findings of this study regarding the use of Fiebig staging to estimate average recency period, and the significant association between early viral load and recency period, would be expected to hold regardless of the specific testing algorithm used.
In summary, our study determined the average recency period in the local HIV-1 subtype C epidemic in Botswana to be 151 days (95% CI from 130 to 172 days; using SOD cutoff of 1.0) post-seroconversion, and provided a detailed picture of the evolution of SOD values in the early stage of HIV-1 subtype C infection. The high inter-subject variability of SOD values presents a significant challenge for accurate estimation of the recency period and HIV incidence in a heterogeneous population. We found that the level of viral replication during the early post-seroconversion period is significantly associated with the time that titers of antiviral antibodies quantified as SOD values reach the threshold.
Figure 2.
Viral load and CD4+ T cell counts in subjects who did not cross the SOD threshold of 1.0. Time 0 corresponds to the estimated time of seroconversion. Note: time and viral load scales differ between subjects. SOD scale is shown at the left; viral load and CD4 scales are shown at the right. Viral RNA (red squares) and proviral DNA (green triangles) are plotted on the same scale. CD4+ T cell counts are shown as blue diamonds. SOD values are indicated by open circles.
Acknowledgments
We are grateful to the subjects from the Tshedimoso study in Botswana. We thank Gaseboloke Mothowaeng, Florence Modise, S'khatele Molefhabangwe, and Sarah Masole for their dedication and outstanding work in the clinic and outreach. We express thanks to Busisiwe Mlotshwa for excellent laboratory support. We greatly appreciate the enthusiasm and strong commitment of Erin McDonald, Melissa Ketunuti, Carl Davis, Kenneth Onyait, and Mary Fran McLane in achieving the overall study goals. We thank the Botswana Ministry of Health, Gaborone City Council clinics, and the Gaborone VCT Tebelopele for their ongoing support and collaboration. We thank Lendsey Melton for excellent editorial assistance. The primary HIV-1 subtype C infection study in Botswana, the Tshedimoso study, was supported and funded by NIH grant R01 AI057027. This work was supported in part by the NIH grants D43 TW000004 (RR) and R37 AI24643 (RW and SL), and also through the AAMC FIC/Ellison Overseas Fellowships in Global Health and Clinical Research (LK).
Footnotes
Author contributions: V.N. and M.E. designed and guided the study, and wrote the paper. E.W. contributed to patient recruitment and provided clinical support for the cohort. L.K., J.G., and R.R. performed the experiments. S.M. and R.M. provided laboratory support. R.W. and S.L. designed and performed statistical analysis, and wrote the paper.
References
- 1.Brookmeyer R, Quinn TC. Estimation of current human immunodeficiency virus incidence rates from a cross-sectional survey using early diagnostic tests. Am J Epidemiol. 1995;141(2):166–172. doi: 10.1093/oxfordjournals.aje.a117404. [DOI] [PubMed] [Google Scholar]
- 2.Janssen RS, Satten GA, Stramer SL, et al. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA. 1998 Jul 1;280(1):42–48. doi: 10.1001/jama.280.1.42. [DOI] [PubMed] [Google Scholar]
- 3.Rawal BD, Degula A, Lebedeva L, et al. Development of a new less-sensitive enzyme immunoassay for detection of early HIV-1 infection. J Acquir Immune Defic Syndr. 2003 Jul 1;33(3):349–355. doi: 10.1097/00126334-200307010-00009. [DOI] [PubMed] [Google Scholar]
- 4.Kothe D, Byers RH, Caudill SP, et al. Performance characteristics of a new less sensitive HIV-1 enzyme immunoassay for use in estimating HIV seroincidence. J Acquir Immune Defic Syndr. 2003 Aug 15;33(5):625–634. doi: 10.1097/00126334-200308150-00012. [DOI] [PubMed] [Google Scholar]
- 5.Parekh BS, Kennedy MS, Dobbs T, et al. Quantitative detection of increasing HIV type 1 antibodies after seroconversion: a simple assay for detecting recent HIV infection and estimating incidence. AIDS Res Hum Retroviruses. 2002 Mar 1;18(4):295–307. doi: 10.1089/088922202753472874. [DOI] [PubMed] [Google Scholar]
- 6.Suligoi B, Galli C, Massi M, et al. Precision and accuracy of a procedure for detecting recent human immunodeficiency virus infections by calculating the antibody avidity index by an automated immunoassay-based method. J Clin Microbiol. 2002 Nov;40(11):4015–4020. doi: 10.1128/JCM.40.11.4015-4020.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soroka SD, Granade TC, Candal D, Parekh BS. Modification of rapid human immunodeficiency virus (HIV) antibody assay protocols for detecting recent HIV seroconversion. Clin Diagn Lab Immunol. 2005 Aug;12(8):918–921. doi: 10.1128/CDLI.12.8.918-921.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson KM, Johnson EI, Croom HA, et al. Incidence immunoassay for distinguishing recent from established HIV-1 infection in therapy-naive populations. AIDS. 2004 Nov 19;18(17):2253–2259. doi: 10.1097/00002030-200411190-00005. [DOI] [PubMed] [Google Scholar]
- 9.Barin F, Meyer L, Lancar R, et al. Development and validation of an immunoassay for identification of recent human immunodeficiency virus type 1 infections and its use on dried serum spots. J Clin Microbiol. 2005 Sep;43(9):4441–4447. doi: 10.1128/JCM.43.9.4441-4447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Ketema F, Sill AM, Kreisel KM, Cleghorn FR, Constantine NT. A simple and inexpensive particle agglutination test to distinguish recent from established HIV-1 infection. Int J Infect Dis. 2007 Sep;11(5):459–465. doi: 10.1016/j.ijid.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Cleghorn FR, Jack N, Murphy JR, et al. Direct and indirect estimates of HIV-1 incidence in a high-prevalence population. Am J Epidemiol. 1998;147(9):834–839. doi: 10.1093/oxfordjournals.aje.a009536. [DOI] [PubMed] [Google Scholar]
- 12.Hargrove JW, Humphrey JH, Mutasa K, et al. Improved HIV-1 incidence estimates using the BED capture enzyme immunoassay. AIDS. 2008 Feb 19;22(4):511–518. doi: 10.1097/QAD.0b013e3282f2a960. [DOI] [PubMed] [Google Scholar]
- 13.Lagakos S, Gable A, editors. Methodological Challenges in Biomedical HIV Prevention Trials. Washington, DC: National Academy Press; 2008. [Google Scholar]
- 14.McDougal JS, Parekh BS, Peterson ML, et al. Comparison of HIV type 1 incidence observed during longitudinal follow-up with incidence estimated by cross-sectional analysis using the BED capture enzyme immunoassay. AIDS Res Hum Retroviruses. 2006 Oct;22(10):945–952. doi: 10.1089/aid.2006.22.945. [DOI] [PubMed] [Google Scholar]
- 15.Karita E, Price M, Hunter E, et al. Investigating the utility of the HIV-1 BED capture enzyme immunoassay using cross-sectional and longitudinal seroconverter specimens from Africa. AIDS. 2007 Feb 19;21(4):403–408. doi: 10.1097/QAD.0b013e32801481b7. [DOI] [PubMed] [Google Scholar]
- 16.Laeyendecker O, Rothman RE, Henson C, et al. The effect of viral suppression on cross-sectional incidence testing in the Johns Hopkins hospital emergency department. J Acquir Immune Defic Syndr. 2008 Jun 1;48(2):211–215. doi: 10.1097/QAI.0b013e3181743980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris MK, Katzenstein DA, Israelski D, Zolopa A, Hendry RM, Hanson CV. Characterization of the HIV-1 specific humoral immune response during highly active antiretroviral therapy (HAART) J Acquir Immune Defic Syndr. 2001 Dec 15;28(5):405–415. doi: 10.1097/00042560-200112150-00001. [DOI] [PubMed] [Google Scholar]
- 18.Gupta SB, Murphy G, Koenig E, et al. Comparison of methods to detect recent HIV type 1 infection in cross-sectionally collected specimens from a cohort of female sex workers in the Dominican Republic. AIDS Res Hum Retroviruses. 2007 Dec;23(12):1475–1480. doi: 10.1089/aid.2006.0240. [DOI] [PubMed] [Google Scholar]
- 19.Buchacz K, Klausner JD, Kerndt PR, et al. HIV incidence among men diagnosed with early syphilis in Atlanta, San Francisco, and Los Angeles, 2004 to 2005. J Acquir Immune Defic Syndr. 2008 Feb 1;47(2):234–240. [PubMed] [Google Scholar]
- 20.Priddy FH, Pilcher CD, Moore RH, et al. Detection of acute HIV infections in an urban HIV counseling and testing population in the United States. J Acquir Immune Defic Syndr. 2007 Feb 1;44(2):196–202. doi: 10.1097/01.qai.0000254323.86897.36. [DOI] [PubMed] [Google Scholar]
- 21.Sakarovitch C, Rouet F, Murphy G, et al. Do tests devised to detect recent HIV-1 infection provide reliable estimates of incidence in Africa? J Acquir Immune Defic Syndr. 2007 May 1;45(1):115–122. doi: 10.1097/QAI.0b013e318050d277. [DOI] [PubMed] [Google Scholar]
- 22.Novitsky V, Woldegabriel E, Wester C, et al. Identification of primary HIV-1C infection in Botswana. NIHMSID # 79283. AIDS Care. 2008 August;20(7):806–811. doi: 10.1080/09540120701694055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novitsky V, Woldegabriel E, Kebaabetswe L, et al. Viral Load and CD4+ T Cell Dynamics in Primary HIV-1 Subtype C Infection. J Acquir Immune Defic Syndr. 2009;50(1):65–76. doi: 10.1097/QAI.0b013e3181900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speers D, Phillips P, Dyer J. Combination assay detecting both human immunodeficiency virus (HIV) p24 antigen and anti-HIV antibodies opens a second diagnostic window. J Clin Microbiol. 2005 Oct;43(10):5397–5399. doi: 10.1128/JCM.43.10.5397-5399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novitsky VA, Gilbert PB, Shea K, et al. Interactive association of proviral load and IFN-gamma-secreting T cell responses in HIV-1C infection. Virology. 2006 Mar 3;349(1):142–155. doi: 10.1016/j.virol.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003 Sep 5;17(13):1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 27.Salazar-Gonzalez JF, Bailes E, Pham KT, et al. Deciphering Human Immunodeficiency Virus Type 1 Transmission and Early Envelope Diversification by Single-Genome Amplification and Sequencing. J. Virol. 2008 April 15;82(8):3952–3970. doi: 10.1128/JVI.02660-07. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young CL, Hu DJ, Byers R, et al. Evaluation of a sensitive/less sensitive testing algorithm using the bioMerieux Vironostika-LS assay for detecting recent HIV-1 subtype B' or E infection in Thailand. AIDS Res Hum Retroviruses. 2003 Jun;19(6):481–486. doi: 10.1089/088922203766774522. [DOI] [PubMed] [Google Scholar]
- 29.Hu DJ, Vanichseni S, Mastro TD, et al. Viral load differences in early infection with two HIV-1 subtypes. AIDS. 2001;15(6):683–691. doi: 10.1097/00002030-200104130-00003. [DOI] [PubMed] [Google Scholar]