Abstract
The role in plants of posttranslational modification of proteins with O-linked N-acetylglucosamine and the evolution and function of O-GlcNAc transferases responsible for this modification are reviewed. Phylogenetic analysis of eukaryotic O-GlcNAc transferases (OGTs) leads us to propose that plants have two distinct OGTs, SEC- and SPY-like, that originated in prokaryotes. Animals and some fungi have a SEC-like enzyme while plants have both. Green algae and some members of the Apicomplexa and amoebozoa have the SPY-like enzyme. Interestingly the progenitor of the Apicomplexa lineage likely had a photosynthetic plastid that persists in a degenerated form in some species, raising the possibility that plant SPY-like OGTs are derived from a photosynthetic endosymbiont. OGTs have multiple tetratricopeptide repeats (TPRs) that within the SEC- and SPY-like classes exhibit evidence of strong selective pressure on specific repeats, suggesting that the function of these repeats is conserved. SPY-like and SEC-like OGTs have both unique and overlapping roles in the plant. The phenotypes of sec and spy single and double mutants indicate that O-GlcNAc modification is essential and that it affects diverse plant processes including response to hormones and environmental signals, circadian rhythms, development, intercellular transport and virus infection. The mechanistic details of how O-GlcNAc modification affects these processes are largely unknown. A major impediment to understanding this is the lack of knowledge of the identities of the modified proteins.
Keywords: O-GlcNAc modification, O-GlcNAc transferase, Arabidopsis, plants, posttranslational regulation, evolution
Background and History
The investigation of O-GlcNAc modification in plants is a product of forward genetic screens that were conducted to identify components of the gibberellin (GA) response pathway. Gibberellins are plant hormones that control many plant processes including germination, growth, flowering and seed development [1, 2]. Mutations and chemicals that block GA biosynthesis in Arabidopsis thaliana (arabidopsis) prevent seed germination. After germination, GA-deficiency causes dwarfism and male sterility. Screens for mutations suppressing all of the GA-deficiency phenotypes identified loss of function alleles of a gene called SPINDLY (SPY) [3–5]. Critically, spy does not suppress these phenotypes by restoring GA biosynthesis and, in plants with normal levels of GA, spy mutations cause a phenotype similar to plants that have been overdosed with GA. Thus, it was proposed that the wild-type SPY protein is a repressor of GA signaling. As will be discussed below, spy plants also have defects in response to light, meristem activity, root growth, circadian rhythms and in response to another hormone, cytokinin, that are not caused by defects in GA signaling.
Higher Photosynthetic Eukaryotes Have Two OGTs
The identification of T-DNA insertion alleles facilitated the cloning of SPY. When animal O-GlcNAc transferase (OGT) genes were cloned [6, 7], it was noted that they encoded proteins with significant overall similarity to SPY; the considerable body of knowledge from animal systems concerning OGT properties, what proteins are modified and the function of O-GlcNAc modification is reviewed elsewhere in this issue [8, 9]. In contrast to animals, which have a single OGT and fungi, which have one or no OGT, mosses and vascular plants contain two OGTs. The second OGT of arabidopsis has been named SECRET AGENT (SEC) [10]. As discussed below, SPY and SEC have distinctive TPR domain structures and catalytic domain sequence motifs (Fig. 1 and 2). The OGT of animals and fungi is SEC-like whereas both SEC- and SPY-like types are found in bacteria. SEC-like OGT also occurs in selected protists, including Giardia, diatoms/brown algae and oomycetes, and certain fungi [11]. Known examples of multiple OGTs in the same genome appear to be the result of lineage-specific gene duplications [e.g., [12]]. Green algae genomes contain only a SPY-like OGT. These observations are consistent with the first photosynthetic eukaryotic organism having both types and the SEC-like OGT being lost from the algae.
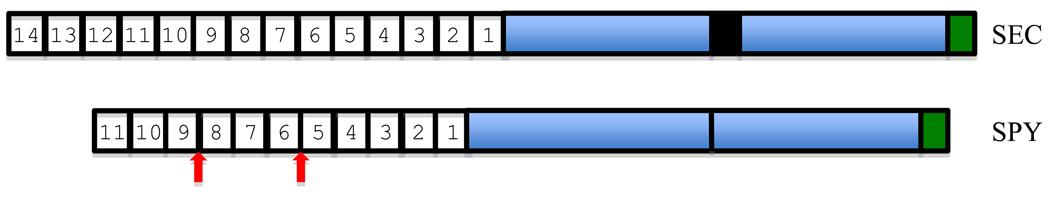
Fig. 1. Structure of SEC and SPY from Arabidopsis.
The numbered boxes indicate individual tetratricopeptide repeats (34 amino acids in length). The blue boxes indicate the two regions that together form the catalytic domain. The black box in SEC indicates the location of a variable length insertion between the catalytic domains. This insertion also occurs in animal and fungal OGTs. The green box indicates the phospholipid-binding region. The vertical arrows indicate the most common locations of amino acid insertions between TPR repeats of SPY and SPY-like enzymes.
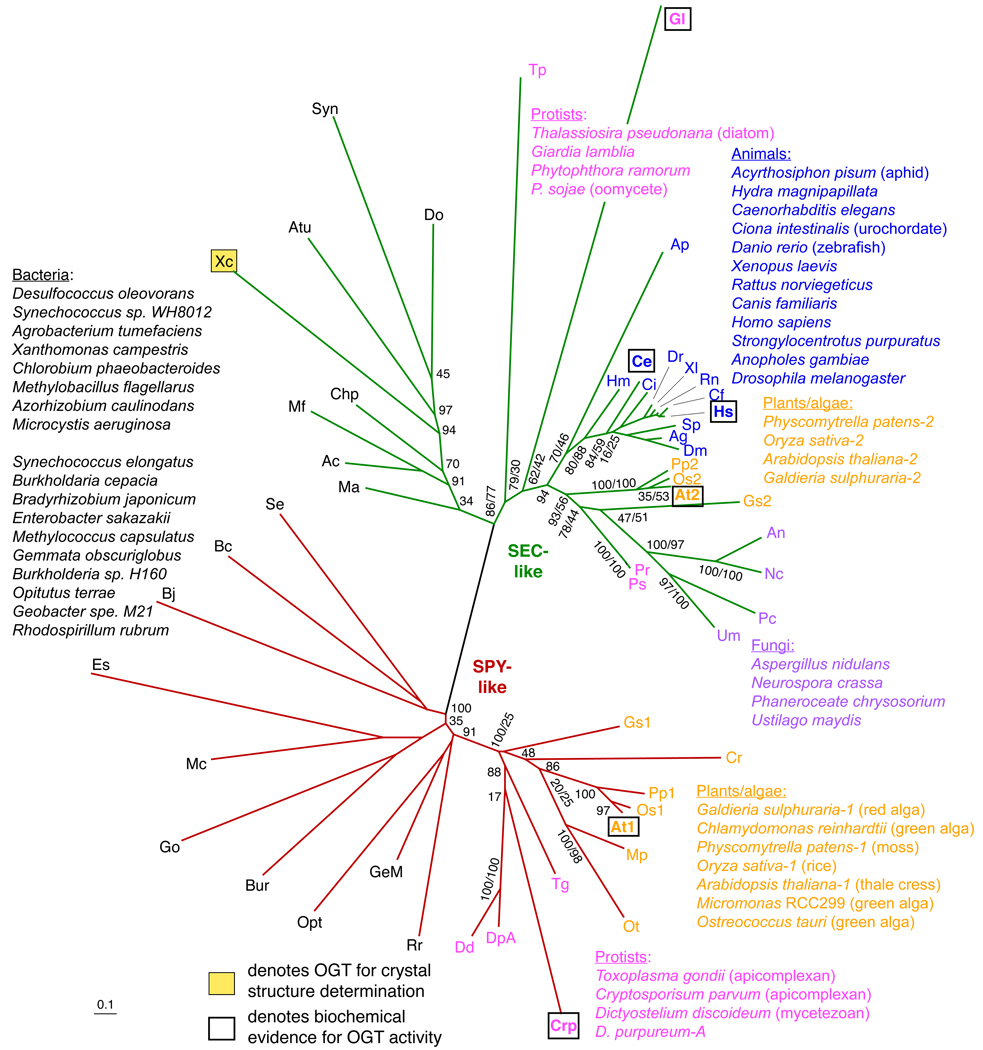
Fig. 2. Phylogenetic relationship of OGT-like catalytic domain sequences.
For the phylogenetic analysis of the aligned sequences (see Supplemental Table 1 and Supplemental Fig. 1), ProtTest 2.2 [70] was used to find the best fitting amino acid substitution model. A LG+G+I+F Le & Gascuel (LG) substitution model [71] was selected with a gamma rate (G) distribution (4 categories, alpha=1.515), a proportion of invariable (I) sites (0.006), and empirically determined amino acid frequencies (F). This model was implemented in a maximum likelihood framework, executed using the program PhyML [72] with 9,766 replicates to search for the best tree. A likelihood ratio test (LRT), as implemented in PhyML, and bootstrapping, were used to calculate support for the tree, and are listed as percent values at the nodes (LRT,bootstrap; or LRT alone). Sequences with experimentally verified OGT activity are bolded and boxed. The shaded box denotes the X. campestris sequence that has been analyzed crystallographically.
Evolutionary Origins of SEC- and SPY-like OGTs
Both plant OGTs have an overall sequence organization (Fig. 1) like that of the animal OGTs. The N-terminal half of the proteins consists of tetratricopeptide repeats (TPRs), which are a protein-protein interaction domain [13]. The second half of the protein is the catalytic region. Recently, OGT-like sequences in the human pathogen protists Giardia lamblia and Cryptosporidium parvum were shown to have in vitro OGT activity [11]. OGT-like sequences that adhere to the domain organization shown in Fig. 1 are generally found across a large number of gram-negative and gram-positive eubacteria and eukaryotic organisms, but not archaea. An alignment of the sequences of their catalytic domains (Suppl. Fig. 1) suggests a high level of conservation of functional amino acids that have been predicted by various mutagenesis studies, or through being located at the active site in the crystal structure of the OGT-like protein from Xanthomonas campestris, implying that OGT activity is conserved across these predicted proteins. A phylogenetic analysis indicates that these sequences can be classified into two major groups, as depicted by the green and red branches (Fig. 2). Known SEC-like and SPY-like sequences (unshaded boxes surrounding species abbreviations in Fig. 2) are found in the green and red clades, respectively. This division is consistent with amino acid similarities, which tend to associate with membership in the SPY or SEC groups at 38 positions distributed throughout the catalytic domain (Supplemental Fig. 1). For example, of the 53 sequences analyzed, all SEC-like sequences have Leu at the equivalent of Hs-747 (Human OGT position 747); whereas none of the SPY-like sequences do (almost all have an Arg); whereas all SPY-like sequences have a Leu at Hs-748; whereas none of the SEC-like sequences do (almost all have Trp). Interestingly, the identity of Hs-748 may be important, as detectable activity is lost when converted to Ala in Hs-OGT.
The two major clades each have the same general structure with prokaryotic sequences branching earliest followed by protists and multicellular eukaryotes, which include only plants in the SPY-clade and both animals and plants in the SEC-clade. Within the eukaryotic subclades, the proposed evolutionary relationship of the sequences approximately follows the evolution of the organism with which they are associated. Thus the OGT-like sequences do not generally appear to be under selective pressure to deviate differentially from their host species, consistent with retention of similar function. Although situations where the phylogeny of OGT-like sequences varies from expectation [14], such as the branching of the aphid before the Hydra sequence, or the association of oomycete (Phytophthora) and fungal sequences, may imply functional specialization of these SEC-like sequences, new amino acid substitution models derived from within eukaryotic and prokaryotic subclades of the SPY- and SEC-like sequences should be examined before drawing such detailed conclusions.
The TPR domains show evidence of coevolution with the catalytic domains. The structure of the TPR domains of the SPY-like and a SEC-like OGTs are distinct in that SPY-like OGTs have insertions between corresponding pairs of TPRs (Table 1, Fig. 1) while in general animal and plant SEC-like OGTs do not have these insertions. Numbering from the C-terminal most TPR toward the N-terminus, the size and location of the insertions is conserved across the SPY-like OGTs of eukaryotes. A phylogenetic analysis of sequences of each of the TPR domains from selected eukaryotes reveals remarkable conservation of domains both close to and distant from the catalytic domain (Suppl. Fig. 2). Thus the proximal and distal TPR domains appear to have co-evolved with their catalytic domain, retaining SEC-like or SPY-like character and positional identity. Conservation of TPR-1 is consistent with structural evidence from X. campestris that it contacts the catalytic domain near the active site [15]. Conservation of other TPRs suggests the existence of conserved interactions with other proteins such as adaptors or substrates, which might have coevolved with the catalytic domain during speciation. While the bacterial OGTs used for the phylogenetic analysis shown in Fig. 2 have TPR domains with between 3 and 13 repeats, a preliminary analysis did not detect evidence that the pattern of TPR domain conservation found among the eukaryotic enzymes extends to the bacterial enzymes.
Table 1.
Presence and position of insertions in TPR domains of selected OGTs. TPR-repeats were predicted [73] as implemented at http://toolkit.tuebingen.mpg.de/tprpred.
| Organism | SPY-like Accession # |
[TPR Number]; Gap Position (Size) |
SEC-like Accession # |
[TPR Number]; Gap Position (Size) |
|---|---|---|---|---|
| Plants/Green Algae | ||||
| Arabidopsis thaliana | NP_187761.1 | [11]; 5 (7), 8 (7) | NP_187074.1 | [14]; No gaps |
| Vitis vinifera | CAO15870.1 | [11]; 5 (7), 8 (7) | XP_002270163.1 | [14]; No gaps |
| Populus trichocarpa | XP_002308458.1 | [11]; 5 (7), 8 (7) | XP_002319130.1 | [14]; No Gaps |
| Oryza sativa | NP_001062501.1 | [11]; 5 (7), 8 (7) | EEE55882.1 | [14]; No Gaps |
| Ricinus communis | EEF31867.1 | [11]; 5 (7), 8 (7) | EEF40435.1 | [13]; No Gaps |
| Physcomitrella patens | XP_001782948.1 | [11]; 5 (8), 8 (7) | XP_001774513.1 | [14]; No Gaps |
| Micromonas sp. RCC299c | ACO61142.1 | [12]; 3 (2), 6 (7), 9(7) | No gene | |
| Ostreococcus tauri | CAL57394.1 | [10]; 5 (7), 8 (13) | No gene | |
| Chlamydomonas reinhardtii | XP_001694146.1 | [10]; 5 (7), 7 (44) | No gene | |
| Apicomplexans | ||||
| Toxoplasma gondii GT1 | EEE26626.1 | [11]; 5 (7), 8 (7) | No gene | |
| Cryptosporidium parvum Iowa II | XP_627974.1 | [7]; 5 (8), 6 (3) | No gene | |
| Dictyosteliida | ||||
| Dictyostelium discoideum AX4 | XP_638298.1 | [8]; 3 (12), 5 (7) | No gene | |
| Dictyostelium purpureum | jgi|Dicpu1|39606| | [7]; 5 (7) | No gene | |
| Other Organisms | ||||
| Giardia lamblia | No gene | XP_001707304 | [10]; 3 (257), 4 (35), 5 (9), 6 (29), 8 (7), 9 (3) | |
| Aspergillus nidulans | No gene | XP_657869 | [4]; 1 (206), 2 (110) |
The overall organization of the catalytic domain tree strongly suggests that the SEC- and SPY-like sequences are related by an ancient gene duplication in the prokaryotic realm. The high degree of conservation of functionally important amino acids (Suppl. Fig. 1) supports other evidence for similar biochemical activity, though this remains to be confirmed for any bacterial example. A recently reported example of an O-β-GlcNAc modification on flagellin protein from Listeria monocytogenes is the product of an unrelated gene [16]. OGT-like sequences are notably absent in many bacteria, protists and fungi suggesting, as noted previously [11], that selective losses occurred from organisms that do not benefit from OGT function. The bacteria that possess SEC-like vs. SPY-like sequences are not obviously related suggesting that each type of OGT is associated with some as yet unknown physiological character. In eukaryotes, SEC-like sequences are broadly distributed in protists, fungi, plants and animals, whereas SPY-like sequences are restricted to mycetozoans, apicomplexans, red algae, green algae, mosses and vascular plants. With the exception of the mycetozoans, these organisms are related by a common ancestor derived from an endosymbiotic relationship with a cyanobacterium, leading to the modern day chloroplast or a relict chloroplast called the apicoplast that is retained in some modern apicomplexans [17]. This suggests that the SPY-like OGTs were introduced into eukaryotes via the endosymbiont that gave rise to the chloroplast, consistent with lack of evidence for both OGT types in known bacterial genomes. While the mycetozoans Dictyostelium is an apparent exception to this model, it is notable that its nuclear genome possesses some (though not all) of the genes associated with starch metabolism and that its CAZy family GT5 and a GH77 4-α-glucanotransferase (transglycosidase)-like genes are closed related to those of glaucophytes and red algae [18] which still possess the endosymbiont. While these observations raise the possibility of an ancient common ancestry of Dictyostelium and chloroplast-containing organisms, entry of SPY-like sequences into the mycetozoans by horizontal gene transfer cannot be excluded. Future studies are needed to investigate the biochemical and cellular functions of the large number of non-metazoan and non-plant OGTs predicted by this analysis, as well as to better understand the functional significance of the ancient SEC/SPY-division, discussed further below.
The spy Alleles Identify Important OGT Domains
All of the spy alleles selected in forward genetic screens were identified as suppressors of GA deficiency. The molecular defects of 20 of these alleles have been determined [4]. Of the eight alleles with mutations affecting the TPR domain, all are predicted to encode proteins with an intact catalytic domain that has only a small in-frame deletion or a single amino acid change in the TPR domain. Numbering from the C-terminus toward the N-terminus, the mutations affect TPR 2, 3 or 5, suggesting that the unaffected TPRs have no role in GA signaling. Since as discussed below SPY has roles in processes beyond GA signaling, it could be informative to conduct screens for new spy alleles based on its role in these other processes and then determine if any of the alleles are affected in TPRs not involved in GA responses.
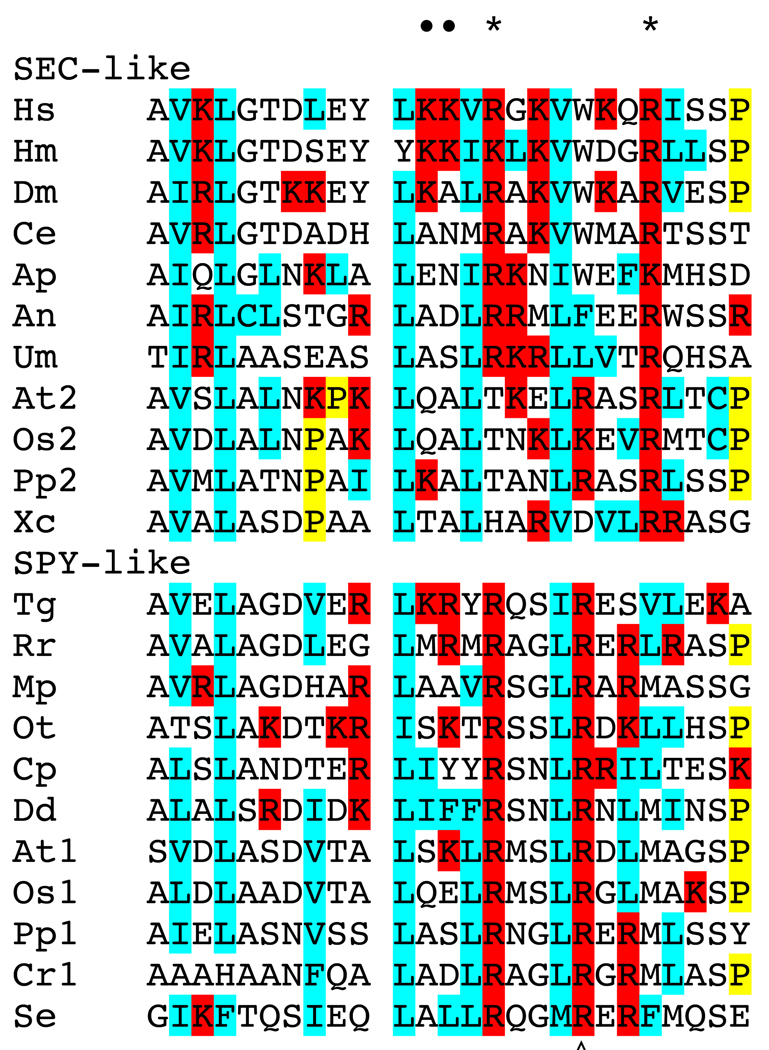
Nine spy alleles are affected in the catalytic domain. Corresponding mutations made for several of these alleles in the human OGT resulted in proteins with reduced enzymatic activity [19]. In addition to the TPR and catalytic domains, a phosphoinositide-binding domain is present at the C-terminus of human OGT [20]. This domain binds PI(3,4,5)P3 most strongly and site-directed mutations that inhibit binding do not inhibit catalytic activity. This domain influences OGT cellular localization and regulatory activity. A non-binding mutant did not localize to the plasma membrane and did not function properly in insulin signaling. Structural analyses of the OGT from Xanthomonas campestris [15, 21, 22] suggest that it has a phospholipid-binding domain and sequence comparisons suggest that this domain is present in most OGTs (Figure 3). Therefore, phospholipid binding and membrane localization are likely to be important to the functioning of many OGTs. The amino acids predicted to have critical roles in phospholipid binding are not highly conserved, suggesting that different OGTs interact with different phospholipids and perhaps localize differently. The amino acids that are likely to be critical for phospholipid binding differ between SPY and SEC, suggesting that they have different binding specificities. The spy-16 and -17 alleles have a missense mutation affecting this domain and cause phenotypic defects similar to other spy alleles, suggesting that phospholipid binding is essential for all SPY functions.
Fig. 3. Alignment of predicted phospholipid binding domain sequences.
Alignment of the phospholipid binding domains of: Hs, Homo sapiens; Hm, Hydra magnipapillata; Dm, Drosophila melanogaster, Ce, Caenorhabditis elegans; Ap, Acyrthosiphon pisum; An, Aspergillus niger; Um, Ustilago maydis; At2, Arabidopsis thaliana SEC; Os2, Oryza sativa SEC; Pp2, Physcomitrella patens SEC; Xc, Xanthomonas campestris; Tg, Toxoplasma gondii; Rr, Rhodospirillum rubrum; Mp, Micromonas RCC299; Ot, Ostreococcus tauri; Cp, Cryptosporidium parvum; Dd, Dictyostelium discoideum; At1, Arabidopsis thalaian SPY, Os1, Oryza sativa SPY; Pp1, Physcomitrella patens SPY; Cr1, Chlamydomonas reinhardtii; Se, Synechococcus elongatus PCC 6301. Amino acids demonstrated by mutagenesis of human OGT (•) or predicted from the Xanthomonas campestris crystal structure (*) to be important for PIP3 binding are indicated above the alignment. The amino acid affected in the spy-16 and -17 alleles is indicated below the alignment (◊). Hydrophobic amino acids are highlighted in blue, basic amino acids are highlighted in red, and Pro in yellow.
It is surprising and perhaps informative that to date no confirmed null spy alleles are known. The characterized alleles are either small in frame deletions, missense mutations or truncate the protein after the catalytic region. The spy-4 allele is a T-DNA inserted into the promoter which greatly reduces SPY RNA abundance but a small amount of transcript is detectable by RtPCR [23]. It is possible that no spy mutation completely blocks enzymatic activity because none of the mutations, including the spy-12 mutation, which eliminates detectable self-modification activity [24], affects amino acids predicted from the XcOGT crystal structure [21, 22] to participate in catalysis. Additionally, one cannot rule out the possibility that SPY has functions that do not require catalytic activity. It is therefore possible that a true null mutant will have a phenotype distinct from any of the characterized mutants.
Processes Affected by Altering OGT Activity
The major challenge for understanding the function of O-GlcNAc modification is to determine the mechanism(s) by which the modification affects plant processes. Currently, the mechanism of action of O-GlcNAc modification is not known for any of the process affected when OGT activity is altered.
SEC and SPY have Overlapping Functions
SPY and SEC appear to have overlapping function in gametes and during embryogenesis [10]. Transmission through gametes is normal when sec or spy alleles are transmitted singly. However, transmission together is greatly reduced through the male and slightly reduced through the female. sec/spy double mutant embryos die prior to completion of seed development. Seed abortion does not occur at a specific stage of development, suggesting that abortion occurs when O-GlcNAc modification of protein(s) drops below a critical threshold. It is also possible that SEC and SPY affect independent processes and that embryo lethality only occurs when both processes are affected. Because of the embryo lethality, the requirements for both SEC and SPY in seedlings or plants have not been determined.
Gibberellin Signaling
Gibberellins (GA) are hormones that regulate numerous plant responses. Loss of SPY function through mutation [3–5, 25, 26] or through dominant negative effects caused by expression of the TPR domain or a heterologous OGT [27–29] activate GA responses indicating that SPY acts to repress GA responses. GA responses occur when GA signaling triggers the proteolytic destruction of a group of transcription factors, called the DELLA proteins, that repress GA responses [30, 31]. The GA bound receptor interacts with the DELLA proteins and the resulting DELLA/GA/receptor complex then interacts with the SCF complex which ubiquitinates the DELLA protein targeting them for destruction by the proteasome. The DELLA domain for which these proteins are named is not required for repression of GA responses. Mutations affecting only the DELLA domain render the protein stable thereby causing a GA-insensitive phenotype. spy suppresses phenotypes caused by mutations in the DELLA domain [5, 25]. Suppression of DELLA domain mutations by spy could occur though promoting the destruction of DELLA proteins or by blocking DELLA activity. The latter seems to be occurring because DELLA protein is more abundant in spy plants [4]. These results suggest that O-GlcNAc modification of DELLA proteins could activate them. Further evidence for this hypothesis comes from rice, where reducing rice SPY expression using antisense and RNAi approaches activates GA signaling and also causes an increase in the phosphorylation of the rice DELLA protein [32]. The increased phosphorylation of rice DELLA is consistent with the model that reducing SPY activity increases phosphorylation of the DELLA by making amino acid(s) that can be either O-GlcNAc modified or phosphorylated available to be phosphorylated. While it is well documented for many animal proteins that the same amino acid can be either phosphorylated or O-GlcNAc modified, and that modification of a given residue affects the alternative modification of neighboring residues [8] [33], it is not known if this also happens in plants.
During germination, GA from the embryo controls the hydrolysis of storage compounds that makes nutrients available to the growing embryo. This process has been well studied in grasses where GA triggers the production and secretion of alpha-amylase from the aleurone layer into the endosperm, the major storage tissue. Consistent with the predicted role as a negative regulator of GA responses, overexpression of barley HvSPY in aleurone protoplasts reduces GA stimulated transcription of alpha-amylase genes [34]. Another hormone, abscisic acid (ABA), blocks the action of GA; HvSPY stimulates the transcription of Dhn1-2, an ABA regulated gene, raising the possibility that HvSPY inhibits GA responses by activating ABA responses. This possibility seems unlikely because HvSPY acts independently of ABA when activating Dhn1-2 transcription [35]. Deletion of promoter elements required for ABA activation of the Dhn1-2 promoter had no effect on activation by HvSPY and moreover, ABA and HvSPY exhibited an additive interaction in the activation of this promoter.
Two hybrid screens for proteins that interact with HvSPY have identified a number of proteins [36] including NAC and myb transcription factors. Expression of these transcription factors in barley aleurone cells inhibits GA-induced activation of the alpha-amylase promoter but did not affect control promoters, suggesting that HvSPY acts by controlling the activity of these transcription factors. It remains to be determined if these transcription factors are O-GlcNAc modified and if modification affects their activity.
SPY Increases Cytokinin Responses
Loss of spy function inhibits a number of cytokinin responses [37, 38], including inhibition of root elongation, suppression of senescence, enhanced production of anthocyanin, trichomes, transcript accumulation and promotion of leaf serration. The interaction between spy and cytokinin response is complicated by the interaction of GA and cytokinins as well as the fact that SPY has both GA dependent and GA independent roles in plant development. Treatment with GA suppresses many cytokinin responses raising the possibility that spy blocks cytokinin action by increasing GA signaling rather than through a direct effect on the cytokinin response pathway but the evidence is most consistent with a direct role for SPY. In general, spy mutations are more effective than GA treatment in suppressing cytokinin action, with only spy having an effect in some cases. In addition, loss of function DELLA mutants respond to cytokinin [38]. Studies of the role of GA, cytokinin and SPY in trichome development also support SPY acting independently of GA [39].
Light and Circadian Rhythms
The arabidopsis protein GIGANTEA (GI) functions in common pathways with SPY. GI and SPY interact physically and genetic interactions between mutations affecting these proteins support this hypothesis [40, 41] but the functional interrelationship between GI and SPY is not clear. GI affects a number of processes including the circadian clock, red light regulation of hypocotyls elongation and long day induction of flowering [42–44]. The delayed flowering and long hypocotyls phenotypes of gi plants are suppressed by spy, suggesting that SPY functions downstream of GI or that spy suppresses gi by a bypass mechanism [40]. Interestingly, like GI, SPY affects circadian rhythms [40, 41]. In spy plants the period of the free running rhythms for cotyledon movement and transpiration is longer. The period of the free running rhythm for cotyledon movement is shorter in plants overexpressing SPY. In addition to affecting the period of the rhythm, gi affects rhythm amplitude, which is not affected in spy plants. Moreover, spy does not suppress the rhythm defects caused by gi.
Root Development
SCARECROW (SCR) is involved in root ground tissue patterning. One defect of scr roots is an occasional premature longitudinal ground cell division leading to early formation of a middle cortex layer. This phenotype is suppressed by treatment with GA [45]. Recently it has been discovered that mutations that reduce GA responsiveness enhance early formation of a middle cortex layer [46]. This work also made the surprising observation that, although spy mutations enhance GA responses, they enhance early formation of a middle cortex layer. This study also discovered that mutations affecting Like Heterochromatin protein 1 (LHP1) promote early formation of the middle cortex layer. LHP1 binds H3 histones trimethylated at lysine 27 and is implicated in epigenetic regulation of gene expression. Since animal OGTs are associated with a Sin3 histone deacetylase repressor complex [47], the effect of the trichostatin A, a histone deacetylase inhibitor, on root development was examined. Trichostatin A causes the precocious formation of the middle cortex layer. These results suggest that SPY, like animal OGTs, may associate with chromatin and regulate gene expression through epigenetic mechanisms involving SPY regulation of histone deacetylase activity but as discussed below SPY appears to function in the cytoplasm when affecting GA and cytokinin responses [38]. Additional support for the hypothesis that O-GlcNAc has a role in root development may come from studies of a rice mutant that is affected in the gene encoding glucosamine-6-phosphate acetyltransferase, OsGNA1 [48]. The mutation reduces UDP-GlcNAc levels to 10% of wild type and causes a temperature-sensitive short root phenotype. Mutant roots were much shorter than wild type when grown at 25°C and but were less affected at 32°C. The mutation reduced respiration and cell division and caused disorganized microtubules. OsGNA1 acts in the de novo UDP-GlcNAc synthesis pathway. The amount of concanavalin A-reacting proteins was reduced in the mutant indicating a reduction in N-linked protein glycosylations. Labeling of proteins with galactose by galactosyl transferase, which selectively labels terminal GlcNAc, was also reduced in the mutant. Based on this result, it was suggested that the amount of O-GlcNAc modification is likely reduced as well. However, plants have many N-linked glycans structures that terminate with GlcNAc [49] and these N-modifications are more abundant than O-GlcNAc modifications [50]. Therefore, it likely that the reduction in labeling of the rice mutant primarily reflects changes in N-glycosylation.
Brassinosteroid Synthesis
Brassinosteroids are hormones that affect many plant processes occurring throughout the life of the plant [51]. Rice plants with reduced levels of spy also exhibited enhanced bending of the lamina joint, a phenotype consistent with increased brassinosteroid signaling [32]. The spy-knockdown plants had more brassinosteroid and exhibited increased expression of brassinosteroid biosynthesis genes, suggesting that in rice SPY suppresses hormone synthesis.
PROCESSES AFFECTED IN sec PLANTS
sec plants have few and relatively minor developmental defects [10, 52]. This could be because, similar to the situation during embryo development, SEC and SPY have overlapping functions and SPY alone is sufficient for most developmental processes. Since spy mutants have a number of developmental defects, SPY is either more active than SEC or SPY has unique activities. SPY and SEC negatively regulate the others’ RNA levels [52]. Therefore, the increased expression of one enzyme due to loss of the other could be sufficient to maintain sufficient enzyme activity for some processes where they have overlapping functions.
sec plants do not have any obvious defects in hormone, light or circadian clock-regulated processes [52]. They do, however, produce leaves at a slightly slower rate and have a shorter shoot at flowering. They flower at the same time as wild type and thus have fewer leaves at flowering. Consistent with an overlapping function during flower development, plants that lack one enzyme and are haploinsufficient for the other have defects in flower development. Although sec plants have no detectable defects in GA responses, there is also a complex and poorly understood interaction between these genotypes and GA during flower development [52]. GA-deficient plants that are homozygous for sec and heterozygous for spy, rather than producing flowers, produce pin-shaped organs. This phenotype is corrected by treatment with GA. Even more surprisingly, haploinsufficiency for sec suppresses the ability of spy mutations to suppress the need for GA during germination. The flower development defects suggest that SEC and SPY have overlapping roles that intersect with GA responses during flower development, but the germination phenotype suggests that SEC and SPY have opposite effects on GA responses during germination.
SEC deficiency impairs infection by Plum Pox Virus (PPV). PPV is a Potyvirus that infects stone fruit trees such as plums and peaches and in the laboratory infects arabidopsis. The virion capsid protein is O-GlcNAc modified [53]. Modification of the capsid is present on virus from spy plants but is not detectable on virus purified from sec plants [54], suggesting that SEC modifies the capsid or that modification of an arabidopsis protein is required for modification of the capsid. The former seems to be the case because SEC O-GlcNAc modifies the capsid protein when they are co-expressed in E. coli [55]. The same amino acids that are modified by SEC in E. coli are modified in arabidopsis [55, 56]. Each modification to capsid protein from plants is a GlcNAc monomer indicating that SEC and animal OGTs make the same modification. Modification of the capsid protein is not required for infection but early in the infection of sec plants, virus movement is slower and the titer is lower [54]. Later in the infection, titer and movement are similar to that seen in wild type. Defects in virus movement, replication, virion assembly or some combination of these processes could be responsible for the defect in infection. It is not clear if the defects in infection are a direct effect of the defect in modification of the capsid and/or a defect in the modification of a plant protein. One study where the known modification sites were mutated to non-modifiable amino acids did not impair infection [56], but it is possible that not all of the modified sites have been identified.
Cell-to-cell Movement
The cytoplasm of most plant cells is interconnected through a complex structure called the plasmodesma. It has been proposed that this structure is related to the nuclear pore [57]. Regulated intercellular movement of macromolecules including proteins and RNA occurs via the plasmodesma. Viruses also move between cells via the plasmodesma. Several lines of evidence suggest that O-GlcNAc protein modification has a role in plasmodesmatal transport. A role for O-GlcNAc in this process is not altogether surprising since the nuclear pore of animals is highly O-GlcNAc modified [9]. While the defects in infection of sec plants by PPV could be due to a defect in cell-to-cell movement, more direct evidence for O-GlcNAc affecting transport through the plasmodesma comes from the work of Taoka et al [58]. Proteins that move from cell-to-cell are called NCAPs for non-cell-autonomous proteins. The phloem component of the vascular system is a rich source of NCAPs. Trafficking of NCAPs is mediated by non-cell-autonomous protein 1 of Nicotiana tabaccum (Nt-NCAPP1; [59]). Taoka et al. found that Nt-NCAPP1 and many pumpkin phloem proteins are phosphorylated and react with anti-O-GlcNAc antibodies. They also found that treatment with phosphatase or hexosaminidase prevented the interaction. More detailed analysis of one NCAP, Cm-PP16-1, found that some serine to alanine mutations affected phosphorylation and GlcNAc modification, suggesting that both modifications could occur at the same residues. Mutations that affected posttranslational modification also blocked interaction with Nt-NCAPP1 and plasmodesmatal trafficking. Although these experiments used antibodies to detect O-GlcNAc, which as discussed below can be problematic, the experiments had sufficient controls to give high confidence that the antibodies were detecting O-GlcNAc on Nt-NCAPP1 and NCAPs. Therefore, O-GlcNAc modification is likely involved in plasmodesmatal trafficking.
What Is The Cellular Site of SPY Action?
SPY is localized both in the nucleus and the cytosol [60]. Genetic experiments suggest that it acts at or downstream of the DELLA proteins, which are transcription factors, suggesting that it acts in the nucleus. Surprisingly, recent experiments suggest that for both GA and cytokinin signaling, SPY is acting in the cytosol [38]. In these experiments, a SPY fusion protein with a nuclear export sequence (NES) rescued the GA and cytokinin response defects of spy mutants but SPY with a nuclear localization (NLS) sequence did not. These experiments do not completely rule out the possibility that SPY is acting in the nucleus. Although SPY:NES fusion protein does not accumulate in the nucleus it likely transiently enters the nucleus because the NES acts by triggering rapid export of proteins from the nucleus. In addition, it is not known if the fusion protein containing the NLS has OGT activity. If SPY acts only in the cytosol during GA and cytokinin signaling, one is left to wonder what, if anything, does nuclear localized SPY do?
What Proteins Are O-GlcNAc Modified?
One of the major limitations to understanding the role of O-GlcNAc modification in plants is the lack of knowledge of the identities of the modified proteins. Less than a handful of proteins have been shown to be O-GlcNAc modified. Detection of modified plant proteins is difficult because several of the methods that are commonly used to detect O-GlcNAc modification are problematical with plants, and thus appropriate control experiments or positive results from several methods are needed for high confidence that a protein is modified. Galactosyl transferase (GalT) is commonly used to selectively transfer labeled or reactive galactose to terminal GlcNAc [61, 62]. Many of the N-linked complex glycans terminate with GlcNAc [49] and thus are labeled by the GalT reaction. When total proteins of arabidopsis are labeled by this method, over 90% of the label is incorporated into structures that are refractory to β-elimination and thus are likely N-linked [50]. Because of structural differences between the N-linked glycans of plants and animals, N-linked plant glycans are resistant to removal by PNGAseF, but are susceptible to PNGaseA. Therefore, for a modification detected by labeling with GalT to be considered O-linked, the modification must be removed by β-elimination and be refractory to glycosidase A. This approach does suffer from the drawback that it does not determine the structure of the modifications. Since as discussed below at least one plant protein, gp40, has a complex O-linked modification with terminal GlcNAc, it is also important to determine the structure of the modification. Commercially available anti-O-GlcNAc antibodies, while a very useful tool for the detection of single O-linked GlcNAc, can react non-specifically with plant proteins [63]. One way to demonstrate the specificity of an antibody reaction is to demonstrate that capping the modification with galactose inhibits antibody binding, or by confirming that non-modified recombinant protein does not react with the antibodies.
Many plants require exposure to prolonged low temperatures (vernalization) for flowering. A recent study with winter wheat vernalization causes a global increases in reactivity with anti-O-GlcNAc antibodies and that devernalization decreases reactivity [64] suggesting a role for O-GlcNAc modification in the vernalization process. It will be interesting to learn if changes in O-GlcNAc modification are confirmed using other methods and if similar changes in response to vernalization are detected in other species.
The only known modified protein that has not already been discussed is the nuclear pore complex protein gp40 from tobacco [65, 66]. Interestingly, this protein is a close homolog of NtNCAPP1, suggesting that O-GlcNAc has roles in both nuclear and plasmodesmatal trafficking. It will be interesting to learn if the modification is made by a SEC-, or SPY-like OGT or other enzyme(s), particularly since the modification is more complex than a single O-GlcNAc.
Future Prospectives
It has been proposed that both substrate and regulatory interactions occur via TPRs. The structural differences between the TPR domains of SEC-like and SPY-like OGTs suggest that they are subject to different regulation, have different substrates, or both. The observation that SPY-like genes occur in organisms that contain chloroplasts, apicoplasts or had chloroplasts in their earlier lineages suggests a functional relationship between SPY and chloroplasts and/or photosynthesis. In modern plants, some steps of the GA biosynthesis pathway occur in the chloroplast. However, like many chloroplast proteins, GA biosynthesis proteins are encoded by nuclear genes [67]. Evidence suggests that these genes have migrated from the plastid to the nucleus, suggesting an evolutionary advantage provided by nuclear regulation of chloroplast function. The GA pathway is not the only novelty brought along by the chloroplast to ancient endosymbiotic cells [68]. In addition to photosynthesis, chloroplasts perform much of the plant’s other biochemistry including synthesis of amino acids, lipids and hormones. While it is not clear if SPY-like genes were transferred from the chloroplast to the nucleus, the enzyme could have been maintained to coordinate chloroplast and cellular activities. Similar to a proposed function of animal OGTs, SPY could be a metabolic sensor that coordinates cellular activity with plastid biochemical activities. It is also possible that this was a historical function that has been lost and that SPY has new functions. How SEC fits into this picture, if at all, is less clear. SEC could augment the role of SPY or it could have unique roles. Since SEC and SPY origins appear to predate the origin of plants, it is possible that SEC has unique functions. The similarity of SEC to animal OGTs suggests it may have similar roles. In addition to being located in the cytosol and nucleus, human OGT is also found in the mitochondria [69]. Perhaps one SEC function is to help coordinate mitochondrial and cellular functions.
Supplementary Material
Acknowledgments
We thank Edward Braun (University of Florida) for his advice on the phylogenetic analyses. Supported by BARD US-3896-06 and NSF MCB-0820666 to NEO, DOE DE-FG01-04ER04 to NEO and LMH, and Oklahoma Center for Advancement of Science and Technology HR04-141 (20041406) and NIH R01 GM37539 to CMW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schwechheimer C. Understanding gibberellic acid signaling--are we there yet? Curr. Opin. Plant Biol. 2008;11:9–15. doi: 10.1016/j.pbi.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Hartweck LM. Gibberellin signaling. Planta. 2008;229:1–13. doi: 10.1007/s00425-008-0830-1. [DOI] [PubMed] [Google Scholar]
- 3.Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 2007;143:987–1000. doi: 10.1104/pp.106.091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson RN, Somerville CR. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 1995;108:495–502. doi: 10.1104/pp.108.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreppel L, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 7.Lubas WA, Frank DW, Krause M, Hanover JA. O-linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- 8.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagen.2009.07.018. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagen.2009.07.017. This Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartweck LM, Scott CL, Olszewski NE. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. Have Overlapping Functions Necessary for Gamete and Seed Development. Genetics. 2002;161:1279–1291. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee S, Robbins PW, Samuelson J. Molecular characterization of nucleocytosolic O-GlcNAc transferases of Giardia lamblia and Cryptosporidium parvum. Glycobiology. 2009;19:331–336. doi: 10.1093/glycob/cwn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webster DM, Teo CF, Sun Y, Wloga D, Gay S, Klonowski KD, Wells L, Dougan ST. O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev Biol. 2009;9:28. doi: 10.1186/1471-213X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Fleites C, He Y, Davies GJ. Structural analyses of enzymes involved in the O-GlcNAc modification. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagen.2009.07.019. This Issue. [DOI] [PubMed] [Google Scholar]
- 16.Shen A, Kamp HD, Grundling A, Higgins DE. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 2006;20:3283–3295. doi: 10.1101/gad.1492606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obornik M, Janouskovec J, Chrudimsky T, Lukes J. Evolution of the apicoplast and its hosts: from heterotrophy to autotrophy and back again. Int J Parasitol. 2009;39:1–12. doi: 10.1016/j.ijpara.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Deschamps P, Colleoni C, Nakamura Y, Suzuki E, Putaux JL, Buleon A, Haebel S, Ritte G, Steup M, Falcon LI, Moreira D, Loffelhardt W, Raj JN, Plancke C, d'Hulst C, Dauvillee D, Ball S. Metabolic symbiosis and the birth of the plant kingdom. Mol Biol Evol. 2008;25:536–548. doi: 10.1093/molbev/msm280. [DOI] [PubMed] [Google Scholar]
- 19.Lazarus BD, Roos MD, Hanover JA. Mutational analysis of the catalytic domain of O-linked N-acetylglucosaminyl transferase. J. Biol. Chem. 2005;280:35537–35544. doi: 10.1074/jbc.M504948200. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 21.Clarke AJ, Hurtado-Guerrero R, Pathak S, Schuttelkopf AW, Borodkin V, Shepherd SM, Ibrahim AF, van Aalten DM. Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 2008;27:2780–2788. doi: 10.1038/emboj.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Fleites C, Macauley MS, He Y, Shen DL, Vocadlo DJ, Davies GJ. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nature structural & molecular biology. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 23.Filardo FF. PhD Thesis. Bundoora, Australia: La Trobe University; 2004. SPY, a negative regulator of GA response, in Arabidopsis thaliana: An investigation of the TPR domain. [Google Scholar]
- 24.Thornton TM. PhD. University of Minnesota; 2001. The role of O-GlcNac modification in gibberellin signal transduction; p. 141. [Google Scholar]
- 25.Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Nat. Acad. Sci. USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobsen SE, Olszewski NE, Meyerowitz EM. SPINDLY's role in the gibberellin response pathway. Symp Soc Exp Biol. 1998;51:73–78. [PubMed] [Google Scholar]
- 27.Izhaki A, Swain SM, Tseng T-s, Borochov A, Olszewski NE, Weiss D. The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J. 2001;28:181–190. doi: 10.1046/j.1365-313x.2001.01144.x. [DOI] [PubMed] [Google Scholar]
- 28.Tseng T-s, Swain SM, Olszewski NE. Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol. 2001;126:1250–1258. doi: 10.1104/pp.126.3.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filardo F, Robertson M, Singh DP, Parish RW, Swain SM. Functional analysis of HvSPY, a negative regulator of GA response, in barley aleurone cells and Arabidopsis. Planta. 2009;229:523–537. doi: 10.1007/s00425-008-0843-9. [DOI] [PubMed] [Google Scholar]
- 30.Achard P, Genschik P. Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 2009;60:1085–1092. doi: 10.1093/jxb/ern301. [DOI] [PubMed] [Google Scholar]
- 31.Schwechheimer C, Willige BC. Shedding light on gibberellic acid signalling. Curr. Opin. Plant Biol. 2009;12:57–62. doi: 10.1016/j.pbi.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 2006;48:390–402. doi: 10.1111/j.1365-313X.2006.02875.x. [DOI] [PubMed] [Google Scholar]
- 33.Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Robertson M, Swain SM, Chandler PM, Olszewski NE. Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell. 1998;10:995–1007. doi: 10.1105/tpc.10.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson M. Increased dehydrin promoter activity caused by HvSPY is independent of the ABA response pathway. Plant J. 2003;34:39–46. doi: 10.1046/j.1365-313x.2003.01697.x. [DOI] [PubMed] [Google Scholar]
- 36.Robertson M. Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol. 2004 136;:2747–2761. doi: 10.1104/pp.104.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D. Cross talk between gibberellin and cytokinin: the arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17:92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maymon I, Greenboim-Wainberg Y, Sagiv S, Kieber JJ, Moshelion M, Olszewski N, Weiss D. Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway. Plant J. 2009;58:979–988. doi: 10.1111/j.1365-313X.2009.03840.x. [DOI] [PubMed] [Google Scholar]
- 39.Gan Y, Liu C, Yu H, Broun P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development. 2007;134:2073–2081. doi: 10.1242/dev.005017. [DOI] [PubMed] [Google Scholar]
- 40.Tseng TS, Salomé PA, McClung CR, Olszewski NE. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering and rhythms in cotyledon movements. Plant Cell. 2004;16:1550–1563. doi: 10.1105/tpc.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sothern RB, Tseng TS, Orcutt SL, Olszewski NE, Koukkari WL. GIGANTEA and SPINDLY genes linked to the clock pathway that controls circadian characteristics of transpiration in Arabidopsis. Chronobiol Int. 2002;19:1005–1022. doi: 10.1081/cbi-120015965. [DOI] [PubMed] [Google Scholar]
- 42.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2000;97:9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 45.Paquette AJ, Benfey PN. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 2005;138:636–640. doi: 10.1104/pp.104.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui H, Benfey PN. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 2009;58:1016–1027. doi: 10.1111/j.1365-313X.2009.03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110:69–80. doi: 10.1016/s0092-8674(02)00810-3. [DOI] [PubMed] [Google Scholar]
- 48.Jiang H, Wang S, Dang L, Chen H, Wu Y, Jiang X, Wu P. A novel short-root gene encodes a glucosamine-6-phosphate acetyltransferase required for maintaining normal root cell shape in rice. Plant Physiol. 2005;138:232–242. doi: 10.1104/pp.104.058248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rayon C, Cabanes-Macheteau M, Loutelier-Bourhis C, Salliot-Maire I, Lemoine J, Reiter WD, Lerouge P, Faye L. Characterization of N-glycans from Arabidopsis. Application to a fucose-deficient mutant. Plant Physiol. 1999;119:725–734. doi: 10.1104/pp.119.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott CL. Thesis (Ph. D.)--University of Minnesota, 2005. Major: Plant biological sciences. 2006. O-N-acetylgluosamine transferases of Arabidopsis thaliana. [Google Scholar]
- 51.Belkhadir Y, Wang X, Chory J. Brassinosteroid signaling pathway. Sci STKE 2006. 2006 doi: 10.1126/stke.3642006cm4. cm4. [DOI] [PubMed] [Google Scholar]
- 52.Hartweck LM, Genger RK, Grey WM, Olszewski NE. SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J. Exp. Bot. 2006;57:865–875. doi: 10.1093/jxb/erj071. [DOI] [PubMed] [Google Scholar]
- 53.Fernandez-Fernandez MR, Camafeita E, Bonay P, Mendez E, Albar JP, Garcia JA. The capsid protein of a plant single-stranded RNA virus is modified by O-linked N-acetylglucosamine. J. Biol. Chem. 2002;277:135–140. doi: 10.1074/jbc.M106883200. [DOI] [PubMed] [Google Scholar]
- 54.Chen D, Juarez S, Hartweck L, Alamillo JM, Simon-Mateo C, Perez JJ, Fernandez-Fernandez MR, Olszewski NE, Garcia JA. Identification of secret agent as the O-GlcNAc transferase that participates in Plum pox virus infection. J Virol. 2005;79:9381–9387. doi: 10.1128/JVI.79.15.9381-9387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott CL, Hartweck LM, de Jesus Perez J, Chen D, Garcia JA, Olszewski NE. SECRET AGENT, an Arabidopsis thaliana O-GlcNAc transferase, modifies the Plum pox virus capsid protein. FEBS Lett. 2006;580:5829–5835. doi: 10.1016/j.febslet.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 56.de Jesus Perez J, Juarez S, Chen D, Scott CL, Hartweck LM, Olszewski NE, Garcia JA. Mapping of two O-GlcNAc modification sites in the capsid protein of the potyvirus Plum pox virus. FEBS Lett. 2006;580:5822–5828. doi: 10.1016/j.febslet.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 57.Lee JY, Yoo BC, Lucas WJ. Parallels between nuclear-pore and plasmodesmal trafficking of information molecules. Planta. 2000;210:177–187. doi: 10.1007/PL00008124. [DOI] [PubMed] [Google Scholar]
- 58.Taoka KI, Ham BK, Xoconostle-Cazares B, Rojas MR, Lucas WJ. Reciprocal Phosphorylation and Glycosylation Recognition Motifs Control NCAPP1 Interaction with Pumpkin Phloem Proteins and Their Cell-to-Cell Movement. Plant Cell. 2007 doi: 10.1105/tpc.107.052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JY, Yoo BC, Rojas MR, Gomez-Ospina N, Staehelin LA, Lucas WJ. Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science. 2003;299:392–396. doi: 10.1126/science.1077813. [DOI] [PubMed] [Google Scholar]
- 60.Swain SM, Tseng T-s, Thornton TM, Gopalraj M, Olszewski NE. SPINDLY Is a Nuclear-Localized Repressor of Gibberellin Signal Transduction Expressed throughout the Plant. Plant Physiol. 2002;129:605–615. doi: 10.1104/pp.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A Chemoenzymatic Approach toward the Rapid and Sensitive Detection of O-GlcNAc Posttranslational Modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 62.Zachara NE, Cole RN, Hart GW, Gao Y. Detection and analysis of proteins modified by O-linked N-acetylglucosamine. Curr Protoc Protein Sci Chapter. 2001;12 doi: 10.1002/0471140864.ps1208s25. Unit 12 18. [DOI] [PubMed] [Google Scholar]
- 63.Kilcoyne M, Shah M, Gerlach JQ, Bhavanandan V, Nagaraj V, Smith AD, Fujiyama K, Sommer U, Costello CE, Olszewski N, Joshi L. O-glycosylation of protein subpopulations in alcohol-extracted rice proteins. J. Plant Physiol. 2009;166:219–232. doi: 10.1016/j.jplph.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 64.Xing L, Li J, Xu Y, Xu Z, Chong K. Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0004854. e4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heese-Peck A, Cole RN, Borkhsenious ON, Hart GW, Raikhel NV. Plant nuclear pore complex proteins are modified by novel oligosaccharides with terminal N-acetylglucosamine. Plant Cell. 1995;7:1459–1471. doi: 10.1105/tpc.7.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heese-Peck A, Raikhel NV. A glycoprotein modified with terminal N-acetylglucosamine and localized at the nuclear rim shows sequence similarity to aldose-1-epimerases. Plant Cell. 1998;10:599–612. doi: 10.1105/tpc.10.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 68.Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 69.Love DC, Kochan J, Cathey RL, Shin SH, Hanover JA. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci. 2003;116:647–654. doi: 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- 70.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 71.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 72.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 73.Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–W339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.