Abstract
Diagnosis of prostate cancer (PCa) typically relies on needle biopsies, which are routinely archived in paraffin after formalin fixation and may contain valuable risk or prognostic information. The objective of this study was to determine the feasibility of mRNA and miRNA expression analysis in laser-capture microdissected (LCM) formalin-fixed paraffin-embedded archived prostate biopsies compared to the gold standard of frozen tissue. We analyzed the expression of compartment-specific and PCa-related genes in epithelial and stromal tissues collected from paired sets of archived prostate biopsies and frozen radical prostatectomy specimens from three patients. Our results showed appropriate compartment-specific and PC-related expression with good within patient agreement between the FFPE-biopsies and the frozen tissue. The potential for both mRNA and microRNA expression profiling in the biopsies was also demonstrated using PCR arrays which showed high correlation between the biopsy and frozen tissue, notwithstanding sensitivity limitations for mRNA detection in the FFPE specimen. This is the first study to compare RNA expression from biopsy and frozen tissues from the same patient and to examine miRNA expression in LCM-collected tissue from prostate biopsies. With careful technique and use of appropriate controls, RNA profiling from archived biopsy material is quite feasible showing high correlation to frozen tissue.
Keywords: prostate cancer, FFPE, RNA expression
INTRODUCTION
Despite advances in the diagnosis and treatment, prostate cancer (PCa) remains the second leading cause of cancer-related deaths in men. The vast majority of PCa are detected by needle biopsies that provide a random and sparse sample of prostate tissue, which is archived in paraffin after formalin fixation. Although RNA is degraded in formalin-fixed paraffin-embedded (FFPE) tissue, recent studies have shown that meaningful gene and microRNA (miRNA) expression data can be obtained from these specimens 1–3. The ability to analyze RNA expression in FFPE-bpx provides a useful tool for risk stratification for negative biopsies, identification of prognostic markers in positive biopsies and could allow gene expression profiling to become an important surrogate endpoint in PCa prevention trials
Prostate biopsies contain a mixture of histological entities (stroma, benign epithelium, cancer, pre-malignancy and atrophy). Therefore, it is imperative to separate well-characterized epithelial areas from the stroma by laser capture microdissection (LCM) prior to RNA analysis. Shukla et al used LCM to obtain reliable gene expression markers for epithelial and stromal compartments in radical prostatectomy (RP) and TURP samples 2. However, only one study to date combined LCM with analysis of the expression of gene transcripts in formalin-fixed prostate biopsies 1. Although that study reported plausible results for expression of genes that are specifically associated with stroma, benign epithelium and PCa, the only direct comparison of FFPE to frozen tissue was made in tissue from a xenograft model and the number of genes was limited.
Here we report validation of RNA expression analysis in FFPE biopsies by comparison to the gold standard, frozen tissue, obtained at radical prostatectomy (RP) from the same patient. We isolated pure populations of normal epithelium, normal stroma and PCa epithelium and then compared matched FFPE and Frozen Tissue for gene expression of several compartment-specific and PCa-related mRNAs. We also examined the expression of several PCa-related miRNAs in normal and malignant epithelium in the matched sample types. Lastly, to determine whether our technique could quantify expression of mRNAs and miRNAs in multi-target panels, we compared fixed and frozen samples from the same patient using PCR arrays.
MATERIALS AND METHODS
Prostate Tissue
Archival formalin-fixed paraffin-embedded (FFPE) prostate needle biopsy (Bpx) specimens were used. Needle biopsy cores (18G), eight per patient, were fixed in formalin immediately and paraffin embedded within 24 hours. Biopsy specimens were collected from February to July 2007 and paired fresh-frozen tissue was acquired at radical prostatectomy (<6 months from biopsy), by a pathologist during the gross examination and dissection of specimens. Tissue was snap frozen and stored at −80°C. Frozen tissue was collected from October 2007 to December 2007. The protocol for acquiring tissues was reviewed and approved by the UIC Institutional Review Board.
Laser Capture Microdissection
For both biopsy and frozen material, two 8μ sections were cut and placed onto Leica RNase-free PEN slides (Leica, Bannockburn, IL, USA). FFPE-Bpx slides were de-paraffinized, lightly stained with 0.5% toluidine blue and dried for 5 minutes on a 37°C slide warmer. FFPE-Bpx tissues were sectioned the day of LCM-collection. Frozen tissue was sectioned and fixed in 100% ethanol one day prior to LCM-collection. LCM collection was done on the Leica LMD-6000; 60–160 acini of epithelium and all of the surrounding stroma were collected. For cancer, all of the foci available on each slide were collected. The tissue was collected into Eppendorf caps containing 50 μl of Digestion Buffer® (from RecoverAll® kit). Microdissected tissue was stored in Digestion Buffer® overnight at −80°C prior to RNA isolation.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from the tissue with RecoverAll® (Applied Biosystems Inc, Foster City, CA). The manufacturer’s protocol was followed with the exception of increased DNase digestion for 60 minutes at 37°C. RNA quantity and quality was determined by OD at 260 and 280 nm on a NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). cDNA was generated from 50 ng of total RNA with Superscript III reverse transcriptase.
mRNA Expression by PreAmp TaqMan® qRT-PCR
The genes of interest included three housekeeping genes (supplemental Table 1): beta-2-microglobulin (B2M), tata-box binding protein (TBP), and hypoxanthine phosphoribosyltransferase 1 (HPRT1); three epithelial-specific genes: prostate specific antigen (PSA), cytokeratin 18 (K18), and NK3-homobox 1(NKX3.1); three stroma-specific genes: desmin, tissue inhibitor of metalloprotease 3 (TIMP3), and insulin-like growth factor 1(IGF1); two genes reported to be up-regulated in PCa: prostate cancer antigen 3 (PCA3), and alpha-methylacyl-CoA racemase (AMACR); and one gene reported to be down-regulated in PCa: NKX3.1. All 11 of the TaqMan® Assays (ABI) were pooled, diluted 1:100 and utilized for 14 cycles of pre-amplification with PreAmp Master Mix (ABI.). cDNA from 50 ng of total RNA was used for a pooled PreAmp reaction. The resulting PreAmp product was diluted 1:20 and served as template for the individual TaqMan® qPCR reactions which were run on the ABI Realtime PCR Machine HT7900 (Applied Biosystems, Inc.). Control PCR samples were run with no template and no reverse transcriptase reactions. Fold-changes were calculated using ddCt method relative to patient #1 and normalized to the expression of HPRT1, B2M and TBP.
microRNA Expression by PreAmp TaqMan® qRT-PCR
5ng of total RNA was reverse transcribed using the Taqman® microRNA Reverse Transcription kit (ABI). RT products were pre-amplified using the PreAmp Master mix as described above. The pre-amplified product was used as the template for the Real-time PCR run on Step-one Plus (ABI). Control PCR samples were run with no template and no reverse transcriptase reactions. Fold-changes were calculated using ddCt method relative to patient #1 and normalized to the expression of RNU44 and RNU48.
Protein Expression by Immunohistochemistry
FFPE biopsy sections (4μ) were baked, deparaffinized and dehydrated. Slides were immunostained using the DakoCytomation Envision+ System-HRP (DAB) kit protocol from DakoCytomation, Inc (Carpinteria, CA). Antigen retrieval was for 20 minutes at 95–99°C in Target Retrieval (Dako) solution. The tissues were blocked in a serum-free protein block (Dako) followed by a peroxidase block (Dako). Anti-NKX3.1 was used at 1:2500 dilution (incubated overnight at 4° C) and was a generous gift from Dr. Charles J. Bieberich (Department of Biological Sciences, University of Maryland Baltimore County). Anti-AMACR (Zeta Corp, Sierra Madre, CA, USA) was used at 1:100 dilution (incubated 1 hour at 37°). HRP-conjugated secondary antibodies were used followed by DAB. The tissues were counterstained with Hematoxylin, rehydrated and mounted.
mRNA expression by TaqMan® Custom PCR Array
Following cDNA synthesis with random decamers, 50 ng of cDNA was used as the input for 14 cycles of preamplification with pooled Taqman® primers for a custom human PCR array (ABI). Following dilution, cDNA was loaded onto the custom array. This 384-format custom array contained 64 genes plated 6 times. We analyzed FFPE-Bpx and frozen-RP normal epithelium in duplicate (128 wells each) and the paired PCa from those samples in singlicate (64 wells each). Ct (crosses threshold; threshold=0.2) values were determined for all samples and genes. Individual amplification plots were manually inspected. The Ct values for the technical duplicates were averaged and the data grouped by Ct value intervals. Pearson correlations between biopsy and frozen Ct values were calculated. For normal versus PCa comparison, fold-changes were calculated by ddCT method. Sensitivity was calculated by (true positives)/(true positives + false negatives) and specificity (true negatives)/(true negatives + false positives), based on frozen-RP results as the gold-standard.
miRNA expression by TaqMan® Low-Density Array (TLDA)
20 ng of RNA was used as the input for cDNA generation. Eight distinct pools of RT primers were used for analysis of 370 distinct miRNAs. Following dilution, 12 cycles of preamplification with the Megaplex pool protocol for the array was performed on the cDNA. Following dilution, the cDNAs were loaded onto two arrays (Human miRNA Array v1.0, ABI). This facilitated analysis of 384 wells; 370 distinct miRNAs analyzed in singlicate and two housekeeping snoRNAs with 8 replicates for each. Ct (crosses threshold) values were determined for all samples and genes. Individual amplification plots were manually inspected. Pearson correlation of Ct values was calculated.
RESULTS
LCM-Collected RNA
Three pairs of prostate specimens were selected from an existing set of radical prostatectomy patients. From each patient, we compared gene expression profiling from their pre-surgical FFPE prostate needle biopsies (FFPE-bpx) to their frozen surgically-removed radical prostatectomy tissue (frozen-RP). The patients were 53–56 years of age with similar Gleason scores (6–7), pre-operative PSA, identical clinical stage and race. LCM was performed to separately collect the normal epithelium, normal stroma and PCa epithelium (Supplemental Figure 1). Areas for LCM were selected by a pathologist avoiding areas of prostatic intraepithelial neoplasia (PIN) or proliferative inflammatory atrophy (PIA). All specimens yielded RNA of sufficient quantity (> 100 ng) and most showed relative high purity with an OD260/280 around 1.6 (Table 1).
Table 1.
LCM-Collected RNA from Prostate Tissue
| SPECIMEN | ng RNA | 260/280 nm | |
|---|---|---|---|
| FFPE-Bpx 1 | N-Epi | 238 | 1.58 |
| N-Str | 198 | 1.23 | |
| PCa-Epi | 275 | 1.49 | |
| FFPE-Bpx 2 | N-Epi | 121 | 1.21 |
| N-Str | 246 | 1.28 | |
| PCa-Epi | 529 | 1.76 | |
| FFPE-Bpx 3 | N-Epi | 561 | 1.42 |
| N-Str | 283 | 1.63 | |
| PCa-Epi | 244 | 1.3 | |
| Frozen-RP 1 | N-Epi | 466 | 1.85 |
| N-Str | 350 | 1.69 | |
| PCa-Epi | 376 | 1.63 | |
| Frozen-RP 2 | N-Epi | 451 | 1.61 |
| N-Str | 736 | 1.36 | |
| PCa-Epi | 288 | 1.16 | |
| Frozen-RP 3 | N-Epi | 294 | 1.75 |
| N-Str | 899 | 1.45 | |
| PCa-Epi | NA | NA | |
N-Epi = normal epithelium, N-Str = normal stroma
PCa-Epi = PCa epithelium, NA = not available
mRNA Expression Analysis
The expression of 11 individual mRNAs was analyzed by PreAmplification-PCR with Taqman® assays (see Supplemental Table 1). We analyzed three housekeeping genes, three epithelial-specific genes, three stroma-specific genes, two genes reported to be up-regulated in PCa and one gene reported to be down-regulated in PCa.
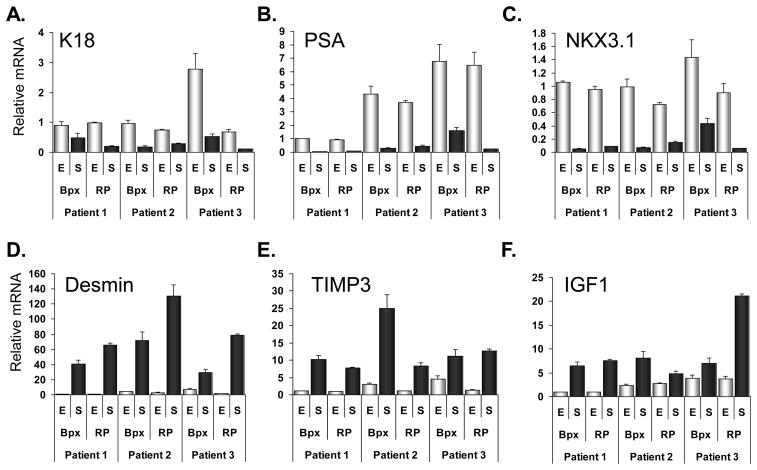
The Pearson correlations between the Ct values of housekeeping genes were calculated using all data points from biopsy and frozen samples (N=36 for each gene)(Table 2). The R-values were all close to 1 indicating linear and unbiased amplification of the cDNA. All three patients showed appropriate expression of epithelium-specific (K18, PSA, NKX3.1) and stroma-specific (IGF1, desmin, TIMP3) mRNAs (Figure 1). In Patients #2 and 3 the IGF1 levels were higher in the normal epithelium than Patient #1, but still lower than the stromal IGF1 levels. In all three patients, there was agreement in the relative mRNA levels between the FFPE-bpx and the RP sample (Figure 1).
Table 2.
Pearson rho* values for housekeeping genes
| B2M | TBP | HPRT1 | |
|---|---|---|---|
| B2M | - | 0.98 | 0.98 |
| TBP | 0.98 | - | 0.99 |
| HPRT1 | 0.98 | 0.99 | - |
R value when comparing Ct values of housekeeping among all data points from biopsy and frozen samples (N=36 for each gene)
Figure 1.
Expression of cell-type-specific mRNAs in FFPE-bpx and frozen-RP specimens using Taqman®PreAmp. Epithelial cell-specific mRNAs K18, PSA and NKX3.1 expression in LCM-collected normal epithelium and stromal tissue (A-C). Stromal cell-specific mRNAs IGF1, Desmin and TIMP3 expression in LCM-collected normal epithelium (E, open bars) and stromal tissue (S, dark bars) (D-F). Results are shown relative to the expression of normal epithelium from Patient 1. Graphs display the mean relative mRNA expression after normalization to the expression of B2M, HPRT1 and TBP. Error bars represent standard error of the averages based on standard deviation of duplicates.
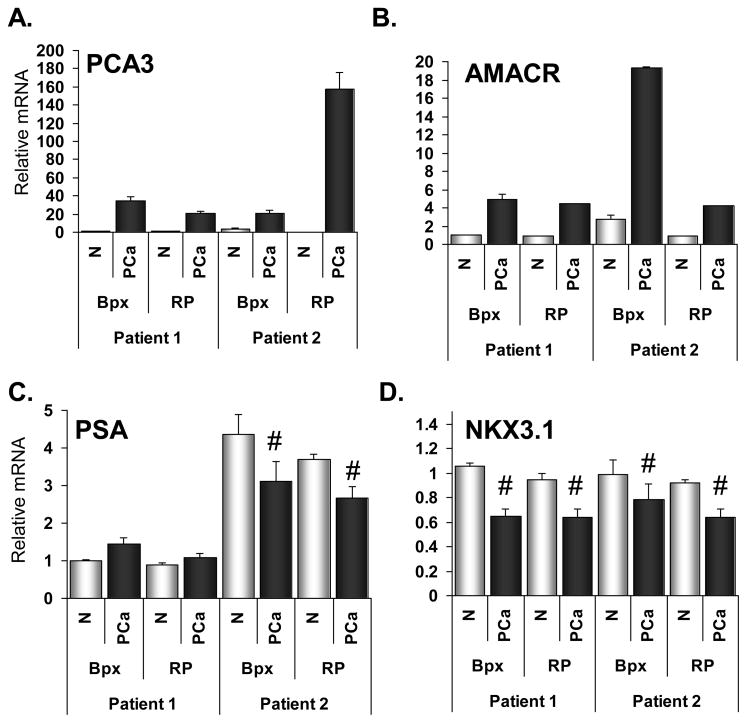
We were able to collect cancerous tissue in FFPE-bpx and RP tissue from two patients (one patient did not have detectable PCa in the frozen specimen). PCA3 and AMACR mRNAs were clearly over-expressed in both tumor samples relative to the normal epithelium (Figure 2). PSA, which may be down-regulated in organ-confined PCa 4, was decreased in only one of the patients (Figure 2). Importantly though, the PSA results were the same for RNA collected from FFPE-bpx and frozen-RP tissues. NKX3.1 mRNA was slightly down-regulated in both tumor samples (Figure 2). Again we observed good agreement in the relative mRNA levels between the biopsy and RP samples.
Figure 2.
Expression of prostate cancer-related mRNAs in FFPE-bpx and frozen-RP specimens using Taqman®PreAmp. Expression of mRNAs that are commonly up-regulated in prostate cancer, PCA3 and AMACR, in LCM-collected normal epithelium (N, open bars) and prostate cancer tissue (PCa, dark bars)(A-B). Expression of mRNAs commonly down-regulated in prostate cancer, PSA and NKX3.1, in LCM-collected normal epithelium and prostate cancer tissue (C-D). Results are shown relative to the expression of normal epithelium from Patient 1. Graphs display the mean relative mRNA expression after normalization to the expression of B2M, HPRT1 and TBP. Error bars represent standard error of the averages based on standard deviation of duplicates. Within each patient, paired t-test between normal epithelium and PCa; # = p < 0.01.
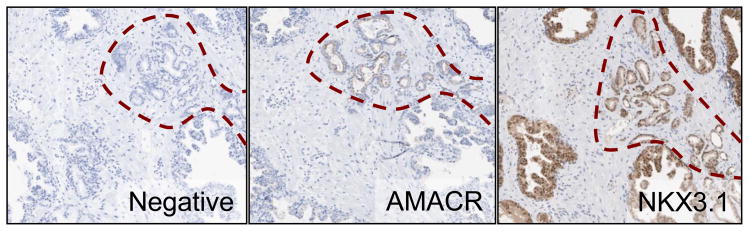
The changes in AMACR and NKX3.1 mRNA were confirmed by immunohistochemical analysis in the biopsies. AMACR protein was not present in normal epithelium or stroma, but was highly expressed in the cytoplasm of PCa epithelium (representative image Figure 3). NKX3.1 protein showed nuclear localization in normal epithelium, which was decreased in PCa epithelium, and was not expressed in stromal tissue (representative image Figure 3). Overall, our findings are consistent with known PCa-related changes in the protein expression of AMACR and NKX3.1 5,6
Figure 3.
Immunohistochemistry staining for AMACR and NKX3.1 protein. Negative control (HRP-secondary antibody only) is shown in left panel. AMACR staining shown in center panel and NKX3.1 staining in right panel. An area of prostate cancer is outlined (red dashes).
miRNA Expression Analysis
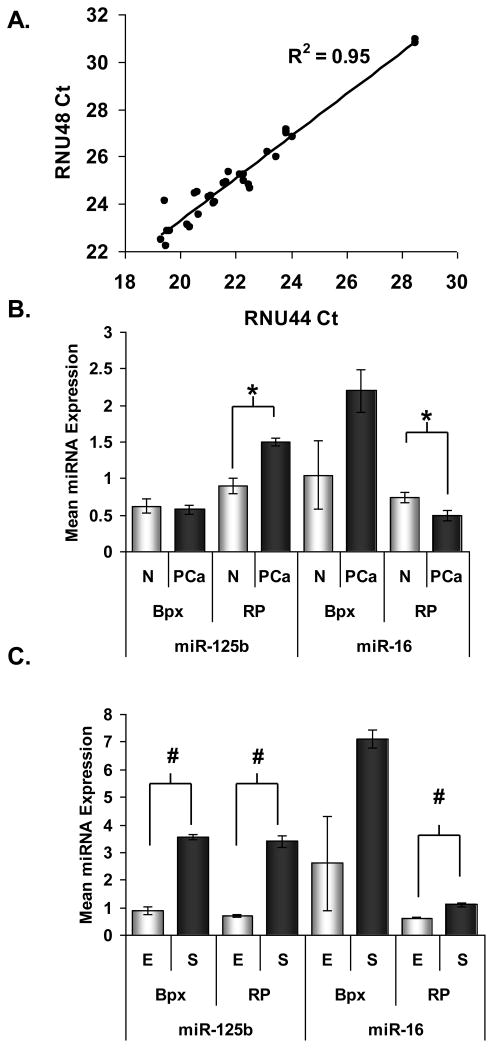
The expression of two PCa related miRNAs and housekeeping small RNAs was analyzed by stem-loop RT-PCR Taqman® Assays and there was excellent correlation (r = 0.98) between the Ct values of the two housekeeping RNAs (Figure 4A) (N=16). MiR-125b and miR-16 were selected because they have been reported to be down-regulated in prostate tumors 7. We did not observe consistent decreased expression of miR-125b and miR-16 in tumor compared to normal epithelium (the means for all specimens are shown in Figure 4B). However, we found miR-125b and miR-16 expression to be 2–3 fold higher in the normal stroma than in the normal epithelium (Figure 4C) and notably, the frozen and FFPE samples were concordant.
Figure 4.
MiRNA expression analysis in LCM-collected tissue from FFPE-bpx and frozen-RP specimens using Taqman®PreAmp. A, Correlation of miRNA housekeeping gene expression in all specimens (N=16). Expression of miR-125b and miR-16 in, B, normal (open bars) and PCa epithelium (dark bars) and, C, in normal epithelium (open bars) versus normal stromal tissue (dark bars). Graphs display the mean relative mRNA expression after normalization to the expression of RNU44 and RNU48. For A, mean of expression for Patients 1 and 2 is shown for normal and PCa epithelium. For B, mean of expression for Patients 1, 2 and 3 is shown for normal epithelium and stroma. Error bars represent standard deviation of 12 observations (n=6, technical duplicates) (except RP normal vs PCa, n=4). Results are shown relative to the expression of normal epithelium from Patient 1. Within each set of specimens (bpx or RP), paired t-test between normal epithelium and stroma; * = p<0.05, # = p < 0.01.
PCR Array analysis of mRNA and miRNA expression
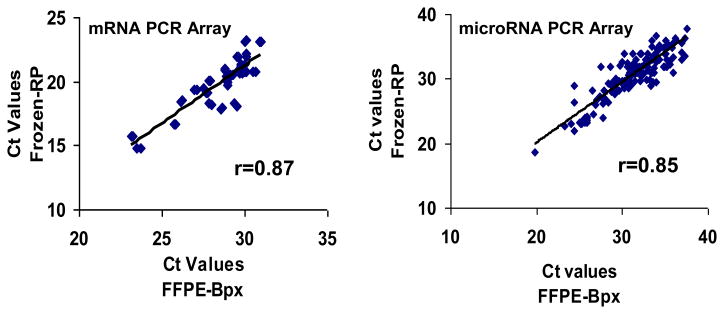
We next tested our ability to simultaneously analyze multiple mRNA and miRNA targets in the FFPE biopsy material using PCR arrays and matched FFPE and frozen samples from a single patient (patient #1). The mRNA array included 64 genes associated with wound-healing, inflammation and hormone signaling, represented by amplicons < 100 bp in length. The level of agreement between duplicate wells was extremely high for both frozen and FFPE samples (CV% < 0.5% for all genes detected). However, the mean Ct for FFPE across all genes was 33.5, compared to 24.5 for frozen tissue, indicating a more than 500-fold greater amount of intact mRNA in the frozen sample. Consistent with the high Ct values, 23/64 (37%) of the gene targets were undetected (Ct >35) in the FFPE samples, versus only one undetectable target in the frozen samples (see Table 3). As expected, the likelihood of detection in FFPE was related to the level of gene expression; among 38 genes with Ct < 25 in the frozen sample, 100% were detectable in the matched FFPE. Among genes that were detected in both samples (n = 41), the overall correlation between FFPE and frozen tissue was very high (r = 0.89). Correlation was greater for highly expressed genes vs. those less highly expressed: r = 0.87 and 0.29 for genes with FFPE sample Ct values of <31 and 31–35, respectively. Figure 5a shows the scatterplot and linear regression line for the agreement between FFPE and frozen normal epithelium for jointly detected genes when the FFPE Ct values are less than 31.
Table 3.
Detection of mRNAs and miRNAs (Ct<35) in Frozen-RP vs. FFPE-Bpx samples of normal prostate epithelium from the same patient.
| mRNA Array† | Detected Frozen-RP | |||
|---|---|---|---|---|
| YES | NO | Totals | ||
| Detected FFPE-Bpx | YES | 41a | 0 | 41 |
| NO | 22 | 1 | 23 | |
| Totals | 63 | 1 | 64 | |
| microRNA Array‡ | Detected Frozen-RP | |||
|---|---|---|---|---|
| YES | NO | Totals | ||
| Detected FFPE-Bpx | YES | 116b | 16 | 132 |
| NO | 40 | 198 | ||
| Totals | 156 | 214 | ||
64 genes analyzed in duplicate
370 miRNA analyzed in singlicate
frozen-FFPE Pearson correlation r=0.89 for jointly detected genes
frozen-FFPE Pearson correlation r=0.85 for jointly detected genes
Figure 5.
Correlation of FFPE-Bpx and Frozen-RP mRNA and miRNA expression profiles by TaqMan® PCR array. Correlation between FFPE-bpx and frozen-RP tissue from Patient #1 in mRNA expression on, A, 41 genes from mRNA PCR array with Cts <31 and, B, miRNAs expressed in both tissue sources (N=116).
Among the 64 genes in the mRNA array, 34 were differentially expressed between frozen-RP samples of normal and PCa tissue, with differential expression defined as at least a two-fold difference in relative expression (Table 4). Ten of these 34 genes (29%) were also found to be differentially expressed in the comparison of normal and PCa tissue from the FFPE biopsies. Among 30 genes that were not differentially expressed in frozen tissue, only 6 (20%) were found to be differential in FFPE.
Table 4.
Detection of mRNAs differentially expresseda in PCa vs normal in the same patient
| mRNA Array | Differential Expression Frozen-RP | |||
|---|---|---|---|---|
| YES | NOb | Totals | ||
| Differential Expression FFPE-Bpx | YES | 10 | 6 | 16 |
| NOb | 24 | 24 | 48 | |
| Totals | 34 | 30 | 64 | |
>2-fold difference in expression, Ct<31
sum of not differentially expressed and not detected
We also compared miRNA expression between FFPE and frozen normal prostate from the same patient using a qPCR array with 370 unique miRNA sequences. All miRNAs were represented by one well, with the exception fo two housekeeping miRNA sequences whichwere evaluated with 8 replicates per array (both CVs < 3%). In contrast to mRNA, the mean Ct for frozen tissue was only 0.6 less than matched FFPE. Consistent with a previous report, approximately half of the miRNAs were not detected, and thus were probably not expressed, in both types of tissue (see Table 3) 8. Agreement between sample types on miRNA detectability was good - 73% of miRNAs detected in frozen tissue were also detected in FFPE (sensitivity = 0.73), while only 8% of FFPE detections were false positives (specificity = 0.92). Among all 116 jointly detected miRNA species, the correlation for Ct values between matched frozen and FFPE samples was 0.85 (see Figure 5b).
DISCUSSION
In this study we directly compared mRNA and miRNA expression from LCM-collected FFPE and frozen tissues from the same patient. It was important to fully validate expression analysis from FFPE-RNA since the RNA is highly fragmented and not an ideal RT-PCR template. We found that small amplicon size (60–75 bp) TaqMan® assays worked well since non-specific primer-dimer products are not detected. Our previous attempts with small amplicon primer pairs and SYBR green resulted in an abundance of primer-dimer products which made quantification impossible (data not shown). We further improved detection sensitivity by running 14 cycles of pre-amplification PCR prior to the quantitative real-time PCR.
We analyzed the expression of 12 well-characterized mRNAs in the FFPE-bpx and frozen-RP samples. The epithelial and stroma-specific genes confirmed that pure populations of cells were collected by LCM and analysis of normal vs PCa also demonstrated appropriate expression of PCA3 and AMACR and NKX3.1. The decrease in small NKX3.1, was consistent and significant in both sample types and confirmed at the protein level by immunohistochemistry.
By comparing the FFPE-bpx and frozen-RP tissue from the same patient, we were able to show that RNA expression in the FFPE-bpx was reflective of the whole prostate as the relative expression was similar to the frozen-RP. Similarity was evident in that the genes with highly differential expression showed the same direction of change. However, there was not always agreement in the amount of change, which is likely due to unavoidable sampling differences between the biopsy and the RP.
The role of miRNAs in cancer is a rapidly emerging area of investigation. Expression profiling has identified miRNA signatures in cancers that associate with diagnosis, staging, progression, prognosis and response to treatment 9. MiRNAs are ideal biomarkers in FFPE-tissue because, unlike mRNA, miRNA integrity is affected very little by formalin fixation 8,10–12. MiRNA expression signatures specific to PCa have been reported 7,13,14; we selected miR-16 and miR-125b for analysis based on a previous report which showed down-regulation in human PCa compared to normal prostate 7. Our results, although based on only two patients, did not confirm the results by Porkka el al. During preparation of this manuscript, we found conflicting findings for miR-16 and miR-125b expression in PCa as they were down-regulated in PCa according to one report 7 and up-regulated according to another 13,14. Methodological differences in expression analysis and tissue preparation likely contributed to the heterogeneity of the published PCa miRNA signatures. The aforementioned studies utilized whole sections from frozen prostate tissue 7,13–16. The prostate is a milieu of stromal cells and epithelial acini and one of the hallmarks of PCa is decreased stromal content. Therefore, profiling whole prostate tissue has potential bias towards an under-representation of stroma-specific miRNAs in the PCa sample. Indeed we found that miR-16 and miR-125b had higher expression in the prostate stroma than in epithelium; a finding supported by Mitchell et al who reported 4-fold higher miR-125b levels in prostate stromal cells than in prostate epithelial cells17. Our data suggest that miRNA profiles generated from whole prostate tissue may display some degree of artifact resulting from decreased stromal tissue in the PCa lesions.
Ideally we would like the ability to profile more than a handful of genes in the LCM-collected biopsy samples. Our results demonstrate feasibility, albeit with several limitations. The biggest hurdle in the mRNA profiling is sensitivity in the meager FFPE-Bpx samples. The results showed excellent correlation when low Ct values (20–31) were analyzed, but much lower correlation at Cts >31. To accommodate the wide range of Cts in our target genes, we utilized five housekeeping genes, which came up at Cts ranging from 23–32 in the FFPE samples. Although mRNA profiling from FFPE-Bpx presents a challenge, as long as there are rigorous controls and careful data analysis, meaningful results can be obtained. We would suggest that proper profiling include the following; at least 3 housekeeping genes with variable expression levels, PCR amplicons <80bp, analysis of Ct values <31 (lower than the typical cutoff of 35). Our data suggests that technical duplicates are not necessary.
When we used these criteria to analyze of gene expression changes in the normal versus PCa, the FFPE-Bpx sample identified the PCa changes with 80% specificity and 30% sensitivity. The sensitivity is likely diminished in the FFPE sample because RNA degradation. However, the specificity was good, suggesting that valid results can be obtained from the biopsy material. It should be noted that our results comparing FFPE and frozen are conservative because they assume that the frozen-RP data represents the “true” data with zero false positives or negatives, which may not be the case because of inherent sampling differences between the needle biopsy and surgically obtained tissue. The ultimate goal is whole transcriptome amplification of biopsy material to provide analysis of all genes by array, which requires μg amounts of RNA compared to the ng amounts achievable from biopsies
In contrast, the miRNA PCR array profiling results show superb correlation between the biopsy and RP specimen without the limitations of the mRNA profiling. The results were correlative throughout all Cts (20–35). Furthermore, the Ct values for the bpx and RP were similar, indicating that the miRNAs are not degraded in the biopsies. Although we utilized the Human microRNA PCR array v1.0 from Applied Biosystems that contained 370 distinct miRNAs there is now a v2.0 PCR array that includes 667 distinct miRNAs. Since the protocol that we used involves dilution of both the RT and preamplification products, the RNA isolated from FFPE-bpx would be more than sufficient to analyze all 667 miRNAs in the current platform.
PCa-associated stroma has been shown to contribute to carcinogenesis 18,19 and has gene expression profiles distinct from normal prostate stroma 20. Unfortunately, there was insufficient PCa-associated stroma in the biopsies to facilitate gene expression analysis.
CONCLUSIONS
We show that despite all of the challenges, with careful attention to technique and a full understanding of the limitations, expression profiling of mRNA gene sets by PCR arrays is readily achievable from archived biopsy material. MiRNA expression profiling from archived biopsies produced equivalent results to data obtained from frozen specimens, further supporting the utility of miRNAs as biomarkers in archived specimens.
Supplementary Material
Acknowledgments
We thank Vijayalakshmi Ananthanaryanan for providing her services as a pathologist in selecting the normal and tumor areas of the tissues, Erika Enk for her assistance in specimen selection, Dr. Debra Tonetti for the preamp primers and mRNA PCR array and Dr. Charles J. Bieberich for the NKX3.1 antibody. This research was supported by NIH-NCI R03 CA131595 (Nonn) and NIH-NCI RO1 CA90759 (Gann).
Footnotes
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rogerson L, Darby S, Jabbar T, Mathers ME, Leung HY, Robson CN, et al. Application of transcript profiling in formalin-fixed paraffin-embedded diagnostic prostate cancer needle biopsies. BJU Int. 2008;102(3):364–370. doi: 10.1111/j.1464-410X.2008.07627.x. [DOI] [PubMed] [Google Scholar]
- 2.Shukla CJ, Pennington CJ, Riddick AC, Sethia KK, Ball RY, Edwards DR. Laser-capture microdissection in prostate cancer research: establishment and validation of a powerful tool for the assessment of tumour-stroma interactions. BJU Int. 2008;101(6):765–774. doi: 10.1111/j.1464-410X.2007.07372.x. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, Mehra R, Rhodes DR, Shah RB, Rubin MA, Bruening E, et al. Whole transcriptome amplification for gene expression profiling and development of molecular archives. Neoplasia. 2006;8(2):153–162. doi: 10.1593/neo.05754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darson MF, Pacelli A, Roche P, Rittenhouse HG, Wolfert RL, Young CY, et al. Human glandular kallikrein 2 (hK2) expression in prostatic intraepithelial neoplasia and adenocarcinoma: a novel prostate cancer marker. Urology. 1997;49(6):857–862. doi: 10.1016/s0090-4295(97)00108-8. [DOI] [PubMed] [Google Scholar]
- 5.Rubin MA, Zerkowski MP, Camp RL, Kuefer R, Hofer MD, Chinnaiyan AM, et al. Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements. Am J Pathol. 2004;164(3):831–840. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelmann EP, Bowen C, Bubendorf L. Expression of NKX3.1 in normal and malignant tissues. Prostate. 2003;55(2):111–117. doi: 10.1002/pros.10210. [DOI] [PubMed] [Google Scholar]
- 7.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67(13):6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 8.Szafranska AE, Davison TS, Shingara J, Doleshal M, Riggenbach JA, Morrison CD, et al. Accurate molecular characterization of formalin-fixed, paraffin-embedded tissues by microRNA expression profiling. J Mol Diagn. 2008;10(5):415–423. doi: 10.2353/jmoldx.2008.080018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6 (11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008;10(3):203–211. doi: 10.2353/jmoldx.2008.070153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoefig KP, Thorns C, Roehle A, Kaehler C, Wesche KO, Repsilber D, et al. Unlocking pathology archives for microRNA-profiling. Anticancer Res. 2008;28(1A):119–123. [PubMed] [Google Scholar]
- 12.Siebolts U, Varnholt H, Drebber U, Dienes HP, Wickenhauser C, Odenthal M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2008 doi: 10.1136/jcp.2008.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Prueitt RL, Yi M, Hudson RS, Wallace TA, Howe TM, Yfantis HG, et al. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate. 2008 doi: 10.1002/pros.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henshall SM, Quinn DI, Lee CS, Head DR, Golovsky D, Brenner PC, et al. Altered expression of androgen receptor in the malignant epithelium and adjacent stroma is associated with early relapse in prostate cancer. Cancer Res. 2001;61(2):423–427. [PubMed] [Google Scholar]
- 19.Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, et al. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res. 2003;9(13):4792–4801. [PubMed] [Google Scholar]
- 20.Dakhova O, Ozen M, Creighton CJ, Li R, Ayala G, Rowley D, et al. Global gene expression analysis of reactive stroma in prostate cancer. Clin Cancer Res. 2009;15(12):3979–3989. doi: 10.1158/1078-0432.CCR-08-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.