Abstract
Some individuals with a genital Chlamydia trachomatis infection develop inflammatory arthritis, but it is unknown whether particular chlamydial serovar(s) engender the disease more often than others. We defined serovar in synovial tissues from arthritis patients infected with this organism. DNA from synovial biopsies of 36 patients with PCR-confirmed synovial C. trachomatis was analyzed. Diagnoses included reactive arthritis, undifferentiated oligoarthritis, rheumatoid arthritis, and osteoarthritis. The chlamydial omp1 and trpA genes were amplified, cloned, and 10 or more clones from each sample were sequenced. The cytotoxin locus also was analyzed. omp1 sequences showed 2 patients having only C. trachomatis A serovar, 1 with only B, and 33 having only C, all ocular serovars. Analyses of trpA and the cytotoxin locus uniformly displayed standard ocular serovar characteristics for each patient. Identification of ocular chlamydial serovars in the synovia of arthritis patients is unexpected. These observations suggest that urogenital chlamydial infections, while consisting primarily of organisms of genital serovars, include some of ocular serovar(s). They further suggest that during such infections unknown selection pressures favor establishment of the latter in the synovium to the exclusion of genital serovar chlamydiae.
Keywords: Chlamydia trachomatis, inflammatory arthritis, infection, genital infection
1. Introduction
Genital infections with the bacterial pathogen Chlamydia trachomatis are a significant problem in the United States, and they must be reported to the Centers for Disease Control (CDC) from all 50 states and the District of Columbia. Data from the CDC indicate that more than 3 million new infections are reported/year, with a stable population of C. trachomatis-infected individuals in the US in excess of 6 million [1]. Importantly, genital chlamydial infections can engender serious sequelae, one of which is chronic reactive (inflammatory) arthritis (ReA) [e.g., 2–6]. Reports indicate that ~ 5% of those with a genital chlamydial infection will develop acute ReA, and about half of these latter will proceed to chronicity [e.g., 7]. Further, published data indicate that 20–40% of individuals with inflammatory arthritis but no documented prior genital infection with C. trachomatis, and who are given a diagnosis of undifferentiated spondyloarthropathy, oligo- or mono-arthritis, are PCR-positive in synovial tissue and/or fluid for C. trachomatis DNA [8,9]. Our studies and those of others have made progress in understanding the molecular and cellular biology of C. trachomatis, and in understanding its pathogenic interaction with its host cells in the synovium and elsewhere [6 for review]. However, we do not know whether organisms of one or more particular serovars/strains of the organism are responsible for arthritogenesis, and if so how organisms of that/those serovars initiate and maintain synovial pathogenesis. We also have a poor understanding of why only some individuals develop arthritis following genital chlamydial infection, and why only a portion of those with acute disease progress to chronicity. It is clear that many aspects of disease induction, severity, and chronicity are functions of the serovar of the infecting Chlamydia, and of as yet unknown properties of the host genetic background [10,11].
In addition to its role in genital infections, C. trachomatis is the etiologic agent for trachoma, a disease that remains the most prevalent cause of treatable blindness worldwide [e.g., 12,13]. As the designation indicates, serovars of this organism have been differentiated serologically via the antigenic structure of the chlamydial major outer membrane protein, encoded by the omp1 gene [14 and see below]. However, many recent studies have utilized DNA sequence information to identify serovars in clinical or other samples [15; see also below]. Chlamydiae of serovars A, B, Ba, and C are ocular (trachoma) agents, while those of serovars D-K are recognized as standard genital agents. A separate biovar includes the chlamydiae responsible for lymphogranulosum venereum (LGV), designated biovars L1–L3 [14,16]. Importantly, organisms of ocular serovars can cause genital infections, and vice-versa, under some conditions [e.g., 17–19]. However, genital infections by ocular serovars are rare [17–19].
Recent reports have demonstrated that ocular and genital chlamydial serovars can be distinguished from one another as groups at several chromosomal loci, in addition to the amino acid or DNA sequence of the omp1 gene. Organisms of ocular and genital serovars possess different deletions in and around the cytotoxin gene (designated gene Ct166); organisms of ocular serovar B have a large deletion spanning gene Ct162 through the trp operon [20]. Further, while genital serovar chlamydiae produce functional products from the trp operon, organisms of ocular serovars have deletions in trpBA that result in nonfunctional products [21,22]. These differences have been postulated to account in part for tissue tropism and variable pathogenicity between organisms of the ocular and genital serovar groups. A recent study further demonstrated that subtle differences in genomic variation in ocular serovars result in variations in IFN-γ sensitivity, growth rate in vitro, virulence, and other characteristics [23].
As mentioned, one longstanding question in relation to synovial pathogenesis in Chlamydia-associated arthritis concerns whether the disease is elicited by organisms of a particular “arthritogenic” serovar, or whether a range of serovars can disseminate to joints and induce disease. Further, it is not clear whether those patients who develop only acute ReA following genital chlamydial infection are infected by serovars different from that/those infecting patients who progress to chronic disease. Here, we present surprising observations relating to the infecting C. trachomatis serovars identified in the synovia of arthritis patients who are PCR-positive in that tissue for DNA of C trachomatis.
2. Results
2.1. Infecting chlamydial serovar determined by the DNA sequence of omp1
Serovars of C. trachomatis often are differentiated and defined on the basis of the predicted amino acid sequence of the omp1 gene product [e.g., 14,15]. We amplified the complete omp1 gene, plus some 5′ and 3′ flanking sequence, using DNA prepared from each of 36 synovial biopsies from individuals known to be PCR-positive for chromosomal DNA from the organism. The resulting DNA sequences were compared to serovar type-sequences in GenBank, and the infecting serovar was deduced from those data. The results shown in Table 1 indicate that of the 36 patient samples so analyzed, 2 were infected with A serovar chlamydiae (patients 7, 12), 1 was infected with organisms of B serovar (patient 36), and the rest were infected with C serovar C. trachomatis. None of the patients analyzed showed any DNA sequences characteristic of genital chlamydial serovars in the synovial DNA preparation used as template for PCR amplification. This finding of exclusively ocular rather than genital serovars in these synovial samples from arthritis patients is unlikely to be the result of laboratory contamination, since none of the groups associated with this work studies or has grown or studied ocular chlamydial serovars for many years. Further, negative control PCR assays run with each amplification reaction for each patient sample were universally negative (see also Patients and Methods, below). To be certain that we were not identifying a minor component serovar in a mix with primarily genital serovars, we sequenced at least 10 (and usually 15 or more) clones from each of the 36 patient samples. In all cases, each clone gave a DNA sequence, and thus a deduced serovar, which matched that of the initially sequenced clone from each patient (Figure 1). Moreover, and interestingly, we found relatively little DNA sequence diversity among the clones sequenced from each patient. This is in contrast to data from other laboratories regarding genital samples assessed for chlamydial serovar by DNA sequence [e.g., 15; see also below].
Table 1.
Characteristics of patients from whom samples were procured
| Patient | Age/sex | Working Diagnosis |
Disease Duration (mo) |
Serovar | trpA | Cytotoxin Region |
|---|---|---|---|---|---|---|
| 1 | 27/F | ReAa | 1 | C | ndb | ocular |
| 2 | 51/M | ReA | na | C | ocular+genital† | ---------c |
| 3 | ?/F | UOd | 6 | C | nd | --------- |
| 4 | 44/M | ReA | 11 | C | ocular | ocular |
| 5 | 47/M | UO | 11 | C | ocular | ocular |
| 6 | ?/F | UO | Na | C | nd | --------- |
| 7 | 38/M | UO | 15 | A | ocular | --------- |
| 8 | 58/M | RAe | 84 | C | nd | ocular |
| 9 | 33/F | UO | 6 | C | ocular | --------- |
| 10 | 25/F | UO | na | C | nd | --------- |
| 11 | 32/M | RA | 7 | C | ocular | ocular |
| 12 | 39/M | RA | 12 | A | ocular | --------- |
| 13 | 32/M | UO | 0.5 | C | ocular | ocular |
| 14 | ?/M | UO | 96 | C | nd | ocular |
| 15 | 44/M | ReA | 2 | C | ocular‡ | --------- |
| 16 | 37/M | ReA | na | C | ocular | ocular |
| 17 | 26/M | ReA | na | C | nd | --------- |
| 18 | 58/F | UO | 75 | C | ocular | --------- |
| 19 | 63/M | ReA | 25 | C | ocular | --------- |
| 20 | 47/F | ReA | 16 | C | nd | --------- |
| 21 | 58/M | ReA | 5 | C | ocular | --------- |
| 22 | 61/F | RA | 7 | C | ocular¶ | --------- |
| 23 | 54/F | ReA | na | C | ocular¶ | --------- |
| 24 | 49/F | ReA | na | C | ocular | --------- |
| 25 | 45/F | ReA | na | C | ocular | --------- |
| 26 | 60/M | OAf | na | C | nd | --------- |
| 27 | 68/M | RA | 4 | C | ocular | --------- |
| 28 | 44/M | ReA | 2 | C | ocular | --------- |
| 29 | 55/F | RA | na | C | nd | ocular |
| 30 | 53/F | ReA | na | C | ocular‡ | ocular |
| 31 | 54/F | UO | na | C | nd | --------- |
| 32 | 57/F | OA | na | C | ocular | --------- |
| 33 | 55/F | RA | na | C | ocular | --------- |
| 34 | 49/F | RA | na | C | ocular | --------- |
| 35 | 82/F | RA | na | C | ocular | --------- |
| 36 | 58/F | ReA | na | B | nd | --------- |
ReA, reactive arthritis
not done due to lack of material
not done due to lack of material, or no PCR product produced
UO, undifferentiated oligoarthritis
RA, rheumatoid arthritis
OA, osteoarthritis
ocular sequences linked to genital sequences; see Figure 3
most trpA DNA sequences of the ocular type but some proportion Group I genital type [22]
most trpA DNA sequences of the ocular type but some proportion Group II genital type [22]
Figure 1.
Predicted omp1 (major outer membrane protein) amino acid sequence deduced from DNA sequences from synovial tissues of 4 representative arthritis patients (#11, 13, 15, 18) compared to the prototype C serovar sequence (C). Nonconserved amino acids are indicated by bold underlined capital letters. Patient samples are from Tampa (patient 18) and from Philadelphia (patients 11, 15, 13).
2.2. Assessment of ocular vs genital chlamydial serovars via the DNA sequence of trpA
Studies from other groups have identified DNA sequence characteristics that can differentiate ocular from genital serovars of C. trachomatis. For example the trpA gene, which encodes the alpha subunit of tryptophan synthase, includes a single base pair (bp) deletion near the 3′ end of the coding sequence (nucleotide 528) in organisms of ocular serovars A, Ba, and C, but not in chlamydiae of genital serovars; also, the ocular serovar organisms have a 3 bp deletion upstream of the single bp deletion (nucleotides 408–410) which results in loss of a phenylalanine [22]. These mutations lead to a non-functional product from trpA. We PCR-amplified this region of the chlamydial chromosome in selected DNA preparations from the synovial biopsies, then subjected the cloned PCR products to DNA sequencing. As with analyses of the omp1 gene above, at least 10 (and usually 15 or more) clones from each sample were screened to insure that we were not identifying a minor component of the population. As summarized in Table 1, all of the samples so assessed showed both the triple and single bp deletions at the specified nucleotide locations characteristic of chlamydiae of ocular serovars (Figure 2). Interestingly, patients 15 and 30, both of whom showed C serovar omp1 sequences exclusively in all clones examined for that gene, gave a mixture of the ocular trpA DNA sequence and 3 clones of trpA sequence characteristic of the Group I genital serovars (G, F, I, H, J; see [22]). Further, patients 22 and 23, both of whom also had only C serovar omp1 sequences, gave a mix of ocular trpA sequences plus 4 clones of the trpA sequence characteristic of Group II genital serovars (again, see [22]). Additional sequencing of several omp1 clones from all of these patients failed to yield a sequence characteristic of either Group I or Group II genital serovar organisms, suggesting that in a few infecting organisms the standard C serovar omp1 sequence is recombined with the genital trpA sequence on the same chromosome. This assertion is supported by the DNA sequence of one trpA clone from patient 2, which showed a combined ocular type sequence with a Group I genital (J serovar type) sequence on the same molecule (Figure 3). This unusual sequence most probably resulted from a recombination event between an ocular and a genital organism in the same inclusion; such recombination events have recently been described by several groups [e.g., 24,25].
Figure 2.
Representative DNA sequences from the trpA gene of C. trachomatis from selected patient samples. These included patients #5 and #32, both of whom had C serovar as judged from omp1 sequence, #7, who had A serovar from the omp1 sequence, and #36, who had B serovar as judged from the omp1 sequence. PCR amplification, cloning, and sequence determination were done as described in Patients and Methods. Below the patient trpA sequences the congruent sequence from serovars A, B, and C, and from the Group I genital serovars G, F, I, H, and J, are included for comparison [22].
Figure 3.
The DNA sequence of one trpA clone from patient 2 displaying recombined elements containing in part the ocular type sequence (underlined) and in part the Group II genital type sequence (italics). PCR amplification, cloning, and sequence determination were done as described in Patients and Methods.
2.3. Assessment of the chromosomal deletion at/around the cytotoxin locus
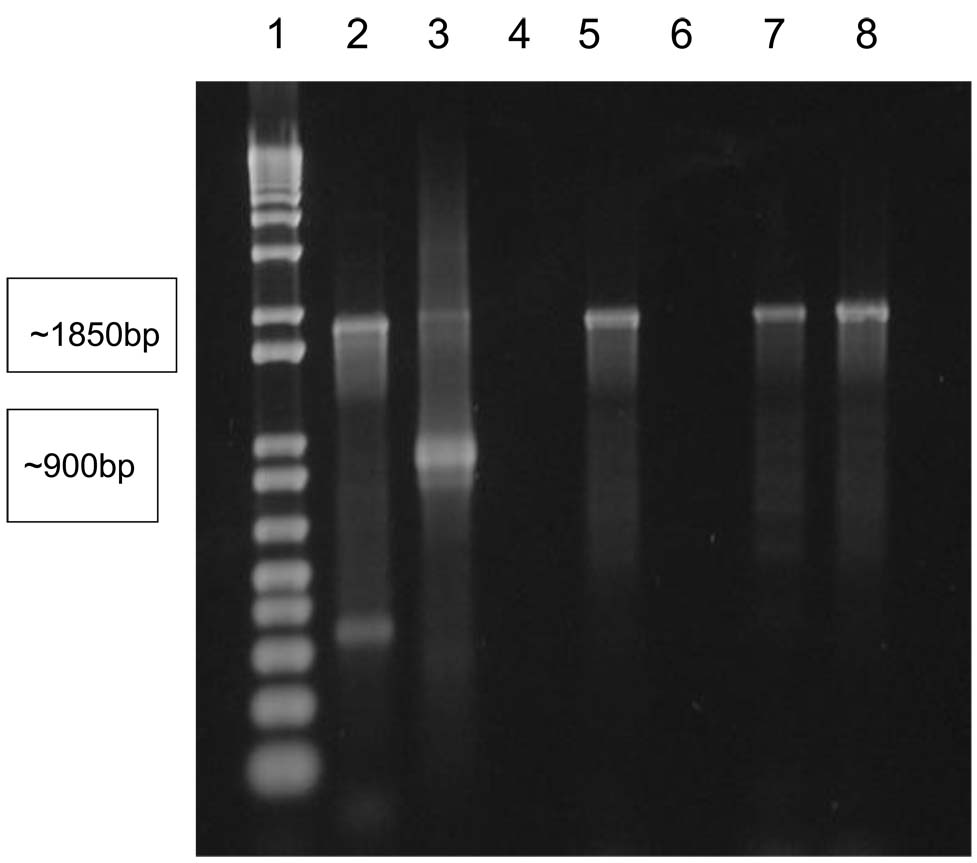
Research from another group has demonstrated that deletions are present in the region upstream of trpR, in and around the cytotoxin locus in the plasticity zone on the chlamydial chromosome [20]. On the chromosome of chlamydiae of ocular serovars A, Ba, and C, the deletion is about 5 kbp in length, while in genital serovars D-G, I, and K the deletion is about 5.8 kbp and is shifted somewhat 3′-ward. In organisms of the genital serovars H and J no deletion is present, and in the organisms of ocular B serovar the entire region is missing. We PCR-amplified the region from gene Ct176 to gene Ct179 in selected patient samples and examined the products produced in standard agarose electrophoretic gels. The data summarized in Table 1 indicate that each of the patient samples so analyzed, and for which a PCR product was produced, provided a pattern characteristic of ocular serovars upon analysis in electrophoretic gels. The data in Figure 4 provide representative results for several of the patient samples so analyzed.
Figure 4.
Representative PCR amplifications to assess the deletion in the plasticity zone on the C. trachomatis chromosome, in the region of the cytotoxin locus. PCR amplification, and display on standard 1% agarose gels, were as described in Patients and Methods. Lanes are: 1, 1 kbp size stds; 2, positive control (A serovar DNA as template); 3, positive control (K serovar DNA as template); 4, negative control (water as template); 5, 7, 8, DNA from patients 1, 11, 29; 6, DNA from an irrelevant (PCR-negative) patient. Ocular serovars produce a PCR product of about 1850 bp, while Group I genital serovars product a PCR product of about 900 bp. See [22].
2.4 Clinical characteristics of the patients analyzed
Individuals with Chlamydia-associated arthritis frequently, but not universally, display one or more of panel of extra-articular features, including conjunctivitis, iritis, keratoderma blennhoragicum, and others [e.g., 4, 26]. A question of interest centers on whether the patients whose synovial DNA preparations were studied displayed any particular set or subset of such features that might be reflective of infection with ocular chlamydial serovars. Examination of available clinical data for these patients showed 3 with axial involvement (patients 11, 14, 18), 1 with conjunctivitis (patient 17) and one with retinal inflammation and detachment (patient 4; several others complained of dry eye), and one with psoriasis (patient 3). Thus, the majority of patients studied displayed no ocular involvement, axial manifestations, or skin involvement. Although it is difficult to judge from the somewhat limited available clinical information, our impression is that no meaningful difference in disease severity exists among patients infected with A vs. B vs. C serovar C. trachomatis.
3. Discussion
In the work presented here, we define the serovars of C. trachomatis infecting synovial biopsies from 36 patients with various forms of arthritis, each of whom was known to be PCR-positive in that tissue for this pathogen from our earlier published reports. All patient samples studied were from our large freezer “library” of samples provided for analyses by collaborating clinicians at widely separated geographic locations in the United States. We were expecting to find one or more commonly identified genital serovars of the organism in these samples, but completely unexpectedly we identified only ocular (trachoma) serovars. Also surprisingly, the DNA sequence diversity in the chlamydial omp1 gene, which is standardly used for serovar definition, was quite low. It is extremely unlikely that these observations result from contamination in the laboratories involved in procurement or processing of the samples, since none of the groups involved grows or studies ocular strains of C. trachomatis, and appropriate negative controls were included with each and every assay run. Moreover, processing and analyses of samples have always taken place in several widely separated laboratories under carefully controlled conditions.
Whether, and if so by what means, chlamydiae of the A, B, and C serovars are in fact uniquely arthritogenic as opposed to those of genital serovars remains to be established. One possible explanation for the apparently exclusive presence of organisms of ocular serovars in the synovia of arthritis patients may be that these organisms simply disseminate from the site of primary infection more efficiently than do all/most genital serovar chlamydiae. Most genital infections with C. trachomatis probably are not clonal, i.e., comprised of a single serovar of the organism. Thus, alternatively and along the same line, it may be that both ocular and genital serovars disseminate equally well from the infected genital tract, but that ocular serovars are positively selected from the infection mix by some as yet unknown means once the organisms reach the synovium. Further, some observations suggest that ocular chlamydial strains transit to the persistent state more easily than do genital strains (Dr. G.I. Byrne, personal communication). It is chamydiae in this latter infection state that are responsible for inflammatory arthritis, and it may be the case that rapid transition to persistence gives C. trachomatis of the ocular strains some advantage in establishing long-term residence in the synovium. Much more study will be required to examine these and other possibilities.
Regardless, one issue that the data presented here probably does explain at least in part relates to the epidemiology of Chlamydia-induced arthritis. That is, as mentioned above it has never been clear why only a small proportion of individuals who acquire a documented genital chlamydial infection develop the acute inflammatory arthritis. Studies from many laboratories have shown that, while ocular serovars can and do cause genital infections, those serovars are only rarely identified in epidemiologic studies of genital chlamydial infection [e.g., 27–29]. Thus, the low percentage of infected patients who develop acute arthritis may reflect little more than the rarity of ocular serovars in the overall pool of organisms involved in causing genital infections. It is of interest to note here as well that ocular features, such as uveitis, iritis, and conjunctivitis, are sometimes identified as extra-articular features of patients with Chlamydia-associated arthritis [e.g., 4, 26]. The presence of chlamydiae of ocular serovars in disseminated sites in such patients may be in part responsible for those ocular effects, although only one of the patients whose DNA was studied here had conjunctivitis. More study will be required to define this issue.
An issue these serovar data do not help to explain is why only about half the number of individuals who develop acute arthritis following a documented genital chlamydial infection progress to chronic disease. The patient samples selected for analysis from our freezer “library” included those with acute and others with chronic arthritis, since we have been interested in the details of host-pathogen interaction in both disease contexts [e.g., 6, 30–32]. The patients studied with acute disease at the time of sample procurement (e.g., patients 1, 13, 15, 28; see also below) displayed no difference in serovar type of infecting organisms compared to the serovar type chlamydiae identified in patients with chronic disease. Thus while more study will be required, our observations provide no evidence for differential serovar infection in patients who develop only acute arthritis vs. those who progress to chronic disease. Further, while detailed clinical assessments are not available at this point for each of the patients studied here, our impression is that disease severity is roughly equivalent in patients infected with each of the ocular serovars identified, as judged from relevant information that is available.
As mentioned above, the samples examined here for serovar were obtained from different, and rather widely separated, regions of the United States, indicating that arthritis presumably due to the synovial presence or organisms of ocular chlamydial strains is not confined to a single geographical region. Those locations are, however, in the eastern USA, and it therefore will be of interest to obtain and analyze samples from patients at clinics located on the west coast and elsewhere. Readers will note that we purposely chose samples from patients known to be PCR-positive for chromosomal DNA but without regard to specific diagnosis; that is, the patients studied here had various forms of arthritis, including but not limited to reactive arthritis due to chlamydial infection. We do not mean to contend that osteoarthritis is caused by chlamydial infection of the joint, of course; rather, we previously demonstrated that 4–5% of patients with that disease, and about the same percentage of perfectly normal control individuals as well, have C. trachomatis DNA in synovial tissues [33]. Moreover, synovial PCR-positivity for the organism in patients with rheumatoid arthritis does not indicate that the infecting organism caused the disease; we are, however, re-examining the possibility that chlamydiae do contribute to genesis of this disease both in the laboratory and in an extensive literature review [34; see also 35]. Our point in assessing the serovars of C. trachomatis found in synovial samples from PCR-positive patients with various arthritides was to determine if one or more specific serovar(s) is/are uniquely responsible for genesis of Chlamydia-induced ReA. In the sense that only ocular serovar chlamydiae are found in the synovium of the patients studied, this does appear to be the case.
It is the case that organisms of C serovar are strongly predominant in samples from the patients studied, and it is not at all clear why chlamydiae of this trachoma serovar are so prevalent in synovial materials. In trachoma studies, C serovar organisms have been identified commonly in isolated indigenous communities in western Australia, in Nepal, and elsewhere [e.g., 36–38]. As indicated above, this serovar indeed has been identified in genital samples, although its occurrence in genital contexts is relatively rare [e.g., 27–29]. Our data indicate that chlamydial infections of the urogenital system are rarely if ever clonal, and that some inocula include a small number of C serovar organisms, or more rarely A and/or B serovar organisms; a selection process favoring those serovars must take place at either the level of dissemination from the genital system or at the level of establishment of stable maintenance of infection in the joint. It is not clear what variations in the C serovar genome, as opposed to the genomes of A or B serovar, enable it to engender inflammatory reactive arthritis in a majority of patients. These are issues that we are investigating.
4. Patients and Methods
4.1. Patients and diagnoses
DNA preparations from synovial tissue samples from 36 patients were chosen from those available in our freezer “library” of samples. Each sample was chosen on the basis of adequate quality and amount of DNA available for the analyses planned, and because each sample had been demonstrated to be PCR-positive for DNA sequences on the chromosome of the organism in various previous studies, using published, well-characterized screening primer systems [e.g., 30, 31]. All samples described here were procured under approved protocols using the standard Parker-Pearson technique [39]. Samples were immediately frozen at −80°C and shipped on dry ice to the Hudson laboratory for analyses. The general characteristics of the 36 patients from whom samples were procured are summarized in Table 1; additional, more detailed clinical information for some patients is not available, since some samples were procured for study as long as 20 yr ago.
The diagnosis for each patient was made according to criteria of the American College of Rheumatology or elsewhere [see below]. ReA occurs 1–6 wk after the initial genital chlamydial infection; clinical symptoms can include inflammatory arthritis, iritis, urethritis, and a characteristic rash on the palms and soles or penis. Most patients have only some of these organ systems involved. These diagnostic criteria require a peripheral arthritis occurring in association with urethritis or cervicitis [26]. The Third International Workshop on ReA requires a peripheral arthritis with sacroiliac involvement and a preceding gastrointestinal or genitourinary infection [40]. The current ACR definition might be too limited in scope, and the latter’s reliance on a preceding infection could lead to under-diagnosis, particularly since asymptomatic chlamydial infections are common [9]. The term undifferentiated oligoarthritis (UO) is non-specific and refers to an inflammatory arthritis involving 2–4 joints. Patients often present in such a manner with no other associated features or diagnostic testing, thereby making a definitive diagnosis difficult. Over time, many of these patients develop features making the correct diagnosis more clinically apparent. Data exist suggesting that C. trachomatis, and the other causative organisms of ReA, can cause an incomplete clinical picture resulting in a diagnosis of UO [8,41].
4.2. Analyses
Total DNA had been prepared from each synovial tissue sample using the hot phenol method extensively described by us [30,31]. Prior to performing the present analyses, the quality of each such preparation was assessed by PCR targeting the host actin gene [30,31], and the overall amount of DNA available for analyses was assessed spectrophotometrically. The omp1 gene, encoding the C. trachomatis major outer membrane protein, was amplified from each patient DNA preparation using a high fidelity polymerase (Platinum Taq™, Invitrogen, Carlsbad, CA USA), and the primers 5′-ggacatcttgtctggctttaact-3′/5′ + 5′-gcgctcaagtagaccgatatagta-3′ (outer), and 5′-ggacgcagtgccgccagaaaag-3′/5′ + 5′-gataagcttactaagaaaagatcc-3′ (inner) [42]. The relevant segment of the chlamydial trpA gene was similarly prepared from DNA preparations of selected samples using the primers 5′-atgatcgggctttagcagaa-3′ + 5′-tttgtgcaagtgcagtcaga-3′ (outer) and 5′-cagcacctttatcacacgga-3′ + 5′-tcacaatgaaaccatctgaat-3′ (inner); the outer primer set was designed by us using GeneRunner™ software (Hastings Software, Westwood NJ USA), and the inner primer set is from Caldwell et al. [22]. The various PCR products were cleaned using the Qia-Quick™ PCR Purification Kit (Qiagen, Valencia, CA USA), then cloned into the pGEM-Teasy™ vector, using conditions specified by the manufacturer (Promega, Madison, WI USA; see also [31]). DNA sequencing was done at the core facility at Wayne State University, and at least 10 clones from each patient and for each gene target were sequenced. Serovar determinations from those sequences were done by BLAST search of the omp1 sequences obtained and computer-based comparison to known serovar-specific sequences. Analysis of the deletions in the trpA gene was done by comparison to data given in ref. [22]. Analysis of the large chromosomal deletion in the region of the toxB gene was done by PCR amplification of sequences flanking the deletion in selected samples using the primers 5′-agaaacttggatcggatcgggtaa-3′ + 5′-ccttgcatggacggtgatatcaa-3′ and 5′-gagctgcaatctatgaagcaagctc-3′ + 5′-gctccatagcacctttaatatgccc-3′. This primer set was designed by us using GeneRunner™ software. Amplification products were displayed on standard 1% agarose electrophoretic gels [e.g., 31]. In some samples, no amplification product was produced after several repeats of the assay (see Table 1); the lack of product in these cases is most probably due to alterations in DNA sequence at the site(s) of primer annealing, thus obviating primer binding. Cycling conditions for each of the PCR reactions used are available upon request [see also 42].
Acknowledgments
This work was supported by research grants AR-42541 (A.P.H.), AR-48331 (J.A.W.-H.), AR-53646 (J.D.C.), and AR-47186 (H.C.G.) from the US National Institutes of Health, and by a research grant from the US Department of Veterans Affairs Medical Research Service (H.R.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.CDC website. ( www.cdc.gov/std/Chlamydia/STDFact-Chlamydia.htm)
- 2.Inman RD, Whittum-Hudson JA, Schumacher HR, Hudson AP. Chlamydia-associated arthritis. Curr Opin Rheumatol. 2000;12:254–262. doi: 10.1097/00002281-200007000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Sieper J. Pathogenesis of reactive arthritis. Curr Rheumatol Rep. 2001;3:412–418. doi: 10.1007/s11926-996-0012-8. [DOI] [PubMed] [Google Scholar]
- 4.Schumacher HR. Chlamydia-associated arthritis. Isr Med Assoc J. 2000;2:532–535. [PubMed] [Google Scholar]
- 5.Rihl M, Köhler L, Klos A, Zeidler H. Persistent infection of Chlamydia in reactive arthritis. Ann Rheum Dis. 2006;65:281–284. doi: 10.1136/ard.2005.044966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gérard HC, Whittum-Hudson JA, Carter JD, Hudson AP. Molecular biology of infectious agents in chronic arthritis. In: Espinoza L, editor. Rheumatic Disease Clinics of North America. Philadelphia PA: Elsevier/Saunders; 2009. pp. 1–19. [DOI] [PubMed] [Google Scholar]
- 7.Kvien TK, Glennås A, Melby K, Granfors K, Andrup O, Karstenssen B, et al. Reactive arthritis: incidence, triggering agents, and clinical presentation. J Rheumatol. 1994;21:115–122. [PubMed] [Google Scholar]
- 8.Zeidler H, Werdier D, Klauder A, Brinkmann S, Viswat M, Mones ML, et al. Undifferentiated arthritis and spondyloarthropathy as a challenge for prospective follow-up. Clin Rheumatol. 1987;6 Suppl 2:112–120. doi: 10.1007/BF02203394. [DOI] [PubMed] [Google Scholar]
- 9.Carter JD, Gérard HC, Espinoza L, Ricca L, Valeriano J, Snelgrove J, et al. Chlamydiae as etiologic agents for chronic undifferentiated spondyloarthropathy. Arthritis Rheum. 2009;60:1311–1316. doi: 10.1002/art.24431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Entrican G, Wattegedera S, Rocchi M, Fleming DC, Kelly RW, Wathne G, et al. Induction of inflammatory host immune responses by organisms belonging to the genera Chlamydia/Chlamydophila. Vet Immunol Immunopathol. 2004;100:179–186. doi: 10.1016/j.vetimm.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Lad SP, Fukuda EY, Li J, de la Maza LM, Li E. Upregulation of the JAK/STAT1 signal pathway during Chlamydia trachomatis infection. J Immunol. 2005;174:7186–7193. doi: 10.4049/jimmunol.174.11.7186. [DOI] [PubMed] [Google Scholar]
- 12.Schachter J. Infection and disease epidemiology. In: Stephens RS, editor. Chlamydia: Intracellular Biology, Pathogenesis, and Immunity. Washington DC: ASM Press; 1999. pp. 139–169. [Google Scholar]
- 13.Whittum-Hudson JA, Hudson AP. Human chlamydial infections: persistence, prevalence, and prospects for the future. Nat, Sci Soc. 2005;13:371–382. [Google Scholar]
- 14.Yuan Y, Zhang Y-X, Watkins NG, Caldwell HD. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 C trachomatis serovars. Infect Immun. 1989;57:1040–1049. doi: 10.1128/iai.57.4.1040-1049.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poole E, Lamont I. C trachomatis serovar differentiation by direct sequence analysis of variable segment 4 region of the major outer membrane protein gene. Infect Immun. 1992;60:1089–1094. doi: 10.1128/iai.60.3.1089-1094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward ME. Mechanisms of Chlamydia-induced disease. In: Stephens RS, editor. Chlamydia -Intracellular biology, Pathogenesis, and Immunity. Washington DC: ASM Press; 1999. pp. 171–210. [Google Scholar]
- 17.Dean D, Kandel RP, Adhikari HK, Hessel T. Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Pathogens. 2008;5:e14. doi: 10.1371/journal.pmed.0050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suchland RJ, Eckert LO, Hawes SE, Stamm WE. Longitudinal assessment of infecting serovars of C trachomatis in Seattle public health clinics: 1988–1996. Sex Transm Dis. 2003;30:357–361. doi: 10.1097/00007435-200304000-00016. [DOI] [PubMed] [Google Scholar]
- 19.Gao X, Chen XS, Yin Y, Zhong MY, Shi MQ, Wei WH, et al. Distribution study of C trachomatis serovars among high-risk women in China performed using PCR-restriction fragment length polymorphism genotyping. J Clin Microbiol. 2007;45:1185–1189. doi: 10.1128/JCM.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson JH, Hughes S, Hogan D, Cieplak G, Sturdevant DE, McClarty G, et al. Polymorphisms in the C trachomatis cytotoxin locus associated with ocular and genital isolates. Infect Immun. 2004;72:7063–7072. doi: 10.1128/IAI.72.12.7063-7072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, et al. Molecular basis defining human C trachomatis tissue tropism. A possible role for tryptophan synthase. J Biol Chem. 2002;277:26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- 22.Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, et al. Polymorphisms in C trachomatis tryptophan synthase genes differentiate between genital and ocular serovars. J Clin Invest. 2003;111:1757–1769. doi: 10.1172/JCI17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kari L, Whitmire WM, Carlson JH, Crane DD, Reveneau N, Nelson DE, et al. Pathogenic diversity among C trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J Infect Dis. 2008;197:449–456. doi: 10.1086/525285. [DOI] [PubMed] [Google Scholar]
- 24.DeMars R, Weinfurther J. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanisms and significance. J Bacteriol. 2008;190:1605–1614. doi: 10.1128/JB.01592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes JP, Bruno WJ, Nunes A, Santos N, Florindo C, Borrgo MJ, et al. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 2007;17:50–60. doi: 10.1101/gr.5674706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klippel JH. Primer on the Rheumatic Diseases. 12th edition. Atlanta: Arthritis Foundation Press; 2007. [Google Scholar]
- 27.Mittal A. Serovar distribution of Chlamydia trachomatis isolates collected from the cervix: use of the polymerase chain reaction and restriction endonuclease digestion. Br J Biomed Sci. 1998;55:179–183. [PubMed] [Google Scholar]
- 28.Persson K, Osser S. Serovars of Chlamydia trachomatis causing postabortion salpingitis. Eur J Clin Microbiol Infect Dis. 1989;8:795–798. doi: 10.1007/BF02185848. [DOI] [PubMed] [Google Scholar]
- 29.Workowski KA, Stevens CE, Suchland RJ, Holmes KK, Eschenback DA, Pettinger MB, et al. Clinical manifestations of genital infection due to Chlamydia trachomatis in women: differences related to serovar. Clin Infect Dis. 1994;19:756–760. doi: 10.1093/clinids/19.4.756. [DOI] [PubMed] [Google Scholar]
- 30.Gérard HC, Krauβ-Opatz B, Rudy D, Rao JP, Zeidler H, Schumacher HR, et al. Expression of Chlamydia trachomatis genes required for DNA synthesis and cell division in active vs. persistent infection. Mol Microbiol. 2001;41:731–741. doi: 10.1046/j.1365-2958.2001.02550.x. [DOI] [PubMed] [Google Scholar]
- 31.Gérard HC, Freise J, Rudy D, Krauβ-Opatz B, Köhler L, Zeidler H, et al. Chlamydia trachomatis genes whose products are related to energy metabolism are expressed differentially in active vs. persistent infection. Microb Infect. 2002;4:13–22. doi: 10.1016/s1286-4579(01)01504-0. [DOI] [PubMed] [Google Scholar]
- 32.Whittum-Hudson JA, Gérard HC, Schumacher HR, Hudson AP. Pathogenesis of Chlamydia-associated arthritis. In: Bavoil P, Wyrick P, editors. Chlamydia- Genomics and Pathogenesis. Horizon Bioscience, Inc.; 2007. pp. 475–504. [Google Scholar]
- 33.Schumacher HR, Arayssi T, Crane M, Lee J, Gérard HC, Hudson AP, et al. Chlamydia trachomatis nucleic acids can be found in synovium of some asymptomatic volunteers. Arthritis Rheum. 1999;42:1281–1284. doi: 10.1002/1529-0131(199906)42:6<1281::AID-ANR27>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 34.Stanich JA, Carter JD, Whittum-Hudson JA, Hudson AP. Open Access Rheumatology: Research and Reviews. Dove Medical Press; Rheumatoid arthritis: disease or syndrome? www.dovepress.com In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pando JA, Yarboro C, El Lallan E, Saaibi D, Branigan PJ, Gérard HC, et al. Prevalence of Chlamydia trachomatis by PCR in the synovium of patients with early rheumatoid arthritis. Arthritis Rheum. 1995;38(9):S287. [Google Scholar]
- 36.Porter M, Mak D, Chidlow G, Harnett GB, Smith DW. The molecular epidemiology of ocular Chlamydia trachomatis infections in western Australia: implications for trachoma control. Am J Trop Med Hyg. 2008;78:514–517. [PubMed] [Google Scholar]
- 37.Stevens MP, Tabrizi, Muller R, Krause V, Garland SM. Characterization of Chlamydia trachomatis omp1 genotypes detected in eye swab samples from remote Australian communities. J Clin Microbiol. 2004;42:2501–2507. doi: 10.1128/JCM.42.6.2501-2507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Lietman, Olinger L, Miao Y, Stephens RS. Genetic diversity of Chlamydia trachomatis and the prevalence of trachoma. Pediatr Infect Dis J. 2004;23:217–220. doi: 10.1097/01.inf.0000115501.60397.a6. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher HR, Kulka JP. Needle biopsy of the synovial membrane: experience with the Parker-Pearson technique. N Engl J Med. 1972;286:416–419. doi: 10.1056/NEJM197202242860807. [DOI] [PubMed] [Google Scholar]
- 40.Kingsley G, Sieper J. Third International Workshop on Reactive Arthritis. 23–26 September 1995, Berlin, Germany. Ann Rheum Dis. 1996;55:564–584. doi: 10.1136/ard.55.8.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fendler C, Laitko S, Sörensen H, Gripenberg-Lerche C, Groh A, Uksila J, Granfors K, Braun J, Sieper J. Frequency of triggering bacteria in patients with reactive arthritis and undifferentiated oligoarthritis and the relative importance of the tests used for diagnosis. Ann Rheum Dis. 2001;60(4):337–343. doi: 10.1136/ard.60.4.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandea CI, Kubota K, Brown TM, Kilmarx PH, Bhullar V, Yanpaisarn S, Chaisilwattana P, Siriwasin W, Black CM. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing of the major outer membrane protein gene (omp1) Sex Transm Inf. 2001;77:419–422. doi: 10.1136/sti.77.6.419. [DOI] [PMC free article] [PubMed] [Google Scholar]