Abstract
Since the discovery of the first gene causing holoprosencephaly (HPE), over 500 patients with mutations in genes associated with non-chromosomal, non-syndromic HPE have been described, with detailed descriptions available in over 300. Comprehensive clinical analysis of these individuals allows examination for the presence of genotype-phenotype correlations. These correlations allow a degree of differentiation between patients with mutations in different HPE-associated genes and for the application of functional studies to determine intragenic correlations. These early correlations are an important advance in the understanding of the clinical aspects of this disease, and in general argue for continued analysis of the genetic and clinical findings of large cohorts of patients with rare diseases in order to better inform both basic biological insight and care and counseling for affected patients and families.
Keywords: Holoprosencephaly, HPE
Introduction
Holoprosencephaly (HPE) can be caused by chromosomal anomalies, result from teratogenic exposure, occur as part of a syndrome, or be due to mutations in one of over 12 known HPE-associated genes. Mutations in SHH, the first such gene identified, were shown to cause HPE in 1996 [Roessler et al., 1996]. Currently, 4 genes (SHH, ZIC2, SIX3, and TGIF) are routinely analyzed on a clinical basis in patients with HPE [Brown et al., 1998; Wallis et al., 1999; Gripp et al., 2000; Dubourg et al., 2007].
In the approximately one-and-a-half decades since the discovery of SHH as a cause of HPE, advances in the understanding of the molecular pathogenesis of the condition, combined with comprehensive clinical analyses of patients allows for analysis of genotype-phenotype correlations. Such correlations are still in the early phases, but it is possible to begin to distinguish between groups of patients with HPE due to mutations in different genes. Additionally, within cohorts of patients with mutations in the same gene, the use of functional analyses allows exploration of intragenic genotype-phenotype correlations. Finally, while the individual genes are considered alone here, it is critical to keep in mind that the complex pathogenesis of HPE appears to involve multiple interacting factors.
In the following discussion, after a brief description of common clinical findings, the known HPE-associated genes will be discussed in turn, and then analyzed as a group. Analysis of patients with mutations in each of these genes raises salient points regarding patients with HPE as a whole, including those in whom the genetic cause is unknown. Further, analysis of the clinical features of affected patients makes three specific conclusions quite clear. First, there is a pressing need for functional analyses in order to better inform clinicians, families, and diagnostic laboratories of the nature of identified variants in order to determine if they are truly mutations. Second, the establishment of robust genotype-phenotype correlations depends on thorough genetic and clinical analyses of a large number of individuals at both ends of the phenotypic spectrum, as well as the active research participation of relatives through their primary care-givers. In the case of a rare disorder such as HPE, the establishment of cooperative international research groups is imperative to increase our understanding of the disease to better inform clinicians and to optimally treat affected patients and families.
General Characteristics
Non-chromosomal, non-syndromic HPE is classically considered an autosomal dominant condition with incomplete penetrance and highly variable expressivity. Studies attempting to explain the wide spectrum of the effects of a single mutation even within a single kindred are still pending, but many lines of analysis, including extrapolation from animal models, point to a complex pattern of inheritance combining multiple interacting genetic and environmental factors [Reviewed in Krauss, 2007 and Schacter and Krauss, 2008; Ming and Muenke, 2001; Lacbawan et al., 2009].
HPE may be recognized in utero because of abnormalities on fetal imaging [reviewed in Volpe et al., 2009]. After birth, HPE is more commonly recognized due to facial findings consistent with the diagnosis and/or abnormalities on neurological examination. Neuroradiological or pathological studies confirm the diagnosis [Plawner et al., 2002; Lazaro et al., 2004; Stashinko et al., 2004; Dubourg et al., 2007; see Hahn and Barnes review on neuroimaging, this issue]. A common facial appearance occurs in most patients with HPE (the notable exception is patients with ZIC2 mutations) [Solomon et al., under review]. In general, the long-standing observation that “the face predicts the brain” holds true: in patients with typical facial abnormalities, the severity of facial findings generally correlates with the degree of brain anomalies and with survival [DeMyer et al., 1964; Muenke and Beachy, 2001; Stashinko et al., 2004; Lacbawan et al., 2009].
At the most severe end of the spectrum, patients may have pronounced microcephaly and cyclopia or synophthalmia below a proboscis. Less-severely affected infants have microcephaly (though hydrocephalus can lead to macrocephaly), hypotelorism, midface hypoplasia with a flat nasal bridge, cleft lip and or/palate, and a single maxillary central incisor (SMCI). Individuals with no abnormalities appreciated on conventional neuroimaging may have subtle facial findings including hypotelorism, a sharp and narrow nasal bridge, and SMCI [Muenke & Beachy, 2001; Lacbawan et al., 2009; Solomon et al., 2009]. These individuals, who are diagnosed with “microform” HPE, are identified due to a severely-affected relative (Fig. 1). It is important to note that even those who have pathogenic mutations in HPE-causing genes and who have clear microform HPE may have no cognitive impairment. Similarly, not all carriers of deleterious mutations manifest clinically detectable facial or cognitive abnormalities (see Table I, Fig. 2) [Muenke lab, unpublished data].
Figure 1.
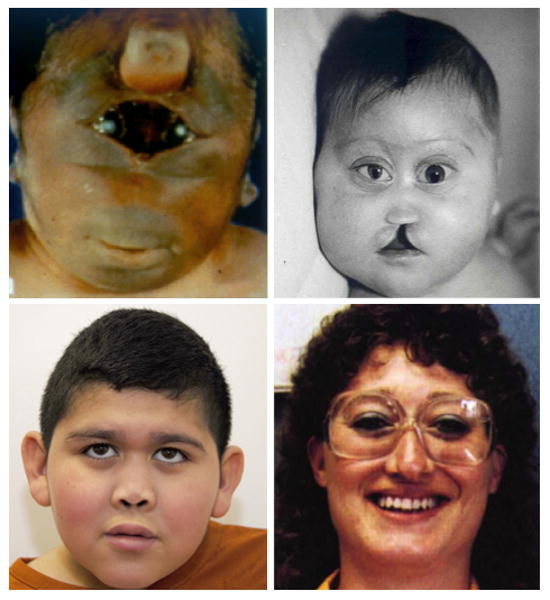
Facial findings in patients with mutations in HPE-associated genes, showing the range of anomalies. Clockwise from top left, photos show: a patient with alobar HPE due to a deletion of TGIF, with synophthalmia and a proboscis [Münke et al., 1988; Roessler et al., 1996]; a patient with alobar HPE due to a mutation in SIX3 demonstrating hypotelorism, a flat nasal bridge, colobomata, and a facial cleft [Lacbawan et al., 2009]; a patient with lobar HPE due to a mutation in SIX3, demonstrating relatively mild but clear signs of HPE, including hypotelorism and a flat nasal bridge [Lacbawan et al., 2009]; a patient with microform HPE due to a mutation in SHH, who has a hypotelorism and a single central incisor [Roessler et al., 1996]. All images reprinted with permission Reproduced from: Roessler et al., Mutations in the human Sonic Hedgehog gene cause holoprosencephaly, 14:357, Copyright (1996), with permission from Nature Publishing Group; Lacbawan et al., Clinical spectrum of SIX3-associated mutations in holoprosencephaly: correlation between genotype, phenotype and function, 46:390, copyright notice 2009, with permission from BMJ Publishing Group, Ltd.
Table I.
All individuals with identified mutations in the 4 most common HPE-associated genes in whom the HPE type (or lack thereof) is known. Individuals presented in this table include 180 propositi and 141 relatives; note that full trio analysis was not available for all propositi. Individuals in whom clinical descriptions did not include HPE type or clinical characterization were not included in the analysis.
| HPE Gene | Brain Anomalies | No Structural Brain Anomalies | Total | ||||
|---|---|---|---|---|---|---|---|
| A | S | L | M | Mic | Phenotypically Normal* | ||
| SHH | 19 | 21 | 7 | 0 | 45 | 11 | 103 |
| ZIC2 | 29 | 44 | 11 | 4 | 6 | 7 | 101 |
| SIX3 | 27 | 22 | 9 | 1 | 13 | 20 | 92 |
| TGIF | 3 | 4 | 4 | 0 | 8 | 6 | 25 |
| Total | 78 | 91 | 31 | 5 | 72 | 44 | 321 |
Individuals are described as phenotypically normal here when no microform features are reported and in whom there was no clinical indication for neuroimaging. All phenotypically normal individuals were relatives of propositi (the vast majority of which were parents of severely affected patients).
A: Alobar; S: Semilobar; L: Lobar; M: MIHV; Mic: Microform
Figure 2.
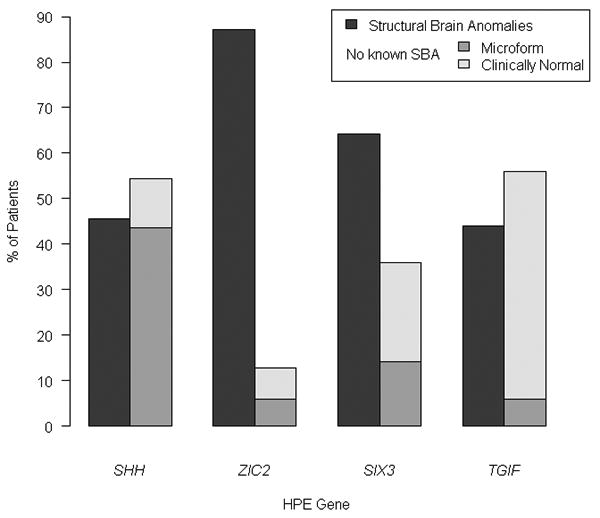
Proportion of HPE cases among all patients with mutations in the four classic HPE genes by the presence (dark bars) or absence of structural brain anomalies (SBA). Note the non-SBA group consists of individuals with mutations and either microform findings (grey bars) or who were apparently normal by standard clinical criteria yet also mutation positive (light bars). Note that the authors suspect that many individuals described as being clinically unaffected may have unappreciated microform features.
In addition to the facial appearance, a characteristic clinical pattern occurs in patients with non-syndromic, non-chromosomal HPE. The most pressing clinical issue commonly is severe neurological impairment, which is universal in patients with structural brain anomalies of the HPE type. The range of impairment is wide; some patients are among the most-severely impaired of any disorder compatible with life, while other patients may be able to walk, eat by mouth, and communicate. To some extent, the degree of impairment can be predicted by the degree and type of brain anomalies, though exceptions abound [Plawner et al., 2002; Hahn et al., 2006; Roesler et al., 2006].
Seizures are frequent in patients with HPE. Seizures may be underappreciated, especially in cognitively very impaired patients, and can be difficult to control [Hahn et al., 2003]. The diagnosis and management of seizures may be further complicated by electrolyte imbalances secondary to diabetes insipidus. Autonomic instability affecting temperature control and cardiac and respiratory function can also extremely challenging to manage.
Posterior pituitary insufficiency manifesting primarily as diabetes insipidus (DI) is common. DI may rarely be the presenting sign of HPE, especially in patients with a relatively normal facial appearance. Anterior pituitary insufficiency and other endocrinological disorders may manifest, but tend to be far less frequent and linked to a narrower subset of genes including GLI2 [Roessler et al., 2003; Lazaro et al., 2004; Hahn et al., 2005; Roessler et al., 2005].
SHH (Sonic Hedgehog; OMIM# 600725)
More families with mutations in SHH have been reported than with mutations in any other gene, accounting for up to 12% of propositi [Roessler et al., 2009a]. Hence, patients with mutations in SHH are sometimes considered the “prototypical” HPE patient. A number of very large families segregating mutations in SHH have been identified, typically only after the diagnosis of a severely-affected propositus. These families epitomize the highly variable expressivity described in many autosomal dominant disorders [Muenke et al., 1994]. Approximately 10-30% of mutations in SHH occur de novo. The presence of structural brain anomalies in patients with SHH mutations is estimated to be approximately 45%, while the penetrance of any manifestations (including microform HPE) is estimated to be approiximately 90% [Muenke lab, unpublished data]. However, penetrance estimates may be skewed for a variety of reasons. For example, penetrance may be underestimated: in multiple separate kindreds, many mutation-positive individuals were noticed to have microform holoprosencephaly, including microcephaly, hypotelorism, and SMCI, only after mutation testing demonstrated the presence of the same mutations found in a severely-affected relative [Ardinger et al., 1988; Roessler et al., 1996]. Conversely, testing of only obviously affected patients would lead to a disproportionate underestimation of mutation prevalence in individuals on the milder end of the phenotypic spectrum.
ZIC2 (Zinc Finger Protein of Cerebellum 2; OMIM# 603073)
Mutations in ZIC2 are estimated to occur in up to 9% of propositi [Roessler et al., 2009b; Solomon et al., under review]. ZIC2 mutations tend to occur de novo much more frequently than mutations in other HPE-associated genes, with mutations occuring de novo in approximately 72% of proposite. In contrast to SHH and SIX3, the other two genes most commonly associated with non-chromosomal, non-syndromic HPE, there are no families identified with mutations in ZIC2 in which the mutation has been ascertained in more than 2 generations. As with other HPE-associated genes, germline mosaicism occurs not infrequently, and must be considered in genetic counseling scenarios when a mutation found in a propositus does not occur in DNA samples obtained from the parents [Lacbawan et al., 2009; Solomon et al., under review]. Unlike other genes, mutations in ZIC2 tend to result in full HPE, and patients with mutations much less frequently demonstrate mild HPE signs. Almost 90% of patients with mutations in ZIC2 have structural brain anomalies, and it is rare that a parent with a mutation will not show clear signs of cognitive impairment [Solomon et al., under review].
Mutations in ZIC2 also give one of two clear examples in which the face does not predict the brain, as opposed to the prevailing idea regarding HPE for over four decades [DeMyer et al., 1964]. Recent analysis of a large cohort of patients with mutations in ZIC2 shows a common facial phenotype consisting of bitemporal narrowness, upslanting palpebral fissures, a flat nasal bridge, a short nose with anteverted nares, a broad and deep philtrum, and large ears [Solomon et al., under review].
SIX3 (Sine Oculis Homeobox, Drosophila, Homolog of, 3; OMIM# 603714)
Mutations in SIX3 are estimated to occur in up to 5% of propositi with HPE [Dubourg et al., 2007; Lacbawan et al., 2009; Muenke lab, unpublished data]. Similar to SHH, large families segregating mutations are typically ascertained only after the identification of a severely affected patient. Also like SHH, multiple kindreds have been described in which many individuals with mutations have subtle microform HPE [Lacbawan et al., 2009; Solomon et al., 2009]. The majority of mutations are inherited, with a de novo mutation rate estimated at 14%. Interestingly, of the 4 genes most commonly associated with HPE, SIX3 is the only one with a statistically significant over-representation of affected females [Keaton et al., 2009; Lacbawan et al., 2009; Solomon et al., under review, Muenke Lab, unpublished data].
While the presence of structural brain anomalies due to mutations in SIX3, at approximately 65% of mutation-positive patients, is intermediate between SHH and ZIC2, it has been posited that mutations in SIX3 result in relatively severe HPE [Ribeiro et al., 2006]. When applied to propositi only, this observation was borne out by a recent large-scale analysis of all known patients with mutations in SIX3 [Lacbawan et al., 2009]. Unlike HPE due to mutations in other genes, patients with HPE brain anomalies due to SIX3 mutations are most likely to have alobar, rather than semilobar HPE.
Finally, SIX3 shows an early instance of the use of functional studies to analyze potential genotype-phenotype correlations. Using results from a functional assay in zebrafish, it was possible to show that predictions of mutation severity in the animal model correlates with human severity of HPE [Domené et al., 2008; Lacbawan et al., 2009]. This study shows the power of functional studies in the interpretation of human mutation data; similar studies of mutations in other genes are critical for improved understanding of the causative effects of specific mutations, which will also directly translate to the clinical realm.
TGIF (Transforming Growth Factor-Beta Induced Factor; OMIM# 602630)
Of the 4 genes commonly tested in clinical laboratories, mutations in TGIF are by far the least common, occurring in approximately 1 to 2% of propositi [Dubourg et al., 2007; Keaton et al., in preparation]. TGIF is the only gene in which a family has been described where two clinically normal parents each contributed a mutant allele of the same gene to a severely affected propositus, though it must be said that only one of these alleles was clearly pathogenic [El-Jaick et al., 2007].
This family raises an important point. In the clinical scenario, finding variations in TGIF and other HPE-associated genes can be challenging to interpret, highlighting a common issue in genetic conditions in which most genetic variations are family-specific. While the deleterious effects of nonsense and frameshift mutations are usually clear, other variations may or may not have functional effects [El-Jaick et al., 2007]. Additionally, the role that alterations in TGIF play in causing HPE may be less completely understood than with other genes, which also muddies issues.
GLI2 (Gli-Krüppel Family Member 2, OMIM# *165230)
GLI2 is a large gene (>4,500 bp coding) in which many uncommon variants have been detected, but relatively few cases have been described in which loss-of-function has been clearly demonstrated. Patients with mutations shown to be loss-of-function (by formal experiments) indicate the presence of a common spectrum of microform HPE (SMCI or midline clefting) and anterior pituitary dysfunction, as well as polydactyly. Reported exceptions to this common GLI2 phenotype highlight the problem of the lack of a commonly accepted mechanism to test the functionality of new variants in a large gene [Roessler et al., 2003; Roessler et al., 2005; Rahimov et al., 2006; Muenke lab, unpublished data]. The fact that patients with mutations in GLI2 appear to have abnormalities (eg, anterior pituitary insufficiency and polydactyly) not seen in most patients with non-chromosomal, non-syndromic HPE emphasizes the importance of a thorough clinical evaluation that takes into account features not typically seen as part of the HPE spectrum.
Other Genes
Patients with mutations in other genes have been described, though these genes are not typically sequenced except on a research basis. Of note, the recent advent of oligonucleotide microarray analysis may reveal more about the consequence of copy number changes affecting these loci, though it may be challenging to interpret the consequences of gain or loss of nearby genes and regulatory regions [Bendavid et al., 2009; Muenke lab, unpublished data].
Patients with HPE-spectrum anomalies and mutations in PTCH1 do not appear to demonstrate findings outside typical HPE [Ming et al., 2002]. Mutations in NODAL may result in HPE, but are more commonly associated with cardiac and laterality defects [Roessler et al 2009d]. Similarly, mutations in FOXH1 (part of the NODAL signaling pathway) may result in cardiac defects or overt HPE [Roessler et al 2008]. Two patients have been reported with mutations in TDGF1 (CRIPTO), one with a midline brain anomaly and one with HPE [de la Cruz et al., 2002]. Finally, patients with loss-of-function mutations in DISP1 may have normal brain structure and development, but may have facial features usually seen in conjunction with frank HPE [Roessler et al 2009c].
Further Statistical Analysis of Combined Results
1) Analysis of structural brain anomalies in all mutation-positive individuals
A statistically significant association was found between the involved gene and whether or not structural brain anomalies (SBA) were present in all patients with mutations (χ2(3) = 42.8, p<0.0001) (Figure 2). After aggregating the information by gene, we compared the proportion of patients with SBA to those without known SBA. This latter group, in whom neuroimaging was not typically indicated nor performed, includes patients with clear microform features and individuals described as phenotypically normal. Two statistically significant differences were found, one in ZIC2 (χ2(1) = 108.4, p<0.0001) and the other in SIX3 (χ2(1) = 13.59, p<0.001), showing that when mutations in these genes are present, it is more likely to find patients with SBA than without SBA. We further compared the proportion of patients among all genes and found that the proportion of patients with SBA is not statistically the same for all genes (χ2(3) = 79.32, p<0.0001). Specifically, mutations in ZIC2 are responsible for ∼3% of SBA cases overall. Likewise, the proportion of patients without SBA differs among genes (χ2(3) = 79.32, p<0.0001), and mutations in SHH are responsible for 48% of patients without SBA.
2) Analysis of variable expressivity
We used the absolute number of patients with molecular changes to present, analyze and plot our data (Tables I and II). Unless specified, χ2-based tests were performed and based-on-simulation p-values were calculated to compare the distribution of HPE types attributed to each gene for both all individuals with mutations (Fig. 3a) and separately for propositi only (Fig. 3b). We considered potential differences in mutation distribution among HPE patients with SBA (thus excluding non-penetrant carriers and microform patients), and looked for differences in the distributional pattern in patients with SBA and those with each HPE type. To do this, we assumed the total SBA group to be the true distribution among the HPE genes and the distribution of each HPE type to be similar, or different, from this core pattern. We used R 2.9.2 Patched (R Development Core Team, 2009) for statistical analysis and graphical representation of data. Patient data were used from the following sources: [TGIF: Keaton et al., in preparation; SIX3: Lacbawan et al., 2009; ZIC2: Solomon et al., under review, SHH: Muenke Lab, unpublished data]
Table II.
HPE type among propositi with mutations in the four most common HPE-associated genes.
| HPE Gene | HPE Type | Total | ||||
|---|---|---|---|---|---|---|
| Alobar | Semilobar | Lobar | MIHV | Microform | ||
| SHH | 15 | 20 | 6 | 0 | 7 | 48 |
| ZIC2 | 27 | 42 | 10 | 4 | 0 | 83 |
| SIX3 | 14 | 14 | 6 | 1 | 2 | 37 |
| TGIF | 3 | 3 | 3 | 0 | 3 | 12 |
| Total | 59 | 79 | 25 | 5 | 12 | 180 |
Figure 3.
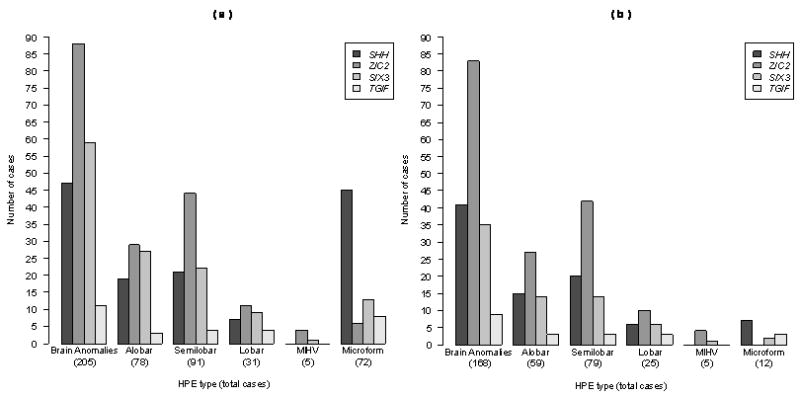
Distribution of mutations by HPE anatomical types among all individuals with clinically apparent penetrant findings (SBA or Microform patients with inclusion requiring (1) the presensce of mutation by sequencing and (2) clinical signs of HPE). (a) All affected mutation-positive individuals; (b) a subset including all propositi. The total number of patients in each group of vertical bars is presented in parentheses.
Side-by-side comparison
We performed a side-by-side comparison of the distribution of HPE types (excluding MIHV due to information paucity) among patients with SBA, plotted separately for all mutation-positive individuals (Fig. 3a) and propositi (Fig. 3b). The distribution of anatomical HPE types among the genes is statistically the same when calculated separately for both all mutation-positive individuals and for propositi alone.
Distribution of mutation-positive patients among HPE types
All mutation-positive individuals
The contribution of each gene to a particular anatomical type was not equal among the three major genes (SHH, ZIC2 and SIX3). This likely reflects the fact that the mutation detection rate in prospective cases is not the same for each gene. Interestingly, ZIC2 was more likely to be associated with semilobar HPE, and patients with microform findings were more likely to have SHH mutations. Statistically significant differences in mutation distribution among the HPE genes (assuming that these genes are equivalent risk factors) were found for the SBA category (χ2(3) = 59.5, p<0.0001) as well as for patients with alobar (χ2(3) = 21.5, p<0.0001), semilobar (χ2(3) = 35.5, p<0.0001) and microform (χ2(3) = 55.4, p<0.0001) HPE. We note that this assumption of equivalence is not supported by the empirical data and mutation detection rate of prospective molecular studies. With the exception of microform HPE, in which mutations in SHH are more common (45/72 patients), mutations in ZIC2, followed by mutations in SIX3, were more frequently observed than mutations in any other gene for the rest of the HPE cohort. This suggests that whereas patients with alobar, semilobar or lobar HPE are likely to have mutations in either ZIC2 or SIX3, patients with microform HPE are more likely to have mutations in SHH.
Propositi
Similarly, the distribution of mutation-positive patients among the HPE genes in the SBA group (χ2(3) = 67.1, p<0.0001) as well as those with alobar (χ2(3) = 19.6, p<0.0001) and semilobar (χ2(3) = 40.9, p<0.0001) HPE are statistically different from the hypothesis that all genes are equal contributors to these disease types. In other words, the contribution to each anatomical class is not equal for the four genes. Whereas propositi with SBA who have either alobar or semilobar HPE are more likely to have a ZIC2 mutation, it is not possible to determine which HPE genes are associated with lobar HPE based on this data. Further, despite the low numbers, SHH mutations seem to be responsible for most microform HPE (7/10 patients).
The spectrum of mutations seen in the structural brain anomalies (SBA) vs. HPE anatomical types
Using the SBA information as the expected number of HPE patients with mutations in each of the four principal HPE genes, we looked for discrepancies as follows: (1) based upon SBA data, we calculated the observed percentage of cases with mutations in each gene; then, (2) we used this percentage to estimate the expected number of mutations within each HPE type; and (3) we performed a χ2-based test to detect any pattern changes. We found that in both all mutation-positive individuals (Figure 3a) and for propositi alone (considered as a group, Figure 3b), the distribution of cases with microform HPE is the only group that significantly differs from these expectations (mutation carriers: χ2(3) = 76.6, p<0.0001; propositi: χ2(3) = 20.3, p<0.001).
Genotype-phenotype correlation
After excluding MIHV due to the paucity of information, we fit a uniform association model (Agresti, 1996; section 7.2.1) for our all mutation-positive and propositi-only data sets separately, considering the HPE types and the HPE genes as ordinal and nominal variables, respectively. For the HPE types, a unit-spaced scores ranging from 1 to 4 were used (1 = microform, 2 = lobar, 3 = semilobar and 4 = alobar HPE). Our results suggest that patients in both groups, patients with the most severe HPE phenotypes are more likely to have mutations in either SHH or ZIC2 (all mutation positive patients: G2 = 80.7, df = 8, p<0.0001; propositi: G2 = 50.7, df = 8, p<0.0001).
Discussion
We present here the first systematic analysis of possible genotype-phenotype correlations in human HPE. The results described above demonstrate that progress has been made in understanding the clinical correlations of HPE-associated mutations. However, this progress is still certainly just a start. Much work remains to more fully understand HPE on a number of levels, including the way in which multiple genetic and environmental mechanisms interact to result in disease.
First, the identification of a mutation does not yet allow satisfying prediction of outcome, either in a research setting or in the clinical realm. Continued work on the molecular basis of HPE-spectrum defects, including robust functional analyses will hopefully address the biological meaning of variations in HPE-associated genes.
Second, in order to better understand the broad HPE spectrum, it is critical to expand our diagnostic approach to include a more thorough analysis of both propositi and family members, especially including mildly-affected individuals. Areas in which data are lacking should be addressed by: a careful examination including attention to features not traditionally seen as part of the HPE spectrum; a robust family history; molecular characterization to include both parents as well as other relatives when applicable, complete physical examination, neuroimaging (with analysis by those familiar with HPE) and neuropsychological evaluation of affected family members.
Third, to accomplish the previous point, it is vital to recognize that HPE is a rare disorder, and most clinical geneticists will only encounter a few affected patients. In order to fully understand the clinical features of this disorder, clinicians must continue to collaborate with researchers studying the condition. Along these lines, international testing centers who commonly perform genetic testing of patients with HPE must continue to recognize the importance of collaboration with other groups. With focused collaborative work between clinicians and researchers who encounter these patients, our understanding of the complex pathogenesis of HPE will directly translate to better care for affected patients and families.
Acknowledgments
The authors would like to extend their deepest gratitude to all patients and families who took part in our research on HPE. JIV would like to thank to Juan Carlos Correa and Ehidy Karime García from the School of Statistics of the National University of Colombia at Medellín, for their helpful comments and suggestions that improved the statistical analysis. This research was supported by the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health and Human Services, United States of America and GIS Maladies Rares GISMR0701/DHOS, France.
Biographies
Benjamin D. Solomon, M.D. is a fellow in the Combined Pediatrics and Medical Genetics Residency Program, based at the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA. He is involved in research on holoprosencephaly and VACTERL association.
Sandra Mercier, M.D. is a researcher in the holoprosencephaly group in the “Génétique des Pathologies Liées au Développement” branch of UMR 6061 CNRS, IGDR, at the University of Rennes 1, France. She is also a senior registrar in the Clinical Genetics Service at CHU Hôpital Sud in Rennes.
Jorge I. Vélez is a fellow in the Medical Genetics Branch of the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA, and a graduate student in the Department of Statistics of The George Washington University, Washington, DC, USA. His research interests include applied statistical genetics, statistical computing, and bioinformatics.
Daniel E. Pineda-Alvarez, M.D. is a medical graduate trained in Colombia and is currently a Clinical Molecular Genetics fellow in the Medical Genetics Branch of the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA. He is a member of the Muenke lab and is involved in investigating the genetics behind holoprosencephaly and attention deficit hyperactivity disorder.
Adrian Wyllie is a senior at the University of the District of Columbia, Washington, DC, USA. He is also a Special Volunteer at the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA. He is interested in holoprosencephaly research and will be attending medical school in 2010.
Nan Zhou is a Biologist in the Medical Genetics Branch of the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA. She specializes in screening four genes which are associated with holoprosencephaly in the CLIA Lab.
Dr. Dubourg is a faculty member in the branch “Génétique des Pathologies Liées au Développement”, UMR 6061 CNRS, IGDR, University of Rennes 1, Faculty of Medicine, Rennes, France, and a hospital member in the Laboratory of Molecular Genetics, CHU Pontchaillou, Rennes, France. Her diagnostic and research interests include holoprosencephaly and mental retardation.
Professor David is the chief of Holoprosencephaly group in the branch of “Genetics of Developmental Pathologies”, UMR 6061 CNRS IGDR, Faculty of Medicine, University of Rennes 1, France. She is the chief of the Molecular Diagnosis laboratory, CHU Pontchaillou, Rennes, France. Her research interest mainly concerns HPE, and her clinical research also focuses on iron overload genetic diseases.
Sylvie Odent, M.D., Ph.D. is a professor of genetics and a member of the holoprosencephaly group in the “Génétique des Pathologies Liées au Développement” branch of UMR 6061 CNRS, IGDR, at the University of Rennes 1, France. She is the chief of the Clinical Genetics Service at CHU Hôpital Sud in Rennes and is a coordinator of a Center of Reference for Rare Diseases, focusing on developmental abnormalities and dysmorphology. Her clinical and research interests mainly concern holoprosencephaly and mental retardation.
Erich Roessler, M.D., Ph.D. is a faculty member of the Medical Genetics Branch of the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA. His research interests include holoprosencephaly and disturbances of organ sidedness, or laterality.
Maximilian Muenke, M.D. is the chief of the Medical Genetics Branch at the Division of Intramural Research in the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA. He has a longstanding interest in elucidating the genetics behind holoprosencephaly, craniofacial malformation syndromes, and attention deficit hyperactivity disorder, as well as an interest in improving knowledge about the formation of the central nervous system.
References
- Agresti A. An Introduction to Categorical Data Analysis. New York: John Wiley & Sons; 1996. [Google Scholar]
- Ardinger HH, Bartley JA. Microcephaly in familial holoprosencephaly. J Craniofac Genet Dev Biol. 1988;8:53–61. [PubMed] [Google Scholar]
- Bendavid C, Rochard L, Dubourg C, Seguin J, Gicquel I, Pasquier L, Vigneron J, Laquerrière A, Marcorelles P, Jeanne-Pasquier C, Rouleau C, Jaillard S, Mosser J, Odent S, David V. Array-CGH analysis indicates a high prevalence of genomic rearrangements in holoprosencephaly: an updated map of candidate loci. Hum Mutat. 2009;30:1175–1182. doi: 10.1002/humu.21016. [DOI] [PubMed] [Google Scholar]
- Brown SA, Warburton D, Brown LY, Yu CY, Roeder ER, Stengel-Rutkowski S, Hennekam RC, Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- de la Cruz JM, Bamford RN, Burdine RD, Roessler E, Barkovich AJ, Donnai D, Schier AF, Muenke M. A loss-of-function mutation in the CFC domain of TDGF1 is associated with human forebrain defects. Hum Genet. 2002;110:422–428. doi: 10.1007/s00439-002-0709-3. [DOI] [PubMed] [Google Scholar]
- DeMeyer W, Zeman W, Palmer CG. The face predicts the brain: diagnostic significance of median facial anomalies for holoprosencephaly (arhinencephaly) Pediatrics. 1964;34:256–263. [PubMed] [Google Scholar]
- Domené S, Roessler E, El-Jaick K, Boorech J, Vélez JI, Bale S, Lacbawan F, Muenke M, Feldman B. Mutations in the human SIX3 gene in holoprosencephaly are loss-of-function. Hum Mol Genet. 2008;17:3919–3928. doi: 10.1093/hmg/ddn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg C, Bendavid C, Pasquier L, Henry C, Odent S, David V. Holoprosencephaly. Orphanet J Rare Dis. 2007;2:8. doi: 10.1186/1750-1172-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Jaick KB, Powers SE, Bartholin L, Myers KR, Hahn J, Orioli IM, Ouspenskaia M, Lacbawan F, Roessler E, Wotton D, Muenke M. Functional analysis of mutations in TGIF associated with holoprosencephaly. Mol Genet Metab. 2007;90:97–111. doi: 10.1016/j.ymgme.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massagué J, Muenke M, Elledge SJ. Mutations in TGIF cause holoprosencephaly and link NODAL signaling to human neural axis determination. Nat Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Barkovich AJ, Stashinko EE, Kinsman SL, Delgado MR, Clegg NJ. Factor analysis of neuroanatomical and clinical characteristics of holoprosencephaly. Brain Dev. 2006;28:413–9. doi: 10.1016/j.braindev.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Delgado MR, Clegg NJ, Sparagana SP, Gerace KL, Barkovich AJ, Olson DM. Electroencephalography in holoprosencephaly: findings in children without epilepsy. Clin Neurophysiol. 2003;114:1908–1917. doi: 10.1016/s1388-2457(03)00169-x. [DOI] [PubMed] [Google Scholar]
- Hahn JS, Hahn SM, Kammann H, Barkovich AJ, Clegg NJ, Delgado MR, Levey E. Endocrine disorders associated with holoprosencephaly. J Pediatr Endocrinol Metab. 2005;18:935–941. doi: 10.1515/jpem.2005.18.10.935. [DOI] [PubMed] [Google Scholar]
- Keaton A, Solomon BD, Pineda-Alvarez DE, El-Jaick KB, Gropman AL, Zafer Y, Meck JM, Bale SJ, Lacbawan F, Roessler E, Muenke M. Correlation between genotype and phenotype in patients with holoprosencephaly and alterations in TGIF. 2009 In Preparation. [Google Scholar]
- Krauss RS. Holoprosencephaly: new models, new insights. Expert Rev Mol Med. 2007;9:1–17. doi: 10.1017/S1462399407000440. [DOI] [PubMed] [Google Scholar]
- Lacbawan F, Solomon BD, Roessler E, El-Jaick K, Domené S, Vélez JI, Zhou N, Hadley D, Balog JZ, Long R, Fryer A, Smith W, Omar S, McLean SD, Clarkson K, Lichty A, Clegg NJ, Delgado MR, Levey E, Stashinko E, Potocki L, Vanallen MI, Clayton-Smith J, Donnai D, Bianchi DW, Juliusson PB, Njølstad PR, Brunner HG, Carey JC, Hehr U, Müsebeck J, Wieacker PF, Postra A, Hennekam RC, van den Boogaard MJ, van Haeringen A, Paulussen A, Herbergs J, Schrander-Stumpel CT, Janecke AR, Chitayat D, Hahn J, McDonald-McGinn DM, Zackai EH, Dobyns WB, Muenke M. Clinical spectrum of SIX3-associated mutations in holoprosencephaly: correlation between genotype, phenotype and function. J Med Genet. 2009;46:389–98. doi: 10.1136/jmg.2008.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro L, Dubourg C, Pasquier L, Le Duff F, Blayau M, Durou MR, de la Pintière AT, Aguilella C, David V, Odent S. Phenotypic and molecular variability of the holoprosencephalic spectrum. Am J Med Genet A. 2004;129A:21–24. doi: 10.1002/ajmg.a.30110. [DOI] [PubMed] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Stratton RF, Sujansky E, Bale SJ, Muenke M. Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet. 2002;110:297–301. doi: 10.1007/s00439-002-0695-5. [DOI] [PubMed] [Google Scholar]
- Ming JE, Muenke M. Multiple hits during early embryonic development: digenic diseases and holoprosencephaly. Am J Hum Genet. 2002;71:1017–1032. doi: 10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenke M, Beachy PA. Holoprosencephaly. In: Scriver CR, Beaudet AL, Sly WS, et al., editors. The Metabolic & Molecular Bases of Inherited Disease. 8e. New York: McGraw-Hill; 2001. pp. 6203–6230. [Google Scholar]
- Muenke M, Gurrieri F, Bay C, Yi DH, Collins AL, Johnson VP, Hennekam RC, Schaefer GB, Weik L, Lubinsky MS, Daack-Hirsch S, Moore CA, Dobyns WB, Murray JC, Price RA. Linkage of a human brain malformation, familial holoprosencephaly, to chromosome 7 and evidence for genetic heterogeneity. Proc Natl Acad Sci U S A. 1994;91:8102–8106. doi: 10.1073/pnas.91.17.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münke M, Page DC, Brown LG, Armson BA, Zackai EH, Mennuti MT, Emanuel BS. Molecular detection of a Yp/18 translocation in a 45,X holoprosencephalic male. Hum Genet. 1988;80:219–223. doi: 10.1007/BF01790089. [DOI] [PubMed] [Google Scholar]
- Plawner LL, Delgado MR, Miller VS, Levey EB, Kinsman SL, Barkovich AJ, Simon EM, Clegg NJ, Sweet VT, Stashinko EE, Hahn JS. Neuroanatomy of holoprosencephaly as predictor of function: beyond the face predicting the brain. Neurology. 2002;59:1058–1066. doi: 10.1212/wnl.59.7.1058. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2009. R: A language and environment for statistical computing. URL http://www.R-project.org. [Google Scholar]
- Rahimov F, Ribeiro LA, de Miranda E, Richieri-Costa A, Murray JC. GLI2 mutations in four Brazilian patients: how wide is the phenotypic spectrum? Am J Med Genet A. 2006;140:2571–2576. doi: 10.1002/ajmg.a.31370. [DOI] [PubMed] [Google Scholar]
- Ribeiro LA, El-Jaick KB, Muenke M, Richieri-Costa A. SIX3 mutations with holoprosencephaly. Am J Med Genet. 2006;140:2577–2583. doi: 10.1002/ajmg.a.31377. [DOI] [PubMed] [Google Scholar]
- Roesler CP, Paterson SJ, Flax J, Hahn JS, Kovar C, Stashinko EE, Jing H, Benasich AA. Links between abnormal brain structure and cognition in holoprosencephaly. Pediatr Neurol. 2006;35:387–394. doi: 10.1016/j.pediatrneurol.2006.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- Roessler E, Du YZ, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz i Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci U S A. 2003;100:13424–13429. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, El-Jaick KB, Dubourg C, Vélez JI, Solomon BD, Pineda-Álvarez DE, Lacbawan F, Zhou N, Ouspenskaia M, Paulussen A, Smeets HJ, Hehr U, Bendavid C, Bale S, Odent S, David V, Muenke M. The mutational spectrum of holoprosencephaly-associated changes within the SHH gene in humans predicts loss-of-function through either key structural alterations of the ligand or its altered synthesis. Hum Mutat. 2009;30:E921–35. doi: 10.1002/humu.21090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ermilov AN, Grange DK, Wang A, Grachtchouk M, Dlugosz AA, Muenke M. A previously unidentified amino-terminal domain regulates transcriptional activity of wild-type and disease-associated human GLI2. Hum Mol Genet. 2005;14:2181–2188. doi: 10.1093/hmg/ddi222. [DOI] [PubMed] [Google Scholar]
- Roessler E, Lacbawan F, Dubourg C, Paulussen A, Herbergs J, Hehr U, Bendavid C, Zhou N, Ouspenskaia M, Bale S, Odent S, David V, Muenke M. The full spectrum of holoprosencephaly-associated mutations within the ZIC2 gene in humans predicts loss-of-function as the predominant disease mechanism. Hum Mutat. 2009;30:E541–554. doi: 10.1002/humu.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ma Y, Ouspenskaia MV, Lacbawan F, Bendavid C, Dubourg C, Beachy PA, Muenke M. Truncating loss-of-function mutations of DISP1 contribute to holoprosencephaly-like microform features in humans. Hum Genet. 2009;125:393–400. doi: 10.1007/s00439-009-0628-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ouspenskaia MV, Karkera JD, Vélez JI, Kantipong A, Lacbawan F, Bowers P, Belmont JW, Towbin JA, Goldmuntz E, Feldman B, Muenke M. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Pei W, Ouspenskaia MV, Karkera JD, Veléz JI, Banerjee-Basu S, Gibney G, Lupo PJ, Mitchell LE, Towbin JA, Bowers P, Belmont JW, Goldmuntz E, Baxevanis AD, Feldman B, Muenke M. Cumulative ligand activity of NODAL mutations and modifiers are linked to human heart defects and holoprosencephaly. Mol Genet Metab. 2009;98:225–234. doi: 10.1016/j.ymgme.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter KA, Krauss RS. Murine models of holoprosencephaly. Curr Top Dev Biol. 2008;84:139–70. doi: 10.1016/S0070-2153(08)00603-0. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Lacbawan F, Jain M, Domené S, Roessler E, Moore C, Dobyns WB, Muenke M. A novel SIX3 mutation segregates with holoprosencephaly in a large family. Am J Med Genet A. 2009;149A:919–925. doi: 10.1002/ajmg.a.32813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon BD, Lacbawan F, Mercier S, Clegg NJ, Delgado MR, Rosenbaum K, Dubourg C, David V, Olney AH, Wehner LE, Hehr U, Bale S, Paulussen A, Smeets HJ, Hardisty E, Tylki-Szymanska A, Pronicka E, Clemens M, McPherson E, Hennekam RCM, Hahn J, Stashinko E, Levey E, Wieczorek D, Roeder E, Imaizumi K, Schell-Apacik CC, Booth CW, Thomas RL, Kenwrick S, Keaton A, Balog JZ, Hadley D, Zhou N, Long R, Vélez JI, Pineda-Alvarez DE, Odent S, Roessler E, Muenke M. Mutations in ZIC2 in human holoprosencephaly: description of a novel ZIC2-specific phenotype and comprehensive analysis of 157 individuals. doi: 10.1136/jmg.2009.073049. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stashinko EE, Clegg NJ, Kammann HA, Sweet VT, Delgado MR, Hahn JS, Levey EB. A retrospective survey of perinatal risk factors of 104 living children with holoprosencephaly. Am J Med Genet A. 2004;128A:114–119. doi: 10.1002/ajmg.a.30070. [DOI] [PubMed] [Google Scholar]
- Volpe P, Campobasso G, De Robertis V, Rembouskos G. Disorders of prosencephalic development. Prenat Diagn. 2009;29:340–354. doi: 10.1002/pd.2208. [DOI] [PubMed] [Google Scholar]
- Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai EH, Rommens J, Muenke M. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet. 1999;22:196–198. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]


