Abstract
The annulus fibrosis electrically insulates the atria and ventricles, allowing the timed sequential beating of these structures that is necessary for efficient heart function. Abnormal development of the annulus fibrosis leads to persistence of accessory electrical pathways from atria to ventricles, providing the anatomical substrate for re-entrant cardiac arrhythmias such as Wolff-Parkinson-White syndrome. To better understand the development of the annulus fibrosis and the etiology of these cardiac arrhythmias, we used Cre-LoxP technology to assess the contribution of epicardium derived cells (EPDCs) to the annulus fibrosis. We found that EPDCs migrated into the region of the forming annulus fibrosis, marked by the protein periostin. These EPDCs also stained positive for procollagen I, suggesting that the EPDCs themselves synthesize proteins of the annulus fibrosis. To further test the hypothesis that EPDCs contribute to cells that synthesize the annulus fibrosis, we purified genetically marked EPDCs from the atrioventricular region and measured gene expression by quantitative PCR. These EPDCs were highly enriched for mRNAs encoding periostin, procollagen I, fibronectin I, vimentin, DDR2, and tenacin C, markers of fibroblasts and components of the annulus fibrosis. In addition, these EPDCs were highly enriched for Snail, Smad1, Slug, and Twist1, markers for epithelial to mesenchymal transition (EMT), and a metallopeptidase, Mmp2, that contributes to cellular migration. Our work provides for the first time definitive evidence that epicardium contributes to formation of the mammalian annulus fibrosis through EMT. Abnormalities of this differentiation process may underlie development of some forms of re-entrant atrioventricular tachycardia.
Keywords: Epicardium, fibroblast, annulus fibrosis, arrhythmia, Wolff-Parkinson-White syndrome
Introduction
Sequential activation of the atria and ventricles, required for efficient heart function, is made possible by the annulus fibrosis, an electrically inert structure that separates the atria and ventricles. Defects in the annulus fibrosis, bridged by threads of cardiac muscle, permit abnormal premature activation of the ventricles and predispose to atrioventricular (AV) reciprocating tachyarrhythmias (Kolditz et al., 2008), as seen in the Wolff-Parkinson-White (WPW) syndrome (Becker et al., 1978). Improved understanding of embryological mechanisms that underlie development of the annulus fibrosis will therefore be important to understand the etiology of abnormal accessory AV connections.
The heart initially forms as a tube of two concentric layers, the endocardium and the myocardium (Buckingham et al., 2005). Cardiac morphogenesis requires that the heart tube receive contributions from other cell populations that initially reside outside the developing heart. One important extra-cardiac cell population originates from the proepicardium, an outgrowth of the septum transversum (Gittenberger-de Groot et al., 1998; Perez-Pomares et al., 2002). Cells from the proepicardium migrate to form an epithelial sheet over the surface of the heart, the epicardium. A subset of epicardial cells undergo an epithelial to mesenchymal transition (EMT) to become epicardium-derived cells (EPDCs). EPDCs migrate into the underlying myocardium, and have been described to differentiate into smooth muscle, endothelial, and fibroblast lineages (Gittenberger-de Groot et al., 1998; Merki et al., 2005; Wilm et al., 2005; Cai et al., 2008; Zhou et al., 2008a). In addition, recent studies indicated that EPDCs also differentiate into cardiomyocytes (Cai et al., 2008; Zhou et al., 2008a).
Using quail-chicken chimeras, Gittenberger-de Groot and co-workers showed that EPDCs are present in the annulus fibrosis and express periostin and collagen III (Gittenberger-de Groot et al., 1998; Lie-Venema et al., 2008). Recent pioneering work by this group showed that block of EPDC migration in avian embryos resulted in abnormal annulus fibrosis formation in which abnormal muscle bundles (accessory pathways) bridged from atria to ventricles. This provided the anatomical substrate for abnormal electrical impulse conduction analogous to that seen in WPW (Kolditz et al., 2007; Kolditz et al., 2008). These studies of avian embryos were the first to reveal the embryological origins that may underlie development of accessory pathways, thus raising interest in epicardium and EPDCs in relation to defective annulus fibrosis formation.
The role of epicardium and EPDCs in the formation of the mammalian annulus fibrosis and the process of migration and cellular transition have not been described. To investigate these questions, we used a Cre-LoxP approach (Soriano, 1999) to genetically trace the fate of EPDCs in the annulus fibrosis of the developing mouse heart. We provide definitive evidence that EPDCs differentiate into fibroblasts that synthesize components of the annulus fibrosis during mamalian heart development. We also identify altered expression of key components of the EMT gene program that likely contribute to formation and differentiation of annulus EPDCs.
Material and Methods
Mice
Wt1CreERT2/+ mice were generated by gene targeting followed by Flp-mediated removal of a Neo resistance casette, as described previously (Zhou et al., 2008a). CreERT2 is a fusion protein composed of Cre recombinase and a modified variant of the estrogen receptor hormone binding domain. CreERT2 recombines floxed targets in the presence, but not the absence of tamoxifen (Feil et al., 1997). We injected 2 mg tamoxifen (Sigma) intraperitoneally to pregnant mice at E10.5 to induce Cre activity. Rosa26fsLz (Soriano, 1999) and Rosa26mTmG (Muzumdar et al., 2007) mice were used as Cre-dependent reporters. These mice express LacZ and membrane localized GFP, respectively, after Cre-mediated recombination. Pregnancies were dated by daily inspection for a vaginal plug. Noon of the day of the plug was defined as E0.5. All mice were used according to protocols approved by the Institutional Animal Care and Use Committee at Children’s Hospital Boston.
Histology
Embryos were collected in PBS on ice and then fixed in 4% PFA at 4°C for 4 hours. After washing in PBS, tissues were treated in 15% and 30% sucrose for 2 hours each and embedded in OCT. 5–8 μm cryostat sections were collected on positively charged slides. X-gal staining was performed first by incubation with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal; 1 mg/ml) at 37°C overnight as previously described (Zhou et al., 2008b). X-gal stained sections were then washed in PBS, blocked in 5% serum/PBS, and subject to immunostaining. Antibody sources were as follows: β-galactosidase (β-gal), MP Biochemicals; Desmin, Santa Cruz; Periostin, Abcam; pro-collagen I, Millipore; GFP, Invitrogen. Antibody staining was visualized by standard indirect immunofluorescence, by the ABC method (Vector Labs), or by tyramide amplification (Perkin-Elmer). Vector SG (gray; Vector Labs) and Permanent Red (Dako) were used for chromogenic detection. Fluorescently stained slides were counter stained with DAPI and imaged with an FV1000 confocal microscope (Olympus). Images shown are representative of findings in a minimum of three independent embryos.
Tissue dissociation and cell sorting
The atrioventricular canal or apical regions of hearts embryonic hearts were isolated by microdissection and dissociated to single cells by collagenase digestion as previously described (Zhou et al., 2008a). In EPDC isolations, tissue from 2–3 embryos was pooled into one sample. Isolated cells were FACS sorted into GFP+ and GFP− populations. Sorted cells were collected into Trizol (Invitrogen) and frozen at −20 °C. On average, we obtained 5000–6000 GFP+ cells per atrioventricular region of an E14.5 or E15.5 heart.
Gene expression analysis
RNA was prepared using the standard Trizol protocol followed by the RNeasy kit with on-column DNase digestion (Qiagen) as previously described (Zhou et al., 2009). Quantitative real-time qRT-PCR was performed on an ABI 7300 Sequence Detector (Applied Biosystems) using Taqman (Gapdh) or Sybr Green (Myh6, Postn, Col1a1, Vimentin, Ddr2, Tnc, Fn1, Slug, Snail, Smad1, Twist1, Mmp2, Scn5a, Gja1, smMHC, Flk1, VE-Cad, PdgfRb) detection methods. See Table 1 for primer sequences. Five to eight samples were used per group.
Table 1.
Primers used for qRTPCR assays in this study.
| Gene | Forward | Backward |
|---|---|---|
| Myh6 | ACATGAAGGAGGAGTTTGGG | GCACTTGGAGCTGTAGGTCA |
| Postn | GACTGCTTCAGGGAGACACA | TGATCGTCTTCTAGGCCCTT |
| Col1a1 | GGAAGAGCGGAGAGTACTGG | TTGCAGTAGACCTTGATGGC |
| vimentin | GACATTGAGATCGCCACCTA | GGCAGAGAAATCCTGCTCTC |
| Ddr2 | CTGTGGGAGACCTTCACCTT | TAGATCTGCCTCCCTTGGTC |
| Tnc | CAGACTCAGCCATCACCAAC | CAGTTAACGCCCTGACTGTG |
| Slug | CACATTCGAACCCACACATT | TATTGCAGTGAGGGCAAGAG |
| Snail | CGTGTGTGGAGTTCACCTTC | GGAGAGAGTCCCAGATGAGG |
| Smad1 | CCAAGCCAGGGACAAATTAT | TGATGAAAGCCCACTTCAGA |
| Twist1 | CGGACAAGCTGAGCAAGAT | GGACCTGGTACAGGAAGTCG |
| Mmp2 | GGGTGGTGGTCATAGCTACTT | TTCCAAACTTCACGCTCTTG |
| Gja1 | AGGGAAGTACCCAACAGCAG | GAACTCCTTGGAGGCTGAAG |
| Scn5a | ACCGTCTTCAGACACTGTGG | ACCTTCCAGGTCACAGAACC |
| Gapdh | detected with a Taqman based Assay (Applied Biosystems) | |
Statistics
All results are expressed as mean ± SEM. Statistical significance was evaluated by Welch’s t-test, using JMP software, version 5.1. P-values of less than 0.05 were considered to be statistically significant.
Results
To delineate the cellular contribution of epicardial derived cells to the annulus fibrosis, we used a Cre-LoxP approach (Soriano, 1999) to genetically label epicardial cells and their descendants. We used an inducible Cre recombinase (CreERT2) (Feil et al., 1997) driven by Wilm’s tumor 1 (Wt1) regulatory elements (Zhou et al., 2008a). The CreERT2 protein, composed of Cre recombinase fused to a modified domain of the estrogen receptor (ESR1), can be detected by ESR1-specific antibody. In control experiments, we showed that hearts of wild-type embryos lack ESR1 immunoreactivity (Suppl. Fig. 1A–B and data not shown). In hearts of E10.5-E16.5 Wt1CreERT2/+ embryos, we found that CreERT2 expression was confined to the epicardium (Fig. 1A–C), consistent with our previous studies (Zhou et al., 2008a; Zhou et al., 2008b). CreERT2-expressing cells were also observed in the dorsal mesocardium and the posterior heart field surrounding the cardinal veins (Fig 1A).
Figure 1. Annulus fibrosis precursors derive from the epicardium.
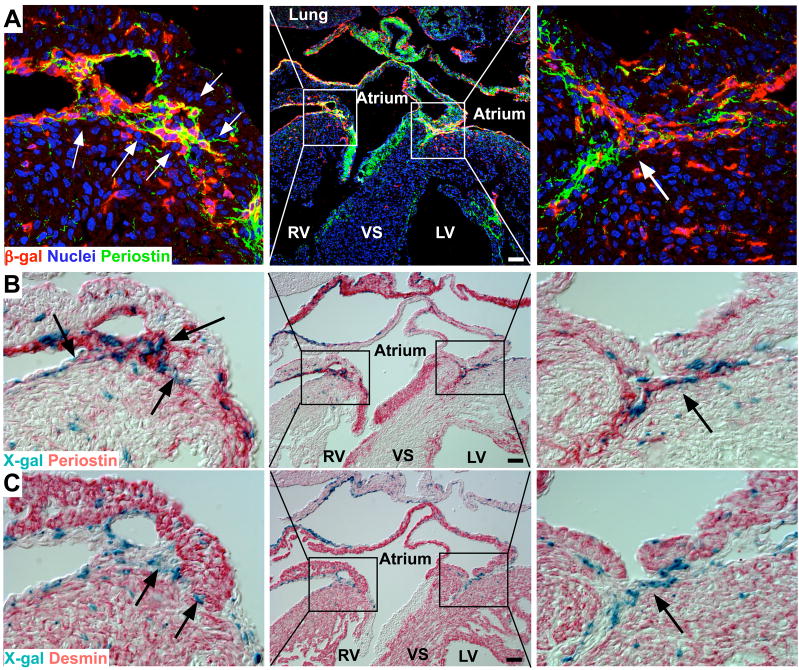
(A–C) Detection of CreERT2 expression in epicardium at E10.5, E13.5, E16.5, using an antibody to the estrogen receptor (ESR1) portion of the CreERT2 fusion protein. Boxes 1, or 2, or 3, are shown at higher magnification. Bar = 100 μm. (D) Schematic depicting the lineage tracing strategy. Tamoxifen (Tam) was injected intraperitoneally at E10.5 to induce Cre recombinase activity. (E) Immunofluorescence staining of periostin (green) and lineage trace marker β-gal (red) at E15.5. A subset of EPDCs forms the periostin+ annulus fibrosis (white arrows). 1, left AV groove; 2, right AV groove. Bar = 100 μm. White arrowheads indicate EPDCs that do not co-express periostin. (F) X-gal (blue; EPDCs) and periostin (red) staining of the consecutive sections. Arrows indicate periostin+ EPDCs in annulus fibrosis. Bar=50 μm. (G) Atrium and ventricular myocardium (desmin, red) were partially separated by EPDCs (blue) in annulus fibrosis (arrows). Bar = 50 μm.
To induce Cre activity and pulse-label Wt1+ epicardial cells, we administered a single dose of the CreERT2 activating agent, tamoxifen (Tam), to gravid females at E10.5. In the absence of Tam treatment, we did not detect activation of the Cre-dependent reporter Rosa26fsLz (Suppl. FIg. 1C). Tam treatment of Wt1CreERT2/+ embryos resulted in irrevocable and heritable Cre-mediated activation of β-galactosidase (β-gal) expression from Rosa26fsLz (Fig. 1D). Epicardium-derived cells could then be identified by expression of the β-gal lineage tracer, even without continued Cre expression. In the E15.5 atrioventricular (AV) junction, we used immunostaining to detect β-gal+ EPDCs within the myocardium (Fig. 1E). To minimize potential artifacts from immunostaining, we also used X-gal staining for enzymatic β-galactosidase activity as an independent method to identify β-gal+ EPDCs (Fig. 1F–G). By both methods, EPDCs were located throughout the myocardium. At the atrioventricular junction, EPDCs were located between atrial and ventricular myocardium (Fig. 1F–G). To evaluate the location of EPDCs relative to the developing annulus fibrosis, we performed co-staining for periostin, an extracellular matrix protein strongly expressed in the annulus fibrosis and other collagen-rich fibrous connective tissues (Kruzynska-Frejtag et al., 2001; Kern et al., 2005; Kolditz et al., 2008). At E15.5, the periostin positive developing annulus fibrosis was located at the atrioventricular junction and incompletely separated atrial and ventricular myocardium. Most of the migrating EPDCs in the AV junction were localized to periostin positive regions, indicating their presence within the annulus fibrosis (Fig. 1E–G, arrows). However, not all labelled EPDCs were periostin positive (Fig. 1E insert 2, white arrowhead). Additionally, EPDC expression of periostin was largely restricted to the annulus, suggesting that local signals regulate EPDC periostin production (Fig. 1E–G, Fig. 2). There were also periostin-rich regions that contained scant EPDCs, such as the AV valve leaflets, indicating that cell types in the AV canal region other than EPDCs also synthesize periostin.
Figure 2. Fatemap of EPDCs in annulus fibrosis using Rosa26mTmG reporter.
(A) Schematic depicting the fate map strategy. Tamoxifen (Tam) was injected at E10.5 to induce Cre recombinase activity. (B) Lineage trace of EPDCs by GFP staining and co-staining with periostin (E15.5) Magnifications of left and right AV grooves are shown in (C) and (D), respectively. Bar = 50 μm.
To provide further evidence of the migration of EPDCs into the annulus fibrosis, we used an independent Cre-activated reporter allele, Rosa26mTmG (Muzumdar et al., 2007), as the readout of Cre recombination can vary depending on the reporter allele (Vooijs et al., 2001; Ma et al., 2008). Before Cre recombination, the Rosa26mTmG allele ubiquitously expresses membrane-localized red fluorescent protein (RFP). After Cre recombination, it expresses membrane-localized GFP. We generated Wt1CreERT2/+; Rosa26mTmG/+ embryos and initiated the lineage trace by injecting tamoxifen at E10.5 (Fig. 2A). At E15.5, GFP-expressing EPDCs were found within the periostin+ annulus fibrosis (Fig. 2B–D), confirming the Rosa26fsLz lineage trace data (Fig 1). We also observed a small number of EPDCs within endocardial cushion-derived structures (Fig 2D), as described in chick models (Gittenberger-de Groot et al., 1998).
We followed the fate of EPDCs through further stages of development of the annulus fibrosis. At E16.5 and E17.5 (Figs. 3–4), periostin staining at the atrioventricular junction more completely separated atrial and ventricular myocardium. Unlike avian embryos (Lie-Venema et al., 2008), periostin was not robustly detectable in most epicardial β-gal positive cells, indicating that murine epicardial progenitors of EPDCs not express periostin (Fig. 3B, white arrowheads). Subepicardial EPDCs that had undergone EMT were likewise periostin negative (Fig. 2B, 3C). In contrast, migrating EPDCs that penetrated the myocardium at the atrioventricular junction became bounded by periostin-containing extracellular matrix (arrows, Fig. 3B and 4A), suggesting that local signals confined to this region stimulate a subset of EPDCs to express periostin. We independently confirmed the above results by X-gal staining and immunohistochemistry (Fig. 4B–C). Our previous work showed that a subset of EPDCs migrate into myocardium and differentiate into cardiomyocytes (Zhou et al., 2008a). The EPDCs in the annulus fibrosis were negative for cardiomyocytes markers (Desmin, TNNT2, Figs 3D and 4C), suggesting EPDCs at the atrioventricular junction largely did not commit to a cardiomyocyte fate.
Figure 3. Fate map of EPDCs in annulus fibrosis at E16.5.
Immunostaining of E16.5 Wt1CreERT2/+;Rosa26mTmG/+ embro heart annulus region by EPDC lineage trace marker β-gal (red). (A, B) Co-immunostaining of atrioventricular junction of two different embryos by periostin (green) showed co-expression with β-gal (arrows). A subset of EPDCs did not express periostin (white arrowheads). (C) Co-immunostaining of ventricular myocardium by perisotin. Periostin was not robustly detected in epicardium (white arrowheads). (D) Co-immunostaining of heart annulus region with cardiomyocyte marker TNNT2 (green). Epi, epicardium; Endo, endocardium. V, ventricle. Bar = 50 μm.
Figure 4. Fate map of EPDCs in annulus fibrosis at E17.5.
(A) At E17.5, EPDCs (β-gal, red) were positive for periostin (green) in annulus fibrosis (white arrows). (B–C) Consecutive sections stained with X-gal and immunostained for periostin or desmin demonstrated EPDCs contribution separation of atrium and ventricles by annulus fibrosis (black arrows). Bar= 50 μm.
Our data indicated that EPDCs are present within the annulus fibrosis. However, because periostin protein is present both intra- and extracellularly (Kolditz et al., 2008), co-immunostaining with periostin did not provide clear evidence to show that these cells directly contribute to synthesis of extracellular matrix components of the annulus. To investigate this question, we used the fibroblast marker pro-collagen I (COL1A1), a precursor of collagen I largely present within cells. Many epicardial cells expressed pro-collagen I (Fig. 5A, white arrowheads). However, most EPDCs no longer expressed this marker (Fig. 5B, blue arrowheads), suggesting that it is downregulated upon transition to mesenchymal cells. Cells at the atrioventricular junction co-expressed pro-collagen I and the β-gal lineage tracer (Fig. 5B–C, white arrows), indicating that EPDCs in this region differentiated into fibroblasts and re-expressed pro-collagen I, contributing to the collagen I content of the annulus fibrosis. As with periostin, EPDCs in the annulus fibrosis, but not within the myocardium, expressed pro-collagen I. Restriction of EPDC expression of periostin and pro-collagen I to the annulus fibrosis suggests that local signals in the atrioventricular junction region stimulate EPDC differentiation into cells that produce annulus fibrosis components.
Figure 5. EPDCs in the annulus fibrosis express the annulus fibrosis component procollagen I.
(A) E15.5 embryo heart was stained with lineage trace marker β-gal (red) and procollagen I (ProColl, green). White arrowheads indicate epicardial expression of pro-collagen I. (B–C) Magnification of left and right AF. Arrows indicate co-expression of β-gal and pro-collagen I in EPDCs at the annulus fibrosis. Blue arrowheads indicate pro-collagen I negative EPDCs within myocardium. Bar = 50 μm.
To further delineate the pattern of gene expression in EPDCs at the atrioventricular junction, we treated E10.5 Wt1CreERT2/+; Rosa26mTmG/+ embryos with Tam and collected E14.5 hearts. Green fluorescence was readily detected in Wt1CreERT2/+; Rosa26mTmG/+ hearts (Fig. 6A), facilitating pooling of embryo hearts with the proper genotype for lineage analysis. We microdissected the atrioventricular junction region of GFP+ hearts, dissociated the tissue with collagenase, and isolated GFP+ and GFP− populations by cell sorting for GFP fluorescence. GFP+ cells include EPDCs and their epicardial precursors. Based on analysis of histological sections (Fig 2C–D), the GFP+ cells were largely composed of EPDCs, while epicardial cells represented a quantitatively smaller fraction. As discussed in detail below, this was further confirmed by qRTPCR (Fig 6 vs Fig 7A–B). qRTPCR demonstrated that GFP+ cells demonstrated strongly reduced levels of the cardiomyocyte marker myosin heavy chain alpha (Myh6) and the cardiac sodium channel (Scn5a) compared compared with GFP− cells (Fig. 6B and Suppl Fig 2), suggesting that the sorting was effective in separating EPDCs from non-EPDCs. This was confirmed by similar measurements at E15.5 and E16.5 (data not shown). Interestingly, expression of the gap junction gene connexin 43 (Gja1) was upregulated in EPDCs compared to non-EPDCs (Suppl Fig 2). Gja1 expression in epicardium and proepicardium has been noted previously (Li et al., 2002).
Figure 6. EMT gene expression profile of annulus fibrosis EPDCs.
(A) Schematic figure showing the dissection of the atrioventricular junction region, dissociation of the region including annulus fibrosis EPDCs, and subsequent FACS isolation of EPDCs for qRTPCR analysis. (B) AF EPDCs were low in cardiomyocyte specific gene expression (Myh6, Tnnt2). (C–E) At E14.5, AF EPDCs were enriched for mesenchymal and fibrosis transcripts (C), EMT-associated transcripts (D), and transcripts encoding the matrix degrading enzyme Mmp2 (E). (F–G) Continued and further upregulated expression of fibrosis, mesenchymal, and EMT-associated transcripts at E15.5-E16.5. n=3. *P < 0.05. Dotted line indicate gene expression of non-EPDCs (GFP- population), assigned a value of 1.
Figure 7. Gene expression signature of Wt1 epicardial progenitors.
(A) Schematic showing dissociation and FACS isolation of GFP+ epicardial cells from Wt1GFPCre/+ E14.5-E15.5 hearts for qRTPCR analysis. GFP (green) was specifically expressed in epicardium. White arrows indicate colocalization with epicardium marker WT1 (red). Bar = 50 μm. (B) Gene expression signature of GFP+ epicardial cells. n=3–4. *P < 0.05. Dotted line indicates gene expression of GFP- non-epicardial cells, assigned a value of 1.
GFP+ EPDCs were highly enriched for mesenchymal/fibroblast markers vimentin (vim) and discoidin domain containing receptor 2 (Ddr2) and tenascin C (Tnc), fibronectin 1 (Fn1), as well as for transcripts encoding the annulus fibrosis components periostin (Postn), and pro-collagen I (Col1a1) (Fig. 6C). Because the qRTPCR assay detects mRNA transcripts rather than potentially secreted protein products, these data providing direct evidence that EPDCs express extracellular matrix components of the annulus fibrosis. Smad1, Slug, Snail, and Twist1, transcription factors that regulate EMT in other contexts (Mani et al., 2008), were also significantly upregulated in GFP+ EPDCs (Fig. 6D), consistent with active formation of EPDCs from atrioventricular junction epicardium via EMT and implicating these factors in the regulation of EPDC EMT. Matrix metalloprotease 2 (Mmp2), which degrades extracellular matrix and promotes cellular migration, were also upregulated in EPDCs (Fig. 6E), suggesting that migrating EPDCs secrete proteins to facilitate their own migration. The fibrosis markers (Postn, Col1a1, Fn1), mesenchymal markers (Vim, Ddr2, Tnc), EMT transcripts (Smad1, Snail, Slug, Twist1) and matrix metalloproteases (Mmp2) were further upregulated in EPDCs at later stages (E15.5-E16.5, Fig. 6F–H), corroborating these results at E14.5 and suggesting sustained expression of an EPDC gene program in the atrioventricular junction during this time window.
In Wt1CreERT2/+; Rosa26mTmG/+ hearts, both EPDCs and their epicardial precursors are GFP+. To investigate the contribution of epicardial progenitors to the measured gene expression profile, we isolated GFP+ epicardial cells from E14.5-E15.5 Wt1GFPCre/+ hearts by collagenase digestion. In Wt1GFPCre/+ hearts, GFP expression was restricted to the epicardium and is co-expressed with the epicardial marker WT1 (Fig. 7A) (Zhou et al., 2008a). Gene expression of dissociated and FACS-purifed GFP+ epicardial cells was analysed by qRTPCR. Compared to GFP+ Wt1CreERT2/+; Rosa26mTmG/+ cells (Fig 6), GFP+ epicardial cells from Wt1GFPCre/+ hearts showed much little to no upregulation of Postn, Fn1, Tnc, and Vim (Fig 7B). Col1a1 was highly upregulated in both populations. The patterns of Col1a1 and Postn expression were consistent with immunohistochemistry (Fig. 1E, Fig. 5A). Among transcriptional regulators of EMT, Snail and Twist1 were not upregulated in epicardial cells (Fig. 7B). Smad1 and Slug was upregulated in epicardial cells (Fig 7B), but to a substantially lower degree compared with AF EPDCs (Fig. 6C and 6F). Mmp2 was also slightly upregulated in epicardial cells, but again the degree of enrichment was less than observed in migrating EPDCs. The divergent of Postn, Fn1, Tnc, Vim, Snail, Twist1, Slug, Smad1, and Mmp2 between EPDCs (GFP+ population from Wt1CreERT2/+; Rosa26mTmG/+ hearts) and epicardial cells (GFP+ population isolated from Wt1GFPCre/+ hearts) demonstrate that our cell sorting strategy enriched for distinct cell populations (EPDCs vs epicardial cells), and highlight differences in gene expression between these populations.
Our immunohistochemistry studies suggested that the fate of EPDCs in the atrioventricular junction region is distinct from the fate of EPDCs in ventricular myocardium. To further investigate this point, we isolated EPDCs from the atrioventricular junction (AF EPDCs) and from the heart apex (Apex EPDCs) by FACS at E15.5 (Wt1CreERT2/+; Rosa26mTmG/+; Tam E10.5; Fig 8). Comparison between AF and apex EPDCs showed that these cell populations express distinct gene programs (Fig. 8). AF EPDCs exhibited higher expression of mesenchymal genes Tnc, Ddr2, and Fn1, upregulation of fibrosis genes Postn and Col1a1, and upregulation of the matrix degrading enzyme Mmp2. EMT-related transcription factors were differentially expressed between AF and apex EPDCs, with AF EPDCs expressing increased Slug and Smad1 (Fig. 7D). The upregulation of Smad1 and Slug transcripts might play a major role in AF EMT. The above findings indicate that AF EPDCs are distinct from apex EPDCs, and suggest that local signals at the atrioventricular junction influence EPDC differentiation into components of the annulus fibrosis.
Figure 8. Gene expression signature of AF versus apical EPDCs.
(A) Schematic showing the dissociation and FACS isolation of AF and apex EDPCs from Wt1CreERT2/+ Rosa26mTmG/+E15.5 hearts for qRTPCR analysis. (B) Many of the fibrosis and EMT-associated trascripts were significantly upregulated in AF vs apex EPDCs. n=3–4. *P < 0.05. Dotted line indicates gene expression of apical EPDCs, assigned a value of 1.
Discussion
The regulated differentiation of EPDCs is essential to form the annulus fibrosis, a ring of fibrous tissue essential for sequential atrial and ventricular contraction. Abnormal development of the annulus fibrosis has been linked to development of abnormal accessory atrioventricular connections, creating the substrate for reciprocating tachycardias such as those seen in the WPW syndrome (Kolditz et al., 2008). In this study, we used genetic lineage tracing to definitively show for the first time that EPDCs contribute to the mammalian annulus fibrosis. Our study indicates that EPDCs migrate into this region through a process of EMT, becoming located within the periostin+ annulus fibrosis. These EPDCs differentiate into fibroblasts and actively contribute to the synthesis of this structure, thereby electrically separating the atria and ventricles. Furthermore, our studies of AF EPDC gene expression demonstrate for the first time that EPDCs in the atrioventricular junction undergo a distinct differentiation program from myocardial EPDCs, suggesting that local signals at the atrioventricular junction direct AF EPDC differentiation.
The molecular regulators that govern EMT of epicardial cells for making AF EPDCs are poorly understood. We found that AF EPDCs are highly enriched in transcripts for EMT regulators Snail, Slug, Smad1 and Twist1, compared with non-EPDCs. Upregulation of these genes is also observed in other contexts associated with EMT (Mani et al., 2008). Interestingly, epicardial cells also exhibited upregulation of Smad1 and Slug, but not Snail or Twist1. This suggests that upregulation of Snail or Twist1 may trigger the EMT process to form EPDCs from epicardial cells.
Epicardial cells undergo EMT throughout the myocardium to form EPDCs. However, our data indicate that there are regional differences in EPDC fate, perhaps as a result of local cues in the atrioventricular junction region. EPDCs were previously reported to differentiate into smooth muscle, endothelial, interstitial cell, and cardiomyocyte fates (Perez-Pomares et al., 2002; Wilm et al., 2005; Cai et al., 2008; Zhou et al., 2008a). By immunohistochemistry, we did not find contribution of EPDCs to the cardiomyocyte lineage in the atrioventricular junction region, and EPDC differentiation to endothelial cells was also low in this region (Fig 3D, 4C, and data not shown). Differences between AF EPDC and apex EPDC fate was further supported by qRTPCR, which revealed that fibrosis-related extracellular matrix genes were strongly upregulated in AF EPDCs. Most striking was the upregulation of periostin in AF EPDCs. This result was corroborated by immunohistochemistry, which likewise demonstrated strong periostin expression restricted to the atrioventricular junction region. Interestingly, the EMT regulators Slug and Smad1 were expressed at higher levels in AF compared to apical EPDCs. This observation suggests that distinct molecular pathways regulate the EMT process in epicardium at the atrioventricular junction versus the myocardial apex and leads us to hypothesize that Slug and Smad1 play important roles in the differentiation of this cell population.
One regional cue that might influence local EPDC fate is periostin, secreted at high levels in endocardial cushions and by EPDCs in the AF region (Lie-Venema et al., 2008; Norris et al., 2008; Snider et al., 2008). Periostin appears to suppress myogenic fates of epicardial cells and endocardial cushion mesenchyme, as expression of an Myh6-GFP transgene was ectopically activated in these locations in periostin null hearts (Norris et al., 2008; Snider et al., 2008). Thus periostin expressed by EPDCs and neighboring endocardial cushions may bias EPDC differentiation away from a myocyte fate and towards a fibroblast fate.
Migrating AF EPDCs may regulate formation of the annulus fibrosis by two mechanisms. First, EPDCs may differentiate into cells that directly contribute to synthesis of the annulus fibrosis. Our immunohistochemistry and gene expression studies definitively demonstrate that EPDCs contribute to the synthesis of the mammalian annulus fibrosis, including periostin and collagen I. Second, EPDCs may have an inductive role, stimulating other cells to differentiate into fibroblasts that contribute to synthesis of the annulus fibrosis. Consistent with these mechanisms, disruption of EPDC formation resulted in defective formation of the annulus fibrosis, producing accessory pathways that are the anatomical substrate for WPW (Kolditz et al., 2007; Kolditz et al., 2008). The epicardium is a rich source of secreted factors, including Wnts (Zamora et al., 2007), erythropoietin (Wu et al., 1999), fibroblast growth factors (Lavine et al., 2005), and retinoic acid (Merki et al., 2005). In future work it will be important to determine if these or other secreted factors play a selective role in directing formation of the annulus fibrosis.
Our study was based on the pulsed labeling of epicardially derived cells by an inducible Cre allele. While the pulse labeling strategy enhances specificity of labeling, it has some limitations. Most importantly, labeling of cells is incomplete, so that the extent of EPDC contribution to the annulus fibrosis is difficult to quantitate using the approach. Thus, partial labeling of periostin-expressing cells of the annulus fibrosis might be due to inefficient Cre labeling, which would lead to an underestimate of the contribution of EPDCs to the annulus fibrosis. Alternatively, unlabeled periostin-expressing cells of the annulus fibrosis might arise from a different source. Additional studies using additional Cre lines or alternative approaches will be required to distinguish these possibilities. However, this quantitative limitation does not undermine our conclusions on the fate of cells that are labeled by Cre. A second limitation is the inference that the pulse-labeled cells arise from the epicardium. Based on our studies of Wt1 and CreERT2 expression in Wt1CreERT2/+ hearts, we conclude that labeled cells within the myocardium arise by EMT of epicardial cells. This conclusion is well supported by independent methods in avian embryos (Kolditz et al., 2008; Lie-Venema et al., 2008), by dye labeling experiments in murine explant culture (Cai et al., 2008; Zhou et al., 2008a), and by other Cre transgenes expressed in the epicardium (Merki et al., 2005; Cai et al., 2008). However, we cannot formally exclude an alternative origin for the labeled cells.
In conclusion, our work advanced our understanding of annulus fibrosis development by definitively demonstrating the cellular contribution of EPDCs. Furthermore, we found that differentiation of EPDCs in the atrioventricular junction region is distinct from differentiation of EPDCs from the apical myocardium. These data suggest that local cues regulate EPDC differentiation.
Supplementary Material
Acknowledgments
This work was funded by grants from NIH (1R01HL094683, WTP), the Harvard Stem Cell Institute (WTP), the American Heart Association (BZ), the Simeon Burt Wolbach Research Fund (BZ), and a charitable donation by Edward Marram and Karen Carpenter. The authors would like to thank A.C. Gittenberger-de Groot and R.W.C. Scherptong for enlightening discussions and their critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- BECKER AE, ANDERSON RH, DURRER D, WELLENS HJ. The anatomical substrates of wolff-parkinson-white syndrome. A clinicopathologic correlation in seven patients. Circulation. 1978;57:870–879. doi: 10.1161/01.cir.57.5.870. [DOI] [PubMed] [Google Scholar]
- BUCKINGHAM M, MEILHAC S, ZAFFRAN S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- CAI CL, MARTIN JC, SUN Y, CUI L, WANG L, OUYANG K, YANG L, BU L, LIANG X, ZHANG X, STALLCUP WB, DENTON CP, MCCULLOCH A, CHEN J, EVANS SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FEIL R, WAGNER J, METZGER D, CHAMBON P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- GITTENBERGER-DE GROOT AC, VRANCKEN PEETERS MP, MENTINK MM, GOURDIE RG, POELMANN RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- KERN CB, HOFFMAN S, MORENO R, DAMON BJ, NORRIS RA, KRUG EL, MARKWALD RR, MJAATVEDT CH. Immunolocalization of chick periostin protein in the developing heart. Anat Rec A Discov Mol Cell Evol Biol. 2005;284:415–423. doi: 10.1002/ar.a.20193. [DOI] [PubMed] [Google Scholar]
- KOLDITZ DP, WIJFFELS MC, BLOM NA, VAN DER LAARSE A, HAHURIJ ND, LIE-VENEMA H, MARKWALD RR, POELMANN RE, SCHALIJ MJ, GITTENBERGER-DE GROOT AC. Epicardium-derived cells in development of annulus fibrosis and persistence of accessory pathways. Circulation. 2008;117:1508–1517. doi: 10.1161/CIRCULATIONAHA.107.726315. [DOI] [PubMed] [Google Scholar]
- KOLDITZ DP, WIJFFELS MC, BLOM NA, VAN DER LAARSE A, MARKWALD RR, SCHALIJ MJ, GITTENBERGER-DE GROOT AC. Persistence of functional atrioventricular accessory pathways in postseptated embryonic avian hearts: implications for morphogenesis and functional maturation of the cardiac conduction system. Circulation. 2007;115:17–26. doi: 10.1161/CIRCULATIONAHA.106.658807. [DOI] [PubMed] [Google Scholar]
- KRUZYNSKA-FREJTAG A, MACHNICKI M, ROGERS R, MARKWALD RR, CONWAY SJ. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech Dev. 2001;103:183–188. doi: 10.1016/s0925-4773(01)00356-2. [DOI] [PubMed] [Google Scholar]
- LAVINE KJ, YU K, WHITE AC, ZHANG X, SMITH C, PARTANEN J, ORNITZ DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- LI WE, WALDO K, LINASK KL, CHEN T, WESSELS A, PARMACEK MS, KIRBY ML, LO CW. An essential role for connexin43 gap junctions in mouse coronary artery development. Development. 2002;129:2031–2042. doi: 10.1242/dev.129.8.2031. [DOI] [PubMed] [Google Scholar]
- LIE-VENEMA H, ERALP I, MARKWALD RR, VAN DEN AKKER NM, WIJFFELS MC, KOLDITZ DP, VAN DER LAARSE A, SCHALIJ MJ, POELMANN RE, BOGERS AJ, GITTENBERGER-DE GROOT AC. Periostin expression by epicardium-derived cells is involved in the development of the atrioventricular valves and fibrous heart skeleton. Differentiation. 2008;76:809–819. doi: 10.1111/j.1432-0436.2007.00262.x. [DOI] [PubMed] [Google Scholar]
- MA Q, ZHOU B, PU WT. Reassessment of Isl1 and Nkx2-5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev Biol. 2008;323:98–104. doi: 10.1016/j.ydbio.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANI SA, GUO W, LIAO MJ, EATON EN, AYYANAN A, ZHOU AY, BROOKS M, REINHARD F, ZHANG CC, SHIPITSIN M, CAMPBELL LL, POLYAK K, BRISKEN C, YANG J, WEINBERG RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MERKI E, ZAMORA M, RAYA A, KAWAKAMI Y, WANG J, ZHANG X, BURCH J, KUBALAK SW, KALIMAN P, BELMONTE JC, CHIEN KR, RUIZ-LOZANO P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUZUMDAR MD, TASIC B, MIYAMICHI K, LI L, LUO L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- NORRIS RA, MORENO-RODRIGUEZ RA, SUGI Y, HOFFMAN S, AMOS J, HART MM, POTTS JD, GOODWIN RL, MARKWALD RR. Periostin regulates atrioventricular valve maturation. Dev Biol. 2008;316:200–213. doi: 10.1016/j.ydbio.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREZ-POMARES JM, PHELPS A, SEDMEROVA M, CARMONA R, GONZALEZ-IRIARTE M, MUNOZ-CHAPULI R, WESSELS A. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs) Dev Biol. 2002;247:307–326. doi: 10.1006/dbio.2002.0706. [DOI] [PubMed] [Google Scholar]
- SNIDER P, HINTON RB, MORENO-RODRIGUEZ RA, WANG J, ROGERS R, LINDSLEY A, LI F, INGRAM DA, MENICK D, FIELD L, FIRULLI AB, MOLKENTIN JD, MARKWALD R, CONWAY SJ. Periostin is required for maturation and extracellular matrix stabilization of noncardiomyocyte lineages of the heart. Circ Res. 2008;102:752–760. doi: 10.1161/CIRCRESAHA.107.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SORIANO P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- VOOIJS M, JONKERS J, BERNS A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILM B, IPENBERG A, HASTIE ND, BURCH JB, BADER DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- WU H, LEE SH, GAO J, LIU X, IRUELA-ARISPE ML. Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development. 1999;126:3597–3605. doi: 10.1242/dev.126.16.3597. [DOI] [PubMed] [Google Scholar]
- ZAMORA M, MANNER J, RUIZ-LOZANO P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci U S A. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU B, MA Q, KONG SW, HU Y, CAMPBELL PH, MCGOWAN FX, ACKERMAN KG, WU B, ZHOU B, TEVOSIAN SG, PU WT. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J Clin Invest. 2009;119:1462–1476. doi: 10.1172/JCI38723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU B, MA Q, RAJAGOPAL S, WU SM, DOMIAN I, RIVERA-FELICIANO J, JIANG D, VON GISE A, IKEDA S, CHIEN KR, PU WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008a;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU B, VON GISE A, MA Q, RIVERA-FELICIANO J, PU WT. Nkx2-5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem Biophys Res Commun. 2008b;375:450–453. doi: 10.1016/j.bbrc.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.