Abstract
Testicular cell adhesion molecule 1 (Tcam1) is a testis-expressed gene that is evolutionarily conserved in most mammalian species. The putative location of TCAM1 on the cell surface makes it an attractive contraceptive target to study. We found that Tcam1 transcription is enriched in the adult testis, and in situ hybridization revealed that Tcam1 is expressed in pachytene to secondary spermatocytes. Immunofluorescence for TCAM1 protein showed strong expression along cell membranes of spermatocytes and weak localization to round spermatids. In light of this evidence, we hypothesized that TCAM1 interacts with an unknown receptor on the surface of Sertoli cells and that this interaction is important for germ cell-Sertoli cell interactions. However, Tcam1 knockout mice that we generated are fertile, and testis weights and sperm counts were not significantly altered. Therefore, we conclude that TCAM1 is not essential for male fertility or germ cell function in Mus musculus.
Keywords: Spermatocyte, Male Fertility, Integral Membrane Protein, Transgenic mice
Introduction
It has been estimated that there are over 2,000 testis-expressed genes that may be essential to male fertility (Schultz et al., 2003). The discovery of the function and relevance of these testis-enriched genes has been a focus of our laboratory for a number of years (Yan et al., 2002; Yan et al., 2004; Greenbaum et al., 2006; Lin et al., 2007; Roy et al., 2007). To study the etiology of genetic causes of infertility and to identify potential contraceptive targets, we have used both in silico approaches and traditional gene expression analyses to identify evolutionarily conserved genes with gonadal-specific expression and interesting functional domains (Lin and Matzuk, 2005). One of the testis-specific genes that we identified in this manner is testicular cell adhesion molecule 1 (Tcam1).
In the literature, Tcam1 has been described in detail in a single report (Sakatani et al., 2000). Sakatani et al. described mouse TCAM1 as a protein composed of a signal peptide sequence, five immunoglobulin (Ig) domains, and a transmembrane plus cytoplasmic domain (Sakatani et al., 2000). Northern blot analysis showed that Tcam1 mRNA was present in the testis at postnatal day 17 (P17) and in adults but not at P12 leading the authors to conclude that Tcam1 was predominantly expressed in pachytene spermatocytes and round spermatids (Sakatani et al., 2000); however, this was never confirmed by in situ hybridization. The authors also showed the close homology of the Ig domains in TCAM1 to those in ICAM1 and ICAM2 (Sakatani et al., 2000).
ICAM1 and ICAM2 are intercellular adhesion molecules that are important mediators of transendothelial migration of leukocytes in response to inflammatory stimuli (van Buul et al., 2007). Overexpression of ICAM1 in vitro promotes transmigration of neutrophils suggesting that the expression of this receptor is sufficient to drive leukocyte transendothelial migration (Sans et al., 2001), and conversely, blocking ICAM1 decreases transendothelial migration in a variety of models (van Buul et al., 2007). The binding of leukocyte-expressed integrins to endothelial-expressed ICAMs induces a variety of intracellular signaling events important for transendothelial migration (van Buul et al., 2007).
In the testis, junctional components of germ cells and Sertoli cells form dynamic interfaces that are constantly remodeled, replacing one component for another as the germ cells mature (Mruk and Cheng, 2004). Some important protein-protein interactions that form these junctions, such as nectin-afadin and integrins-ADAMs, are similar to protein-protein interactions that are important for contact between immune cells and their targets cells (Seals and Courtneidge, 2003; Mruk and Cheng, 2004; Fuchs and Colonna, 2006). Since ICAMs represent a similar class of molecules that are important for immune cell binding to target cells, we hypothesized that TCAM1 acts in a homologous way to ICAMs in transendothelial migration to aid germ cell migration and maturation within the seminiferous tubule. In this report, we sought to confirm and expand the previous literature on Tcam1 using additional techniques including in situ hybridization, immunofluorescence, and knockout mouse technology.
Materials and Methods
Alignments
Predicted amino acid sequences of TCAM1 for cow (NP 001033300.1), rat (NP 067705.1), and mouse (NP 083743.2) based on mRNA sequences were obtained from Entrez Gene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene). Predicted amino acid sequences of TCAM1 for dog (ENSCAFG00000012690) and rhesus monkey (ENSMMUG00000009283) were based on hypothetical mRNA sequences obtained from Ensembl (http://www.ensembl.org). Protein sequences were aligned using the multiple alignment algorithm, CLUSTAL V (MegAlign, DNAStar; (Higgins and Sharp, 1989)).
RNA Isolation and Semi-Quantitative RT-PCR
RNA was extracted using TRIzol reagent according to the manufacturer's protocol (Invitrogen) and was converted to cDNA using SuperScriptIII primed by random hexamers (Invitrogen). Tcam1 expression was examined at the exon 7-8 junction (Fwd, 5′-CTCCGTCAGCAAAGACATCA-3′; Rev, 5′-CATGCCAGGCTATTTCTGGT-3′) for multi-tissue semi-quantitative RT-PCR and the exon 2-3 junction (Fwd, 5′-AATGCTTCTGTTGGGTGTCTG-3′; Rev, 5′-GAGGGTAAGGGTGAGGCTCT-3′) to ensure efficacy of targeting. Hprt1 (Fwd, 5′-CCTGGTTAAGCAGTACAGCC-3′; Rev, 5′-TACTAGGCAGATGGCCACAG-3′) and Gapdh (Fwd, 5′-AACTTTGGCATTGTGGAAGG-3′; Rev, 5′-ACACATTGGGGGTAGGAACA-3′) were utilized as endogenous loading controls. All product sizes and sequences were validated and reflected the expected results.
Histology, In Situ Hybridization, and Immunofluorescence
Tissues were fixed in Bouin's fixative or 4% paraformaldehyde prior to paraffin embedding. Tissue embedding, sectioning, and staining for periodic acid Schiff and hematoxylin were performed by the Histology Core of the Department of Pathology of Baylor College of Medicine. In situ hybridization was performed on paraformaldehyde-fixed tissues as described in (Albrecht et al., 1997) with a 619bp probe encompassing parts of exon 7 and 8 (primers described above). Immunofluorescence was performed on Bouin's-fixed tissues as described in (Greenbaum et al., 2006). Samples were blocked (3% bovine serum albumin/10% serum in PBS) and then incubated with affinity-purified primary antibody against TCAM1 (Cocalicos, 1:100) followed by fluorescent secondary antibody for one hour at room temperature (Molecular Probes, 1:500). Fluorescent samples were mounted with VECTASHIELD containing DAPI (Vector).
Animal Care and Sample Collection
Mice were housed with unlimited access to food and water and exposure to 12 h:12 h light:dark cycles in accordance with the standards of the Association for Assessment and Accreditation of Laboratory Animal Care and the Baylor College of Medicine Institutional Animal Care and Use Committee. After anesthetization, the mice were euthanized, and the desired tissues were harvested. In general, one testis was fixed for histological analysis and the other was frozen for DNA (-20°C) or RNA (-80°C) analysis.
Generation of Tcam1 Knockout Mice and Genotyping
An 11.4kb portion of BAC clone bMQ-410j22 (Wellcome Trust Sanger Institute, (Adams et al., 2005)) was retrieved into pBluescript SK containing diptheria toxin for negative selection (pDTA.3 kindly provided by Dr. Pumin Zhang). A 332bp portion of the plasmid containing exon 2 of Tcam1 was replaced by loxP-Pgk-Neo-loxP (PL452, NCI Preclinical Repository) using a recombineering strategy (http://recombineering.ncifcrf.gov/, (Liu et al., 2003)). The targeting vector was linearized and electroporated into embryonic stem cells (AK7 kindly provided by Dr. Philip Soriano) derived from 129S4/SvJae mice (Friedrich and Soriano, 1993). Clones were screened by Southern blot analysis for proper recombination on both 5′ and 3′ sides, and selected clones were expanded and injected into recipient C57BL/6J (B6) blastocysts. Chimeric males were bred to females of both the B6 and 129S5/SvEvBrd (129S5) stains to obtain mice heterozygous for the Tcam1 targeted mutant allele (Tcam1tm1Zuk) on two genetic backgrounds. Tail DNA was utilized for PCR genotyping which was performed according to the manufacturer's protocol (New England Biolabs, Ipswich, MA) in a multiplex reaction to obtain 450bp (null) and 790bp (wild-type) products. Primers (Fwd, 5′-AGGGACAGTAAAGGAGGGTGA-3′; wtRev, 5′-TGGGAGCCCTCCTCTACACA-3′; nullRev, 5′-TGGCTGGACGTAAACTCCTC-3′) were designed using Primer3 online software version 0.4.0 (http://primer3.sourceforge.net/, (Rozen and Skaletsky, 2000)).
Fertility Analysis
Mutant mice and control littermates were mated to age-matched wild-type B6;129S5 females at 6 weeks of age. Each mating pair was monitored to obtain the number of litters and pups born per litter over a 6 month period. These values were used to obtain the average values for litters/month and pups/litter for each genotype.
Sperm Counts and Motility Analysis
Total epididymal sperm counts were obtained as in (Roy et al., 2007) by mincing caput and cauda epididymides in prewarmed M16 medium (Sigma-Aldrich, St. Louis, MO) and allowing sperm to swim out during a 30 minute incubation at 37°C under 5% CO2 in air. Sperm were diluted in water to cause immobility prior to counting with a hemocytometer. Sperm motility was assessed by diluting only the caudal spermatozoa in M16 medium to achieve a concentration of roughly 106/ml, transferring to a MicroCell fixed-depth chamber (Conception Technologies, San Diego, CA) on a 37°C stage, and scoring 100 sperm manually as either motile or immotile. Both sperm counts and sperm motility procedures were performed twice for each sample and then averaged.
Statistical Analysis
Statistical analysis utilized JMP 7.0.1 software (SAS institute). Statistical significance was determined by one-tailed T test assuming unequal variance for two sample comparison and by one-way analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) test for multiple sample comparisons. Comparison of linear regression analyses was determined by standard least squares means analysis. Groups were considered not significantly different from one another if p>0.05.
Results
TCAM1 exhibits evolutionary conservation and is predicted to be an integral membrane protein
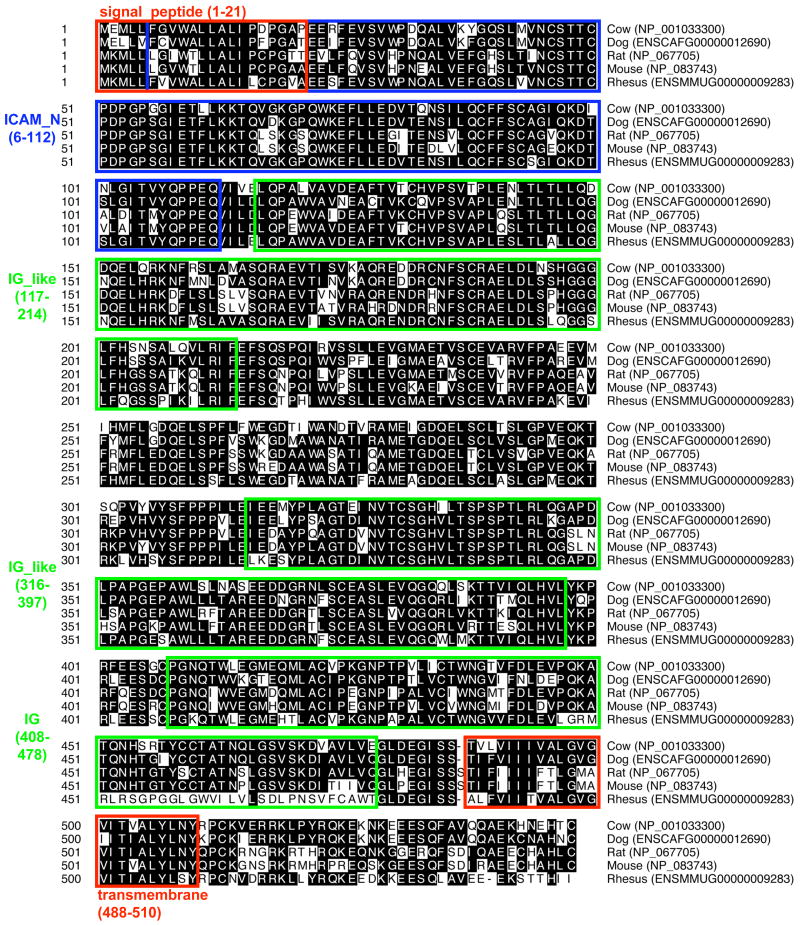
Comparative genomic analysis revealed that a duplication of Icam2 occurred during the evolution of Eutheria, or placental mammals, leading to the establishment of the Tcam1 gene, which is located approximately 90kb away from Icam2 on mouse chromosome 11 (Kent et al., 2002; Hulsen et al., 2006). There is a high degree of conservation in the Tcam1 gene among placental mammals ranging from cow to rhesus monkey (Fig. 1). Computational prediction programs also show conservation of the predicted motifs of protein interaction domains (i.e., ICAM_N) and integral membrane domains such as the signal peptide and transmembrane domains (Fig. 1). Although not included in this alignment, the gene prediction program, N-SCAN (Gross and Brent, 2006), identified a putative human TCAM1 protein that was increased in length by roughly 80 amino acids but contained all the same motifs as its mammalian orthologs and exhibited 52.7% homology to the mouse ortholog.
Fig. 1.
Alignment and protein structure of TCAM1. Amino acids matching the consensus sequence are highlighted in black. Protein domain structure is annotated based on the mouse TCAM1 sequence (NP 083743.2) using SMART (http://smart.embl.de, (Schultz et al., 1998)).
Tcam1 is highly testis-expressed
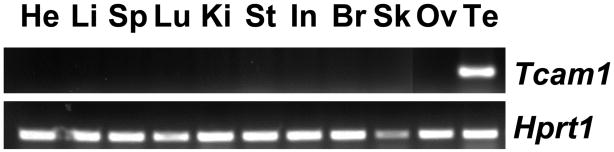
Initially, we identified murine Tcam1 using publicly available online methods to enrich for “testis-specific” genes via in silico subtraction of cDNA tissue libraries (cDNA Digital Gene Expression Displayer, http://cgap.nci.nih.gov/Tissues/GXS). When murine Tcam1 was used for BLAST search in the NCBI EST database, identified ESTs from bulls, dogs, boars, and squirrels were also enriched in testis (data not shown). To ensure that our gene of interest was truly testis-specific, we interrogated the online multi-tissue database GNF symatlas (http://symatlas.gnf.org, (Su et al., 2002)) for our initial screening and found that the Tcam1 probe (gnf1m23688_at in Mouse GeneAtlas GNF1M, gcRMA) was abundantly expressed in the testis, showing greater than 100-fold expression above the median of expression in all tissues. To further confirm testis-specificity, we isolated RNA from a variety of mouse tissues and performed semi-quantitative RT-PCR (Fig. 2). Unlike the housekeeping gene Hprt1, which was found in all tissues screened, Tcam1 was only detected in the testis (Fig. 2). These data, together with the previous in silico analysis, encouraged us to more carefully study TCAM1 expression and function in vivo.
Fig. 2.
Mouse Tcam1 mRNA is testis-enriched. Multi-tissue semi-quantitative RT-PCR analysis of mouse Tcam1 amplifies only with testis cDNA (24 cycles). Hprt1 serves as a loading control. He: heart, Li: liver, Sp: spleen, Lu: lung, Ki: kidney, St: stomach, In: intestine, Br: brain, Sk: skeletal muscle, Ov: ovary, Te: testis.
Tcam1 exhibits germ-cell specific expression patterns
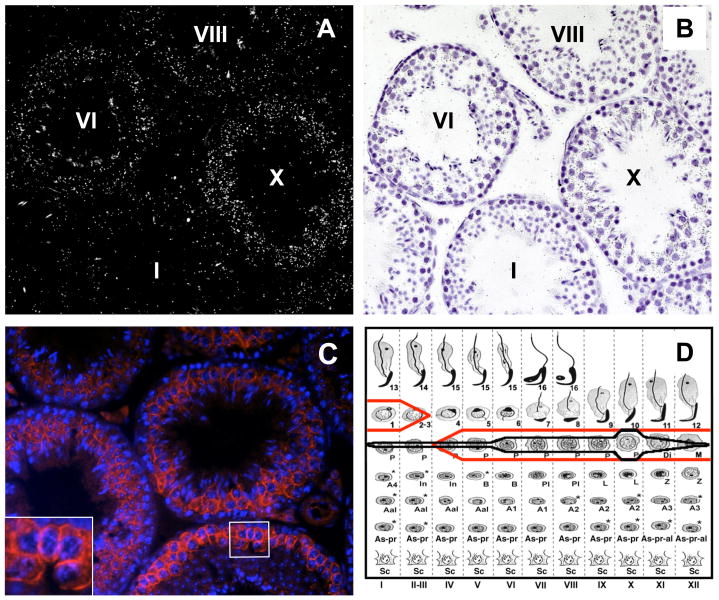
Although we suspected that Tcam1 was germ cell-specific based on prior studies (Sakatani et al., 2000), we wanted to confirm and expand these data using in situ hybridization. By performing this analysis, we found that Tcam1 mRNA is present in all stages of pachytene spermatocytes but peaks during the late stages with maximal expression in stage X tubules (Fig. 3A, B). Expression of Tcam1 wanes in diplotene and secondary spermatocytes and is undetectable in early round spermatids (Fig. 3D, black line). Tcam1 did not appear to be expressed in any of the somatic cells of the testis.
Fig. 3.
Expression of Tcam1 mRNA and protein in the testis. In situ hybridization of Tcam1 as shown by darkfield (A) and lightfield (B) microscopy reveals that Tcam1 is expressed in male germ cells, mostly in pachytene spermatocytes. Expression of Tcam1 mRNA has a maximal point in the late pachytene spermatocytes of stage X tubules (D, black line). Immunofluorescence analysis of TCAM1 shows that it localizes along the germ cell membrane (C, inset) of mid-pachytene spermatocytes to early round spermatids (D, red line). Non-specific signals are a result of autofluorescence of spermatozoa and interstitial cells (data not shown).
TCAM1 is highly expressed on the membrane of pachytene spermatocytes
To determine if TCAM1 protein expression followed the mRNA patterns, we developed an antibody against amino acids 206-395 of the mouse TCAM1 protein (Fig. 1). We chose this antigen based on its lack of homology to other ICAM family members and due to its purported high antigenicity (Protean, DNASTAR). Using immunofluorescence analysis, we were able to visualize the presence of TCAM1 to the vicinity of the membrane of late pachytene spermatocytes and, to a weaker extent, early round spermatids (Fig. 3C). These results correlated with our in situ hybridization data (Fig. 3D, red line) and suggested that TCAM1 was indeed an integral membrane protein as predicted by amino acid sequence analysis. Although Leydig cell autofluorescence is obscuring the possibility that this cell type is expressing TCAM1, we did not observe mRNA expression in Leydig cells by in situ hybridization, so it is unlikely that these cells are expressing TCAM1.
Generation of Tcam1 knockout mice
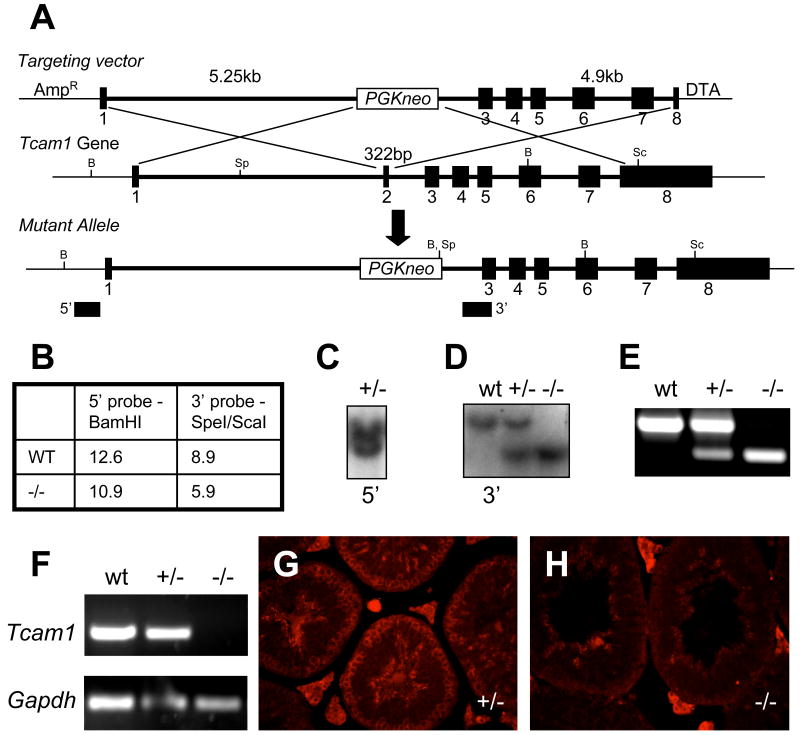
To define the essential functions of TCAM1 in vivo, we generated a targeted deletion of the second exon of Tcam1 (Fig. 4A). This gene targeting event was designed to remove the initiation ATG codon and the regions encoding the signal peptide, which would effectively create a null protein since the signal peptide is essential for proper membrane localization of integral membrane proteins (Matzuk et al., 1995; Lodish, 2008). We electroporated our targeting construct into AK7 embryonic stem cells derived from 129S4/SvJae mice (Friedrich and Soriano, 1993). Proper targeting of clones was ensured by Southern blot analysis using 5′ external and 3′ internal probes (Fig. 4B-D). These Tcam1 ES cell clones were subsequently injected into recipient C57BL/6J blastocysts. High percentage chimeric males (as estimated by percent agouti coat color) were mated to C57BL/6J or 129S5/SvEvBrd wild-type females to obtain progeny that were heterozygous for the Tcam1tm1Zuk allele. These F1 heterozygotes were intercrossed and produced progeny in the appropriate Mendelian ratios (27 wild-type:67 heterozygotes:38 homozygotes). Homozygous mice had no apparent defects in viability as we had predicted, due to the germ cell-specificity of TCAM1 expression. We ensured that our knockout allele was targeted correctly by performing RT-PCR for the exon 2-3 junction of Tcam1 (Fig. 4F) and immunofluorescence for the protein (Fig. 4G,H). Both analyses revealed that the Tcam1tm1Zuk allele created a functionally null Tcam1 allele (herein referred to as Tcam1-). Since we were able to detect TCAM1 mRNA and protein in heterozygous animals, we decided to use these mice as our controls in subsequent studies since we were most interested in the effect of complete loss of TCAM1 on mammalian testis function.
Fig. 4.
Generation of Tcam1 knockout mice. We utilized a recombineering strategy (A) to construct our mutant allele. Using our Southern blot strategy (B), we screened our ES cell clones for recombination by looking for heterozygote colonies. Germline transmission of the null allele in F1 progeny was confirmed by Southern blot with 5′ external (C) and 3′ internal (D) probes. PCR genotyping was developed to genotype F2 progeny (E). Deletion of Tcam1 was confirmed by screening testis cDNA at the exon 2-3 junction (F). The creation of a null allele was confirmed by immunofluorescence analysis of TCAM1 (control, G; null, H). B: BamHI, Sp: SpeI, Sc: ScaI.
Tcam1 null males show no apparent defects in germ cell maturation or fertility
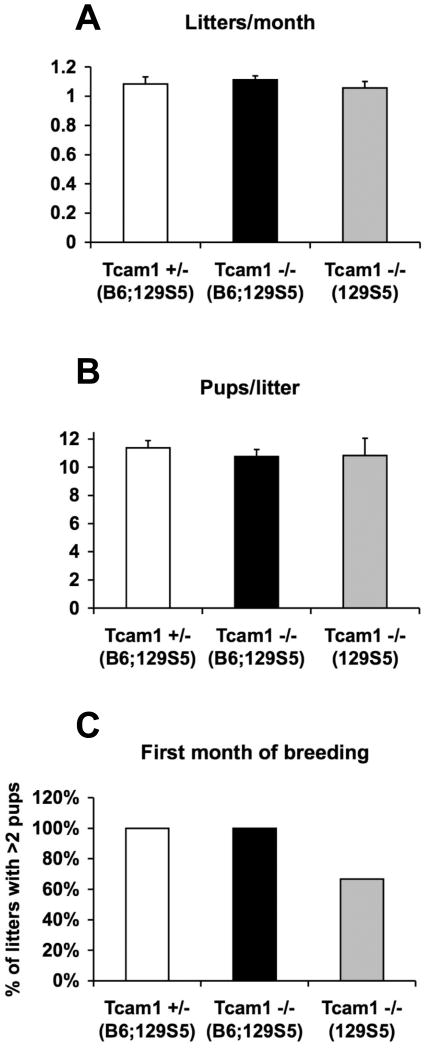
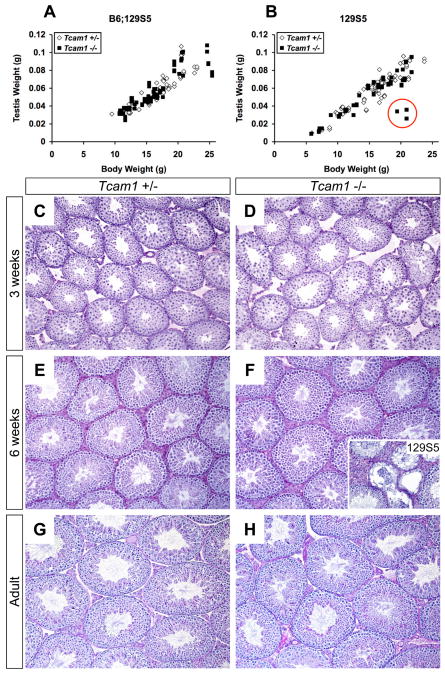
When Tcam1+/- and Tcam1-/- males reached 6 weeks of age, they were placed in mating cages with either wild-type or heterozygous B6;129S5-females for 6 months and their progeny were recorded. Tcam1+/- males and Tcam1-/- males on a mixed genetic background (B6;129S5) or a congenic background (129S5) exhibited normal fecundities during this period (Fig. 5). In addition, there were no significant differences in litters per month or pups per litter between any of these groups. Since mice can exhibit normal fertility even when sperm counts are as little as 10% of normal levels (Russell, 1990), we also tested the sperm parameters in these groups. As reported in Table 1, eight month-old Tcam1+/- and Tcam1-/- males had normal sperm counts and motility on two genetic backgrounds. These mice also exhibited normal testis histology with a full complement of maturing germ cells in the seminiferous epithelium (Fig. 6).
Fig. 5.
Fertility analysis of null and heterozygous animals. Mating cages were monitored for a period of 6 months to ascertain the average number of litters/month (A) and pups/litter (B). Although there were no significant differences between groups when calculated in this manner (p>0.05 by one-way ANOVA), we did notice that 2 out of the 6 129S5-Tcam-/- mating cages showed a delay in full fertility producing 1 or no pups in their first litter (C). Values in the graphs represent the mean of each sample group (n=4, 9, and 6 for each group respectively), and error bars indicate the standard error of the mean.
Table 1.
Testis and Sperm Parameters of 8 Month-Old Control and Tcam1-Null Males
| Strain | Genotype | Testis Weight (mg)N.S. | Sperm | |
|---|---|---|---|---|
| Count (107/epi)N.S. | Motility (%)N.S. | |||
| B6;129S5 | Tcam1+/- | 127.5±3.6 (6) | 3.42±0.28 (3) | 56.7±7.8 (3) |
| B6;129S5 | Tcam1-/- | 122.8±3.0 (6) | 3.68±0.32 (3) | 52.3±1.4 (3) |
| 129S5 | Tcam1+/- | 116.5±2.9 (6) | 3.51±0.90 (3) | 48.7±2.8 (3) |
| 129S5 | Tcam1-/- | 118.7±4.4 (6) | 4.25±1.35 (3) | 52.1 ±2.2 (3) |
Values are means ± standard errors of the mean
Values in parentheses are the number of samples used per group
N.S., Not Statistically Significant (p>0.05) by one-way ANOVA epi, epididymis
Fig. 6.
Developmental analysis of Tcam1 null and heterozygous animals. Testes were harvested from B6;129S5 (A) and 129S5 (B) mice ranging in age from 2 to 7 weeks. Body weights versus testis weights were plotted for both genotypes, and the linear regression analysis of each group was compared. The 129S5-Tcam1-/- group differed significantly (p<0.002) from the heterozygous control group due to the presence of three outliers (B, red circle). Histological analysis of representative B6;129S5 testes at 3 weeks-old (C, D), 6 weeks-old (E, F), and adult (G, H) show comparable germ cell development in Tcam1 heterozygous (C, E, G) and null (D, F, H) mice. In contrast, one of the 129S5-Tcam1-/- outliers shows variable progression of spermatogenesis and germ cell sloughing (F, inset). C-H, 100× magnification; F, inset, 200× magnification.
Although Tcam1+/- and Tcam1-/- males had normal fertility and sperm parameters as adults, we did notice that 129S5-Tcam1-/- males had variable subfertility in the first month of breeding (2 out of 6 mating pairs had less than two pups in the first month), so we sought to examine if there was any juvenile phenotype in these animals. Histological analysis of heterozygous and homozygous B6;129S5 testes at 3 and 6 weeks of age showed grossly comparable germ cell compositions and tubular sizes (Fig. 6C-F); however, since we did not perform specific germ cell counts, we cannot exclude the possibility of subtle defects in germ cell maturation. Testis weights were measured in animals ranging from 2 weeks of age (when pachytene spermatocytes first appear) to 7 weeks of age. Linear regression analyses of testis weights from each genotype revealed no significant difference in the B6;129S5 background (Fig. 6A, p>0.05) but not the 129S5 background (Fig. 6B, p<0.002). This result was mainly due to three outliers (Fig. 6B, red circle) as this difference was no longer significant when these outliers were removed. This observation correlated with the variable subfertility that we saw in the first month of breeding of 129S5 homozygous males, so the possibility remains that TCAM1 is important to testis development in certain cellular contexts.
Discussion
Our results show that Tcam1 transcription peaks in late pachytene stages and ceases prior to the formation of round spermatids. TCAM1 expression is localized to the cell membrane of pachytene spermatocytes and early stage round spermatids. Despite strong evidence that Tcam1 is evolutionarily conserved and testis-expressed in a variety of species, Tcam1-null mice have no consistent deficits in germ cell maturation or adult fertility in two genetic backgrounds suggesting that it is not essential for fertility in mice.
This study provides several possibilities for the actual function of TCAM1. First, it is possible that TCAM1 truly has no function, and its appearance is the result of an accidental duplication of ICAM2. Although the evolutionary conservation of Tcam1 argues against this possibility, we discovered (after completion of this study) that human TCAM1 gene is a testis-expressed mRNA pseudogene (GenBank AB026156.1). Comparison of the mouse and human TCAM1 loci reveals that partial duplications of the growth hormone locus may have led to the disruption of TCAM1 in humans. Second, TCAM1 may be redundant with another cell adhesion molecule during spermatogenesis. This may explain why there was variable subfertility in the first litter of some of these knockout mice that quickly resolved. Third, TCAM1 may only be required in certain environmental contexts, such as the resumption of spermatogenesis from seasonal regression (Bronson, 1985), and the duplications of the growth hormone gene that disrupted the human TCAM1 locus may have been more beneficial to fitness than TCAM1 in other species, thus explaining its loss in humans during evolution. Last, TCAM1 may be playing an instructive rather than permissive role in spermatogenesis. Although loss of TCAM1 does not block germ cells from progressing through spermatogenesis, perhaps expression of TCAM1 is instructing them to progress. A limitation of our study is that this theory has not been adequately addressed. One might speculate that aberrantly overexpressing TCAM1 in an earlier germ cell lineage may instruct germ cells to enter meiosis II prematurely.
There are a number of implications of this study that relate directly to the study of spermatogenesis. Our results suggest that TCAM1 may be useful as a marker of pachytene spermatocytes and could be used as a cell surface marker for flow cytometry and/or cell sorting. Currently, elutriation methods of Sertoli cell enrichment experience contamination from pachytene spermatocytes that are eliminated by culturing methods (Zhao et al., 2007). If our anti-TCAM1 antibody is used in magnetic activated cell sorting (MACS), this would provide a culture-free method to remove pachytene spermatocytes from Sertoli cells prepared in this fashion. The Tcam1 locus may also be useful for knock-in of Cre recombinase or fluorescent proteins. The use of this locus for Cre recombinase is especially attractive since Tcam1 peaks in expression immediately prior to meiosis II and not much is known about this process.
Our in silico analysis and expression data provided strong evidence to pursue TCAM1 in vivo as an important mediator of germ cell maturation. The lack of a consistent phenotype in our model suggests that TCAM1 is not essential for germ cell maturation in mice.
Acknowledgments
We thank Philip Soriano who provided the AK7 ES cells, Pumin Zhang who provided the pDTA.3 plasmid, and Angshumoy Roy who provided invaluable help with testis staging and sperm motility assay experiments. We also thank Mark Edson for help with ES cells, Ankur Nagaraja for collection of the multi-tissue RNA library, Stephanie Pangas for help with in situ hybridization, and Tokuko Iwamori for help with manuscript preparation. These studies were supported in part by National Institutes of Health grant HD33438 (to M.M.M.). R.L.N. is supported in part by the Edward J. and Josephine G. Hudson Scholar Fund and the Medical Scientist Training Program at Baylor College of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, Humphray S, Plumb B, Liu P, Rogers J, Bradley A. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics. 2005;86:753–758. doi: 10.1016/j.ygeno.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Eichele G, Helms JA, Lu HC. Visualization of gene expression patterns by in situ hybridization. In: Daston GP, editor. Molecular and Cellular Methods in Developmental Toxicology. CRC Press; Boca Raton, FL: 1997. pp. 23–48. [Google Scholar]
- Bronson FH. Mammalian reproduction: an ecological perspective. Biol Reprod. 1985;32:1–26. doi: 10.1095/biolreprod32.1.1. [DOI] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Insertional mutagenesis by retroviruses and promoter traps in embryonic stem cells. Methods Enzymol. 1993;225:681–701. doi: 10.1016/0076-6879(93)25044-3. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Colonna M. The role of NK cell recognition of nectin and nectin-like proteins in tumor immunosurveillance. Semin Cancer Biol. 2006;16:359–366. doi: 10.1016/j.semcancer.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Greenbaum MP, Yan W, Wu MH, Lin YN, Agno JE, Sharma M, Braun RE, Rajkovic A, Matzuk MM. TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci U S A. 2006;103:4982–4987. doi: 10.1073/pnas.0505123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SS, Brent MR. Using multiple alignments to improve gene prediction. J Comput Biol. 2006;13:379–393. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Groenen PM. PhyloPat: phylogenetic pattern analysis of eukaryotic genes. BMC Bioinformatics. 2006;7:398. doi: 10.1186/1471-2105-7-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YN, Matzuk MM. High-throughput discovery of germ-cell-specific genes. Semin Reprod Med. 2005;23:201–212. doi: 10.1055/s-2005-872448. [DOI] [PubMed] [Google Scholar]
- Lin YN, Roy A, Yan W, Burns KH, Matzuk MM. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell Biol. 2007;27:6794–6805. doi: 10.1128/MCB.01029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish HF. Molecular cell biology. W.H. Freeman; New York: 2008. [Google Scholar]
- Matzuk MM, Kumar TR, Bradley A. Different phenotypes for mice deficient in either activins or activin receptor type II. Nature. 1995;374:356–360. doi: 10.1038/374356a0. [DOI] [PubMed] [Google Scholar]
- Mruk DD, Cheng CY. Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr Rev. 2004;25:747–806. doi: 10.1210/er.2003-0022. [DOI] [PubMed] [Google Scholar]
- Roy A, Lin YN, Agno JE, DeMayo FJ, Matzuk MM. Absence of tektin 4 causes asthenozoospermia and subfertility in male mice. Faseb J. 2007;21:1013–1025. doi: 10.1096/fj.06-7035com. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Russell LD. Histological and histopathological evaluation of the testis. Cache River Press; Clearwater, Fl: 1990. [Google Scholar]
- Sakatani S, Takahashi R, Okuda Y, Aizawa A, Otsuka A, Komatsu A, Ono M. Structure, expression, and conserved physical linkage of mouse testicular cell adhesion molecule-1 (TCAM-1) gene. Genome. 2000;43:957–962. doi: 10.1139/g00-071. [DOI] [PubMed] [Google Scholar]
- Sans E, Delachanal E, Duperray A. Analysis of the roles of ICAM-1 in neutrophil transmigration using a reconstituted mammalian cell expression model: implication of ICAM-1 cytoplasmic domain and Rho-dependent signaling pathway. J Immunol. 2001;166:544–551. doi: 10.4049/jimmunol.166.1.544. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Buul JD, Kanters E, Hordijk PL. Endothelial signaling by Ig-like cell adhesion molecules. Arterioscler Thromb Vasc Biol. 2007;27:1870–1876. doi: 10.1161/ATVBAHA.107.145821. [DOI] [PubMed] [Google Scholar]
- Yan W, Rajkovic A, Viveiros MM, Burns KH, Eppig JJ, Matzuk MM. Identification of Gasz, an evolutionarily conserved gene expressed exclusively in germ cells and encoding a protein with four ankyrin repeats, a sterile-alpha motif, and a basic leucine zipper. Mol Endocrinol. 2002;16:1168–1184. doi: 10.1210/mend.16.6.0864. [DOI] [PubMed] [Google Scholar]
- Yan W, Ma L, Burns KH, Matzuk MM. Haploinsufficiency of kelch-like protein homolog 10 causes infertility in male mice. Proc Natl Acad Sci U S A. 2004;101:7793–7798. doi: 10.1073/pnas.0308025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Rohozinski J, Sharma M, Ju J, Braun RE, Bishop CE, Meistrich ML. Utp14b: a unique retrogene within a gene that has acquired multiple promoters and a specific function in spermatogenesis. Dev Biol. 2007;304:848–859. doi: 10.1016/j.ydbio.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]