Abstract
This paper describes improvements of and extensions to the Mayo Clinic 2006 smoking status classification system. The new system aims at addressing some of the limitations of the previous one. The performance improvements were mainly achieved through remodeling the negation detection for non-smoker, temporal resolution to distinguish a past and current smoker, and improved detection of the smoking status category of unknown. In addition, we introduced a rule-based component for patient-level smoking status assignments in which the individual smoking statuses of all clinical documents for a given patient are aggregated and analyzed to produce the final patient smoking status. The enhanced system builds upon components from Mayo’s clinical Text Analysis and Knowledge Extraction System developed within IBM’s Unstructured Information Management Architecture framework. This reusability minimized the development effort. The extended system is in use to identify smoking status risk factors for a peripheral artery disease NHGRI study.
Introduction
The Mayo Clinic Natural Language Processing (NLP) system for smoking status identification was first developed for the 2006 Shared Task on Natural Language Challenges for Clinical Data within the Informatics for Integrating Biology and the Bedside (I2B2) [1]. The Smoking Status Discovery challenge presented a task that called for classifying patient records into five pre-determined categories - past smoker (P), current smoker (C), smoker (S), non-smoker (N), and unknown (U), where a past and current smoker are distinguished based on temporal expressions in the patient’s medical records. A past smoker label is assigned if a patient has not smoked for at least one year; a current smoker label is assigned if a patient was a smoker within the past year. A more detailed description can be found in the reference [1]. In the 2006 I2B2 challenge the classification task is at the document-level, i.e. one of the five categories is assigned to each medical record. Uzuner et al. [2] summarized characteristics and results of the systems developed for this challenge. Most top performing systems filtered out “unknown” documents before further classification [3–5]. Many top systems assigned an “unknown” label if they did not find smoking-related information in the document [5–8]. The majority of the systems used machine learning approaches for the classification [3, 4, 7, 9, 10], some used rule-based methods [11], some employed both of them [5, 6, 8], and others used their own methods [12, 13].
Our 2006 I2B2 entry system for patient smoking status discovery [6] employed both machine learning and rule-based methods. It was built within an open source framework, IBM’s Unstructured Information Management Architecture (UIMA) a and used text analysis components from the Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES) [14]. Of note, cTAKES is to be released open-source in mid March, 2009 under the Open Health NLP Consortium (OHNLP) b umbrella. This approach allowed us to build a shareable system with a modest amount of effort [6].
Our 2006 I2B2 entry system for patient smoking status discovery had some limitations as applied to the real cases [6]. They stem mainly from errors in the negation detection for non-smoker, errors in the temporal resolution component that distinguishes a past smoker from a current smoker, and errors in the unknown smoking status assignment. Our current work aims at addressing these sources of errors by: 1) adding non-smoker lexical markers to the non-smoker dictionary, and modifying the negation rules, 2) improving the temporal resolution features for the machine learning component that identifies a past and current smoker, 3) applying a keywords search to discover the unknown cases, 4) utilizing the metadata information of clinical documents such as section headings, e.g. a family history section could contain smoking-related sentences of patient’s family member but not related to the patient himself, and these false indications potentially could hinder the correct identification of the patient’s smoking status. We present our enhancements in the Methods section followed by the Evaluation results on Mayo Clinic datasets.
The 2006 I2B2 Clinical NLP challenge aimed at assigning a smoking status to each clinical document. We call this document-level classification. However, a patient usually has quite a few clinical documents over a period of time. Hence, we expanded our previous system to perform the task of patient-level smoking status assignment over a multitude of documents. An automated way for a patient-level smoking status identification reduces the cost of human annotation for a variety of applications – patient cohort identification, risk factors identification, decision support systems.
Methods
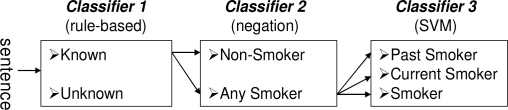
We cast the document-level label assignment as a sentence classification problem followed by a rule-based logic [6]. Furthermore, we also assumed that a patient-level class can be identified from the results of document-level classifications. Our system was built on IBM’s UIMA engineering framework with text analysis components from Mayo’s cTAKES (e.g. tokenizer, sentence boundary detector, document classifier). The UIMA framework provides an efficient way to add new components such as classifiers. Our sentence-level classification consists of three layered classifiers (Figure 1). Classifier 1 identifies two categories—unknown and known (i.e., smoking-related) sentences (past smoker, current smoker, smoker, and non-smoker) based on a keywords search approach. All smoking-related sentences are passed to Classifier 2. Classifier 2 uses negation detection to identify non-smoker inspired by the negEx [15] algorithm. The remaining sentences other than the ones with a non-smoker label are passed to Classifier 3 to identify a past smoker, a current smoker, and a smoker using a machine learning algorithm.
Figure 1.
High level architecture of sentence-level classification.
Sentence-Level Classification
Classifier 1: Known and Unknown Classification
This classification is based on the presence or absence of a set of smoking-related keywords such as smoke, tobacco, cigarette, nicotine which were manually selected from a training set. If keyword(s) appears in the sentence, known label is assigned. If not unknown label is assigned. Our original 2006 I2B2 entry used support vector machines (SVM) [16, 17] where the feature set was limited to select words and trained on a small I2B2-provided training set. Such a classifier is difficult to extend especially given the fact that access to the I2B2 data was only for the duration of the challenge. For example, if the word “nicotine” is not on the feature list, the machine learner will fail to correctly assign a smoking-related class to sentences containing this word. The current keyword-based method is extendable and not limited to a specific training set.
Classifier 2: Non-smoker Classification
This classifier first finds smoking-related anchor words, for example smoke, tobacco, cigarette, in a sentence and then checks if those words are negated. If negated, the sentence is assigned a non-smoker label. For example, in the sentence “She does not smoke.”, the anchor word “smoke” is negated by “not”, therefore this sentence is assigned a nonsmoker label. We extended the cTAKES negation detection component to include semantic negation represented by words such as nonsmoker, non-smoker.
Simple negation fails in some complicated cases such as “Tobacco: no, quit 10 years ago.” This sentence indicates a past smoker despite the presence of a negation marker. To overcome this issue, the system was extended to include words that override negation e.g., quit, stop, discontinue. If those words appear in the sentence along with a negation marker, the negation detection component is not triggered and the sentence is passed to Classifier 3 for further consideration.
Classifier 3: Past Smoker, Current Smoker, and Smoker Classification
This classification employs a machine learner, SVM [16, 17] with manually selected temporal resolution words and date indications as the features. Temporal resolution is an important factor to distinguish a past from a current smoker. Features we used were: 1) temporal resolution unigram and bigram words (e.g., day, month, days ago, months ago, years ago, remote, distant, current, past, etc.), 2) date indication captured by regular expression (e.g., 2009, 2/26/2009, 3-11-2005, 1990s, etc.), 3) smoking-related verbs without normalization to keep tense information (e.g., smoke, smokes, smoked, quit, quits, etc.), 4) copula tense markers (e.g., is (a) smoker, was (a) smoker – here the article was removed during preprocessing), , 5) bigrams for verb infinitives (to quit, to stop, to discontinue). These infinitival phrases could imply the true smoking status different from the semantics of the word itself. For example, the sentences “Patient is advised to quit smoking” and “He is planning to quit smoking” do not mean those patients already quit smoking. Just on the contrary, these sentences imply that the patient is still smoking. We observed that this kind of meaning often comes with the infinitival form along with specific words such as “quit”. This bigram “to quit” is a useful indicator to denote the actual smoking status.
After these features were extracted, they were arranged in a binary vector indicating their presence or absence in the sentence and a SVM model was built using Weka [18]. Our previous 2006 model also used SVMs with selected temporal resolution features. In that earlier version, only unigram words with high weight values in the SVM training were chosen [6]. Our current model carefully selected temporal resolution words at the unigram and bigram level, and also used additional discriminative features (features described in 2), 4), 5)).
Document-Level Classification
After the sentence-level classification is completed, all sentence labels in a given document except for sentence(s) in the family history section are processed through a precedence rules logic to assign the document-level smoking status. Current smoker (C) has the highest precedence, followed by past smoker (P), smoker (S), non-smoker (N), and unknown (U). The detailed rules are in the following:
If (exist any sentence classified as C)
Label that doc as C
Else If (exist a sentence classified as P)
Label that doc as P
Else If (exist a sentence classified as S)
Label that doc as S
Else If (exist a sentence classified as N)
Label that doc as N
Else (i.e., all sentences are classified as U)
Label that doc as U
Patient-Level Classification
This is a new component that was not implemented in our 2006 system. Each patient usually has a number of documents. Once all documents for a given patient are assigned a smoking status label, then a final summarization logic is applied to produce the patient-level smoking status label. Our logic is a combination of precedence rules and document-level class frequency. For a current and past smoker assignment the category with the highest frequency of document-level smoking status is assigned as the final patient-level label. This is based on our observations that the most frequent smoking status indicated by the patient is likely to be the true status. For the other classes the precedence logic was applied. The detailed rules of patient-level smoking status are described in the following:
If (exist a doc classified as C or P)
If (exist C but no P)
Label that patient as C
Else If (exist P but no C)
Label that patient as P
Else (i.e., exist both C and P)
If (freq of C >= freq of P)
Label that patient as C
Else Label that patient as P
Else If (exist a doc classified as S)
Label that patient as S
Else If (exist a doc classified as N)
Label that patient as N
Else (i.e., all docs are classified as U)
Label that patient as U
Data Sets
For the document-level classification, we used 390 documents from Mayo Clinic patients. Of note, no I2B2 data was used as it was available only for the duration of the challenge. We manually assigned each document a smoking status label following the I2B2 2006 challenge guidelines. The document-level distribution is: 56 past smokers, 34 current smokers, 4 smokers, 66 non-smokers, and 230 unknowns. Since we used keyword search for unknown vs. smoking-related classes and negation detection for a nonsmoker class, we only needed a training set for the classifier 3 that used SVMs to identify a past smoker, a current smoker, and a smoker. For classifier 3, we randomly selected 2/3 of the P, C and S smoking status documents for training and held out the remainder for testing. From the training set, we manually extracted smoking-related sentences and annotated their smoking status. We used these sentences to train our sentence-level classifier 3. Note that the test set for document-level classification consists of 1/3 documents from P, C, S classes plus all of N and U documents (This is because only 2/3 P, C, S documents are used for the training set).
For the patient-level classification, we used 36 patients for a total of 831 documents (16 patients of P, 5 patients of C, 13 patients of N, 2 patients of U). Note that we used the manually assigned document-level smoking status labels for this experiment instead of our system’s document-level classification outputs. The reason for it is two-fold: 1) evaluation of the patient-level summarization logic by itself independent of the lower level system components, 2) not enough data to set aside as a patient-level classification test set because many of P, C, and S documents in the patient-level data were already used for training the sentence-level classifier.
Evaluation
Precision, recall, and F-measure are used as our evaluation measurement. Precision is a ratio of retrieved examples that are correct. Recall is a ratio of correct examples that are retrieved. F-measure is the weighted harmonic mean of precision and recall defined as: 2(precision × recall)/(precision + recall). Macro and micro averages are also presented. Macro average is obtained by first calculating each class metric and then taking the average of these. Micro average is obtained by using a global count of each class and averaging these sums.
Results and Discussion
We report the performance of both document-level and patient-level classification tasks. Table 1 shows the contingency tables for the document-level classification. Table 2 shows the evaluation scores. Our system produced a macro average F-measure of 0.719 and a micro average F-measure of 0.967 (our 2006 system produced a macro average F-measure of 0.646 and a micro average F-measure of 0.912 on the same test set). All unknown (U) documents were correctly identified with an F-measure of 1 justifying the keyword search approach. The non-smoker (N) class also produced high F-measure (0.961). One source of errors was the presence of multiple smoking-related sentences in the document. For example, “He denies any drug or tobacco use...when he was sitting around a campfire and thinks he inhaled some smoke...”. Here, the first sentence was identified as a non-smoker, but the second sentence is a challenge for a machine learner. The system assigned the second sentence an incorrect current smoker label. Another source of error is negation scope. In some cases, the negation word is too far from the smoking-related anchor word and negation did not trigger. For example “She denies hx of heart problems, cp, drug use, smoking.”. Here “smoking” is too far from “denies” to be negated (we use a 7-word window before and after the anchor word to detect negation). Opening the window is likely to increase the false positive rate. The most difficult task was to identify a past smoker (P), a current smoker (C), and a smoker (S). This classification is heavily dependent on a temporal resolution module. The main source of errors was related to missing relevant features in the machine learner. For example, the test set had this text describing the smoking status - “a distant smoking history”. However, the word “distant” did not appear in the training set and so a machine learner failed to correctly identify this case. Our system failed to identify all generic smoker (S) documents.
Table 1.
Contingency table of document-level classification in the test set (Column is a system output and row is gold standard).
| P | C | S | N | U | |
| P | 15 | 4 | |||
| C | 12 | ||||
| S | 2 | 0 | |||
| N | 1 | 4 | 61 | ||
| U | 230 |
Table 2.
Evaluation of document-level classification in the test set.
| Precision | Recall | F-measure | |
|---|---|---|---|
| P | 0.938 | 0.789 | 0.857 |
| C | 0.545 | 1.000 | 0.706 |
| S | 0.000 | 0.000 | 0.000 |
| N | 1.000 | 0.924 | 0.961 |
| U | 1.000 | 1.000 | 1.000 |
| Macro Ave | 0.697 | 0.743 | 0.719 |
| Micro Ave | 0.967 | 0.967 | 0.967 |
The main reason is lack of data set for this class (we have only two documents for training).
The poor performance in the generic smoker class causes to degrade macro average in our system. Without the smoker class documents, the macro average F-measure is 0.899 and micro average F-measure is 0.970 on the test set. Our highest performance is on the unknown category. If we exclude both the smoker and the unknown class the remaining three classes produce macro average F-measure of 0.864 and micro average F-measure of 0.898 on the test set.
For the patient-level classification, our rules correctly identified all patients’ smoking statuses. This result was based on the assumption that the document-level classification is all correct (note that we used manually labeled documents in this experiment). Our rules used both precedence and frequency of the document-level smoking status. Initially, we considered using the note date to determine the final patient-level class with one possibility being to assign the class label of the most recent document as the final patient-level label. However, in our data set, we observed that the latest document did not always contain all necessary information to correctly determine patient’s final smoking status. For example, in some patients the most recent document contain only non-smoker related information while preceding documents indicate a smoking history. Therefore, we employed the precedence and frequency rules that produce the most likely true patient-level smoking status. In the future, we will use temporal resolution from the context of the documents to implement a more robust patient-level summarization.
The system we described in this paper is used to retrieve the smoking status information for a cohort of 3000 patients for an NHGRI study on Peripheral Artery Disease.
Conclusion
In this paper, we described the extended Mayo Clinic NLP system for document- and patient- level smoking status identification. We tackled these tasks by casting them into simpler problems such as sentence-level, document-level, and finally patient-level classifications. Each step relied on the previous level classification results. Our system used text analysis components from the Mayo clinical Text Analysis and Knowledge Extraction System which was built within IBM’s UIMA engineering framework. This framework allowed us to efficiently add new components as well as reuse and rebuild the existing models.
Acknowledgments
The work was funded under NHGRI grant U01 HG 04599, EMR Phenotype and Community Engaged Genomic Associations (PI Chute, Kullo).
Footnotes
References
- [1].Uzuner O, Szolovits PS, Kohane I. i2b2 workshop on natural language processing challenges for clinical records. Proceedings of the Fall Symposium of the American Medical Informatics Association; 2006. [Google Scholar]
- [2].Uzuner O, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15:14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aramaki E, Imai T, Miyo K, Ohe K. Patient status classification by using rule based sentence extraction and BM25 kNN-based classifier. i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- [4].Clark C, Good K, Jezierny L, Macpherson M, Wilson B, Chajewska U. Identifying smokers with a medical extraction system. J Am Med Inform Assoc. 2008;15:36–9. doi: 10.1197/jamia.M2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cohen A. Five-way smoking status classification using text hot-spot identification and error-correcting output codes. J Am Med Inform Assoc. 2008;15:32–5. doi: 10.1197/jamia.M2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo Clinic NLP system for patient smoking status identification. J Am Med Inform Assoc. 2008;15:25–8. doi: 10.1197/jamia.M2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Szarvas G, Farkas R, Iván S, Kocsor A, Fekete RB. Automatic extraction of semantic content from medical discharge records. i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- [8].Wicentowski R, Sydes M. Using implicit information to identify smoking status in smoke-blind medical discharge summaries. J Am Med Inform Assoc. 2008;15:29–31. doi: 10.1197/jamia.M2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Carrero F, Hidalgo JG, Puertas E, Maña M, Mata J. Quick prototyping of high performance text classifiers. i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- [10].Pedersen T. Determining smoker status using supervised and unsupervised learning with lexical features. i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- [11].Guillen R. Automated de-identification and categorization of medical records. i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- [12].Heinze DT, Morsch ML, Potter BC, Sheffer RE. A-Life Medical I2B2 NLP smoking challenge system architecture and methodology. J Am Med Inform Assoc. 2008;15:40–3. doi: 10.1197/jamia.M2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rekdal M. Identifying smoking status using Argus MLP. i2b2 Workshop on Challenges in Natural Language Processing for Clinical Data; 2006. [Google Scholar]
- [14].Savova GK, Kipper-Schuler K, Buntrock JD, Chute CG. UIMA-based clinical information extraction system. LREC 2008: Towards enhanced interoperability for large HLT systems: UIMA for NLP. 2008 [Google Scholar]
- [15].Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- [16].Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KRK. Improvements to Platt’s SMO algorithm for SVM classifier design. Neural Computation. 2001;13(3):637–49. [Google Scholar]
- [17].Platt J. Advances in Kernel Methods - Support Vector Learning. MIT Press; 1998. Machines using sequential minimal optimization. [Google Scholar]
- [18].Witten IH, Frank E. Data Mining: Practical machine learning tools and techniques. 2nd Edition ed. Morgan Kaufmann; San Francisco: 2005. [Google Scholar]