Abstract
The identification of relevant literature from within large collections is often a challenging endeavor. In the context of indexed resources, such as MEDLINE, it has been shown that keywords from a controlled vocabulary (e.g., MeSH) can be used in combination to retrieve relevant search results. One effective strategy for identifying potential search terms is to examine a collection of documents for frequently occurring terms. In this way, “Tag clouds” are a popular mechanism for ascertaining terms associated with a collection of documents. Here, we present the Literature and Genomic Electronic Resource Catalogue (LigerCat) system for exploring biomedical literature through the selection of terms within a “MeSH cloud” that is generated based on an initial query using journal, article, or gene data. The resultant interface is encapsulated within a Web interface: http://ligercat.ubio.org. The system is also available for installation under an MIT license.
INTRODUCTION
The organization of information objects using text phrases has become a popular means for both describing and navigating data within large collections. Del.icio.us1 pioneered the concept of “social bookmark” based searches, which enable one to search content using tag phrases that are applied to a given data object by other users. Resources like Connotea2, 3 enable the use social bookmarks to organize and navigate scientific data objects (e.g., journal articles or books).
Traditionally, social bookmarking approaches do not make explicit use of controlled vocabularies. Instead, user-generated terms are associated to given information object. These individual terms can be combined into “folksonomies,” which enable a high-level view of an entire collection. Such “tag clouds” are most commonly realized as an alphabetical list of tags with varying font sizes correlating to the popularity of a given tag, where larger font sizes are associated with more popular tags (e.g., for Connotea, see: http://www.connotea.org/cloud). Users can then browse the collection according to any of the tags contained in this “tag cloud4.”
Within the context of biomedical information retrieval systems, the use of controlled vocabulary terms has been shown as a reliable method to retrieve relevant articles5–7. Terms from controlled vocabularies are generally applied to data objects through a process of expert curation. In the case of MEDLINE, the MeSH controlled vocabulary is used and has been shown to enable the retrieval of relevant information from MEDLINE6, 7. Information retrieval systems, such as the PubMed interface to MEDLINE, additionally enable users to combine keyword phrases using Boolean logic (e.g., AND, OR, NOT).
The determination of which MeSH descriptors will yield the best results for a particular research domain can be difficult8, 9. This may be due in part to the size of MeSH (~29,000 descriptors) or lack of complete expertise of all the publications associated with a particular topic10. Additionally, the utility of a given MeSH descriptor is necessarily linked to its meaningfulness (e.g., a MeSH descriptor associated with all of MEDLINE is less meaningful than a term that is associated with a specific topic). The utility of developing clusters of MeSH descriptors as searching sets has been shown to be a valuable mechanism to identify relevant articles, and subsequently potentially new knowledge8, 11.
Social bookmark information retrieval systems are built on a “single click” paradigm12. That is, one clicks on a particular term within a tag cloud, and the user is shown the list of documents annotated with the selected tag. Social bookmark based information retrieval thus returns results with high sensitivity relative to the selected tag. In contrast, biomedical information retrieval systems (e.g., PubMed) depend on the ability to combine search terms (e.g., MeSH descriptors) in order to increase the specificity of searches. A balance between sensitivity and specificity based on search terms can be achieved through the combination of MeSH descriptors relative to a particular topic13.
Here, we describe the development of the Literature and Genetic Electronic Resource and Catalogue (LigerCat) system. LigerCat treats MeSH descriptors associated with MEDLINE citations as annotation “tags.” The LigerCat system enables the identification of relevant MeSH descriptors associated with NLM Journal collection queries, PubMed-style article queries, or molecular sequence queries. The identified MeSH descriptors are organized into a tag cloud inspired “MeSH cloud.” Of significance, LigerCat MeSH clouds enable the real-time filtering and sorting of relevant MEDLINE results by enabling the selection of multiple MeSH ‘tags.’
METHODS & RESULTS
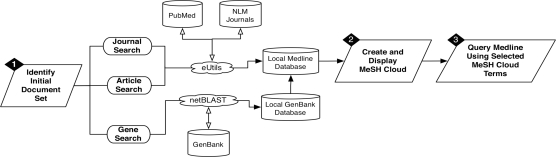
The general workflow of LigerCat is to: (1) identify the collection of documents associated with a particular topic; (2) represent the retrieved document collection as a MeSH cloud; and, (3) combine individual MeSH descriptors to create a MEDLINE search query to retrieve relevant literature. LigerCat makes use of local versions of MEDLINE (which was obtained through a license agreement with the National Library of Medicine) and GenBank as well as dynamic Web service calls using the Entrez Programming Utilities (eUtils14) and a network version of the Basic Local Alignment Search Tool (netBLAST15). A graphical overview of the LigerCat process is depicted in Figure 1.
Figure 1:
Overview of LigerCat process. (1) MEDLINE results are examined based on an initial text query (to the NLM Journals or MEDLINE database) or molecular sequence query (to a local GENBANK database that we have linked to MEDLINE results); (2) the results are generated and then (3) used to query MEDLINE via a multi-selection enabled MeSH Cloud.
Step 1: Identify Initial Collection of Documents
LigerCat currently has three modalities of searches: (1) Journal-based; (2) Article-based; and, (3) Gene-based. Regardless of the modality used for the initial query, a combination of local databases and eUtils-mediated queries are used to arrive at a full list of MEDLINE articles that are then used to create the MeSH cloud (shown in Figure 1). A brief description of each initial search method approach follows.
Journal Search
Domain specific journals represent natural collections of articles that are associated with particular topics. Previous work has discussed the utility of using journal-based collections to define related sets of articles16. Queries entered into the LigerCat “Journals” tab are used to search the NLM Journals database17.
Searches are enhanced using words in titles of journals. This approach has shown promise to categorize biomedical literature18. In our implementation, stop words (e.g., “the,” “journal,” “association”) are removed from the list of journal titles returned from the initial search, and then the top 15% of remaining title words are determined. The initial search is then enhanced with these additional title words. Across all the journals indexed in MEDLINE, we observed that on average this enhanced approach increased results by approximately 25% over just using subject terms associated with journals (as listed on the NLM Website19). In a specific example, searching the NLM Journals database for journals using the term “Aging” returns 41 journals; the enhanced approach implemented in LigerCat returns 119. A manual inspection of the 119 indicates that all of the journal titles are relevant to the biology of aging.
LigerCat presents users with the resulting list of journal titles, and the user selects those that they wish to include in the MeSH cloud generation process. Using a local MEDLINE database, all PubMed identifiers (PMIDs) are then retrieved for the selected list of journals.
Article Search
Traditional PubMed searches are fundamental to the identification of relevant biomedical knowledge. Through the “Articles” tab in LigerCat, users enter a standard PubMed query. These queries can be entered as if they were at the PubMed homepage20. A series of eUtils-mediated searches are then used to retrieve the list of PMIDs.
Gene Search
The third modality of search aims to identify relevant literature that is associated with a molecular sequence. Within the context of GenBank, 71% of sequences with citation information are associated with at least one MEDLINE citation21. Each PMID reference associated with GenBank records are extracted and stored within a local GenBank database. Through the LigerCat “Genes” tab, the user can enter a FASTA-formatted sequence of a single molecule (e.g., a gene). Related sequences are then identified using netBLAST. Based on the returned results, sequences are examined above a minimally stringent E-value (1.0). A list of PMIDs is then derived from the local GenBank database.
Step 2: Represent Initial Query Results as MeSH Cloud
For each PMID that is part of the result set from Step 1, the local MEDLINE database is queried to identify associated MeSH descriptors. Each descriptor is ranked on a scale of 0.0(highest)-1.0(lowest) that accounts for the frequency of each descriptor (MeSH_freq) and the number of times this frequency occurs in the entire PMID result set (occfreq). For each MeSH descriptor (meshd), a tag_score is thus calculated using the following equation that accounts for the importance of a given MeSH descriptor given its occurrence in all of MEDLINE:
The top 75 MeSH descriptors (i.e., the 75 lowest tag_score values) that have a tag_score value of less than 0.01 are chosen as “tags” for the displayed MeSH cloud. The value of 75 was chosen based on the number of terms that can be visually depicted in the current version of LigerCat and still be meaningful within the context of a Web browser.
For each MeSH descriptor identified as a “tag” for inclusion in the final MeSH cloud, the frequency of its occurrence is determined by summing the frequencies for the given tag across all the retrieved results (freqquery). The resulting frequency for the tag is then normalized using a database uniqueness score. This score (uniqdb) is calculated based on how common a given MeSH descriptor is in the source database on a scale of 0.0 (never occurs in database) to 1.0 (has most occurrences in database). Within the context of LigerCat, MEDLINE is used as the source database for “Journals” or “Articles” searches; GenBank is used for “Genes” searches. The resulting normalized frequency (freqnorm) is then calculated using the following equation:
This normalization process reduces the “popularity” of tags that, while occurring frequently in a resulting PMID result set, are not meaningful in the greater view of the source database. For example, the MeSH descriptor “Humans” is associated with a significant portion of MEDLINE (uniqdb score of 1.0) and should thus be significantly down-weighted compared to other MeSH descriptors associated with a particular topic. The goal of the normalization process is to enable users to focus on tags that are of significant relevance to the original query without the artifactual up-weighting of tags that are just generally popular across all of MEDLINE or GenBank.
Step 3: Browse MEDLINE Using MeSH Cloud
LigerCat presents the resulting ranked MeSH descriptors to users as “MeSH clouds,” which are visually very similar to canonical “tag clouds.”
However, unlike traditional tag clouds, LigerCat enables the selection of multiple terms within the tag cloud. Furthermore, in contrast to typical MEDLINE searches, LigerCat enables real-time modification of MeSH-based queries through the selection of terms selected from within a MeSH cloud. The selected MeSH descriptors are joined using the “AND” Boolean operator. As tags are selected (by clicking on an unselected tag) or deselected (by clicking on a selected tag), a dynamic query to MEDLINE shows the number of MEDLINE results that are returned from PubMed using eUtils. Users can thus determine how much effect a given MeSH descriptor may have on a query based on the number of results returned. In this way, different combinations of MeSH terms can be added to or removed from a single query, thus enabling one to explore how particular combinations of MeSH descriptors yield a tractable set of specific results. For example, starting with just the MeSH descriptor “Aged” returns 1,680,590 results; adding the descriptor “Animal” reduces this set to 1,676,606 results; adding “Dementia” returns 40,667 results; adding “Genetics” returns 396 results; finally, adding “Risk Factors” returns 83 articles.
Based on the selected tags of MeSH descriptors, the user can invoke a search to MEDLINE via PubMed. An important feature of the search is that while the initial MeSH cloud is based on the original query, the final search to PubMed is across all of MEDLINE. As such, the LigerCat system enables the identification of relevant descriptors from MeSH clouds associated with groups of articles associated with a particular topic that be used to expand or focus a particular search.
DISCUSSION
The LigerCat interface is a traditional information retrieval system in the sense that it enables one to either select just one term (akin to a traditional tag cloud) or connect multiple terms using the Boolean “AND” operator (akin to PubMed searches that combine terms with “AND”). However, unlike traditional tag clouds, clicking on a term within a tag cloud will not display a set of results; LigerCat only reports how many results will be reported based on adding the tag to the search query. Thus, while the developed interface violates the “single click” paradigm of traditional tag cloud implementations, it results in added functionality that enables more specificity in the returned documents. As a result, we feel that LigerCat represents a true synergy between conventional multi-term (e.g., Boolean) and contemporary social bookmarking single-term information retrieval tools. The current version of LigerCat connects multiple selected tag terms using the “AND” Boolean operator. We anticipate that future versions of the system will enable other Boolean operators (e.g., “OR” and “NOT”).
Conventional searches to MEDLINE often incorporate some knowledge of MeSH descriptors. The PubMed interface does reconcile query terms to MeSH descriptors (e.g., “Alzheimers” is automatically reconciled to the MeSH descriptor, “Alzheimer Disease”). Still, a significant disadvantage to this approach is that one is often required to know what terms should be used to yield relevant results (e.g., what MeSH descriptors are associated with “geriatrics”?).
LigerCat incorporates aspects of both the typical information retrieval and social bookmarking inspired approaches to identify relevant content. In the current implementation, the number of tags shown is limited to a pre-defined value (75 tags); there are plans to enable users to customize the maximum number of tags displayed. Additionally, we have plans to present different organizations the presented MeSH tags (e.g., instead of alphabetical, present the tag clouds according to semantic category).
Social bookmarking approaches to information retrieval posit that the information retrieval process can leverage community based tagging of information objects22. A limitation to this approach of information retrieval is that the store of data objects must first be annotated. LigerCat treats MeSH descriptors associated with MEDLINE indexed articles as annotations. By treating the keyword metadata (MeSH descriptors) as tags that have been “applied” to a MEDLINE citation, collections of MEDLINE citations can be used to generate tag clouds. In this way, LigerCat also sidesteps a common criticism of tag clouds by using tags that have been applied by a trusted source (the National Library of Medicine).
LigerCat makes use of a hybrid approach of local databases and Web accessible resources at the National Library of Medicine (NLM Journals, PubMed, and GenBank). This synergy between live and archived data is leveraged both for speed (many of the queries and MeSH profile data are precompiled) and to maintain currency. Additionally, the use of the eUtils for mediating MEDLINE queries enables the leveraging of the synonym mapping features that are part of the Entrez interface (e.g., a search for ‘movement disorders’ will include results from the search ‘motor impairments’ as would be the case in the PubMed interface). As new items are discovered through the system, they are dynamically added to the database and the MeSH ranks are appropriately adjusted. The queries invoked to NLM are mediated through a load-balancing mechanism that off-loads tasks to a background queue. This is necessary in part because eUtils require that there is a minimum 3-second interval between queries to the live data and the volume of data that may need to be processed to create the resultant MeSH cloud.
Conventional searches to MEDLINE are based on literature queries, such as the journal or article based search paradigms described above. LigerCat is also able to invoke MEDLINE searches based on molecular sequences as a “Genes” search. In the current instantiation, users can enter a single nucleotide or amino acid sequence as a FASTA formatted sequence. Future versions may allow the submission of GenBank Accession numbers as well as multiple sequences (e.g., from a gene family).
There are existing approaches to identify MeSH descriptors associated with gene symbols (e.g., Gene2MeSH23). In contrast to situations where a gene symbol is known, LigerCat Genes searches are initiated with molecular sequence data. Thus, in addition to the traditional BLAST to GenBank to identify similar sequences, LigerCat Genes may enable molecular biologists to explore relevant literature related to the query sequence. An interesting future study may be to create and explore MeSH clouds for the numerous “hypothetical” proteins that are associated with many genomes.
Based on limited examinations of LigerCat Genes searches, we observed that some MeSH descriptors are shown that occur within certain classes of sequences that are not adequately filtered. For example, when invoking a LigerCat Genes search using an Amyloid Precursor Protein (gi: 41406053; Figure 2), terms associated with general sequencing initiatives are in the MeSH cloud as popular tags (e.g., “DNA, Complementary”). It is important to point out that there are still a number of relevant MeSH descriptors displayed in the MeSH cloud (e.g., “Amyloid beta-Protein Precursor”). We are currently developing a statistical model to further down-weight MeSH descriptors that are associated with high-volume sequence projects. Another approach might include limited sequence searches to purely gene-based resources, as opposed to the entirety of GenBank. The present study focused exclusively on the determination of MeSH descriptors to identify relevant articles in MEDLINE. A similar strategy can be used to also identify significant text phrases that are also associated with a particular topic. The combination of MeSH descriptors and strategically chosen text phrases may yield even more complete sets of relevant articles. To address this, LigerCat is currently being configured to also allow users to select significant text phrases that are associated with articles within the context of a given search. These text phrases will also be ranked and presented in the same manner as the MeSH descriptors.
Figure 2:
MeSH cloud based on Genes search using Amyloid Beta Precursor Protein (gi: 41406053).
Once a set of MeSH descriptors is created and the user is confident in the results returned, it may be of interest to the user to be alerted when new articles are found using the selected set of MeSH descriptors. This can be done directly through the Real Simple Syndication (RSS) implementation in PubMed. We also feel that users may wish to be alerted of changes to a given MeSH cloud (e.g., when new tags emerge). To this end, we hope to implement a system where one can store MeSH clouds for different types of searches and be sent alerts via RSS or direct email of changes to any or all of the MeSH descriptor profiles.
LigerCat demonstrates the feasibility of creating MeSH clouds, which are analogous to tag clouds. In this study, we did not explicitly evaluate the utility of using MeSH clouds to identify relevant literature. A systematic evaluation is planned to assess the utility of MeSH cloud based information retrieval, versus current modes to identify relevant literature in MEDLINE. Nonetheless, we feel that the functionality demonstrated by the MeSH clouds as created by the LigerCat application represents a potentially new paradigm for searching MEDLINE.
CONCLUSION
MeSH descriptor based queries are an efficient and reliable means to identify relevant literature in large collections like MEDLINE. LigerCat is a Web based tool to browse through MEDLINE content based on “MeSH clouds.” These MeSH clouds are generated based on an initial journal, article, or gene-based query. Of significance, MeSH clouds enable the simultaneous selection of multiple tags that can be combined into MEDLINE queries. The resulting system thus incorporates advantages from both traditional information retrieval systems and those associated with social bookmarking.
LigerCat was developed using Ruby on Rails and is accessible at: http://ligercat.ubio.org. Instructions for download and installation of a local version of LigerCat are also available at the Web site.
Acknowledgments
This work is funded in part thanks to grants from the Ellison Medical Foundation and the National Institutes of Health (R01LM009725-01A1).
REFERENCES
- 1.http://www.del.icio.us
- 2.http://www.connotea.org
- 3.Lund B, Hammond T, Flack M, Hannay T. Social Bookmarking Tools (II): A Case Study - Connotea. D-Lib. 2005;11:1–15. [Google Scholar]
- 4.Begelman G, Keller P, Smadja F. Automated Tag Clustering: Improving Search and Exploration in the Tag Space. WWW2006. 2006 [Google Scholar]
- 5.Chang AA, Heskett KM, Davidson TM. Searching the Literature Using Medical Subject Headings Versus Text Word with PubMed. Laryngoscope. 2006;116:336–240. doi: 10.1097/01.mlg.0000195371.72887.a2. [DOI] [PubMed] [Google Scholar]
- 6.Lowe HJ, Barnett GO. Understanding and Using the Medical Subject Headings (MeSH) Vocabulary to Perform Literature Searches. JAMA. 1994;271:1103–8. [PubMed] [Google Scholar]
- 7.O'Rourke A. Another Fine MeSH: Clinical Medicine Meets Information Science. Journal of Information Science. 1999;25:275–81. [Google Scholar]
- 8.Srinivasan P. MeSHmap: A Text Mining Tool for MEDLINE. Proc AMIA Symp. 2001:642–6. [PMC free article] [PubMed] [Google Scholar]
- 9.Struble CA, Dharmanolla C. Clustering MeSH Representations of Biomedical Literature. HLT-NAACL 2004 Workshop: Biolink. 2004;2004:41–8. [Google Scholar]
- 10.Aronson AR, Mork JG, Gay CW, Humphrey SM, Rogers WJ. The NLM Indexing Initiative's Medical Text Indexer. Medinfo. 2004;11:268–72. [PubMed] [Google Scholar]
- 11.Srinivasan P. Text Mining: Generating Hypotheses from MEDLINE. Journal of the American Society for Information Science. 2003;55:396–413. [Google Scholar]
- 12.Hassan-Montero Y, Herrero-Solana V. Improving Tag Clouds as Visual Information Retrieval Interfaces. International Conference on Multidisciplinary Information Sciences and Techologies (InSciT2006). 2006.
- 13.Wilczynski NL, Haynes RB. Robustness of Empirical Search Strategies for Clinical Content in MEDLINE. Proc AMIA Symp. 2002:904–8. [PMC free article] [PubMed] [Google Scholar]
- 14.http://eutils.ncbi.nlm.nih.gov
- 15.Madden TL, Tatusov RL, Zhang J. Applications of network BLAST server. Methods Enzymol. 1996;266:131–41. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 16.Humphrey SM. Automatic Indexing of Documents from Journal Descriptors: A Preliminary Investigation. Journal of the American Society for Information Science. 1999;50(8):661–74. doi: 10.1002/(SICI)1097-4571(1999)50:8<661::AID-ASI4>3.0.CO;2-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=journals
- 18.Demner-Fushman D, Hauser S, Thoma G. The Role of Title, Metadata and Abstract in Identifying Clincally Relevant Journal Articles. Proc AMIA Symp. 2005:191–5. [PMC free article] [PubMed] [Google Scholar]
- 19.http://www.nlm.nih.gov/bsd/journals/subjects.html
- 20.http://www.pubmed.gov
- 21.Miller H, Norton CN, Sarkar IN. GenBank and PubMed: How connected are they? BMC Res Notes. 2009;2:101. doi: 10.1186/1756-0500-2-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond T, Hannay T, Lund B, Scott J. Social Bookmarking Tools(I) D-Lib. 2005;11(4) [Google Scholar]
- 23.Ade AS, States DJ, Wright ZC.Gene2MeSH Ann Arbor (MI): National Center for Integrative Biomedical Informatics,University of Michigan; 2007 [cited 2009]; Available from: http://gene2mesh.ncibi.org