Abstract
Clinical information systems offer an opportunity to provide clinicians with medical reference materials during clinical encounters when the information is most beneficial. Implementation of this “Infobutton” concept has been described by a number of institutions with locally developed clinical information systems and electronic medical records. This article describes the development of an infobutton-like application called ClinRefLink embedded within a commercial clinical information system. ClinRefLink is somewhat unique in that it offers clinicians the option to perform reference searches based on clinical entities identified within narrative documents. In the first 30 days after implementation, 1018 reference searches were performed. The characteristics of the clinicians and the clinical context of the search terms are described. These data support the value of clinical term extraction from narrative documents as a component of an infobutton system.
Introduction
The rapid expansion of and changes to biomedical knowledge presents a tremendous challenge to clinicians. Providing clinicians with convenient, online access to clinical reference materials at the time and place where care is being rendered is one way to help clinicians provide the highest level of care to their patients. This concept has been termed “infobuttons”, defined as “context-specific links from one information system (usually a clinical information system such as an electronic health record) to some other resource that provides information that might be relevant to the initial context)”1. Providing direct access to reference information within the context of a clinical information system has been shown to improve the care of patients2,3.
Ideally, the infobuttons should be implemented such that they provide navigation tools in very close proximity to the clinical term or entity about which the clinicians may wish to access information. This can be done by means of presenting the term onscreen as a hyperlink or placement of a virtual button next to the term of interest. While this approach is possible in the case of a locally developed clinical information system2,3,4, this may not be available in a commercial application and other approaches may be necessary.
Typically, infobuttons are tied to discrete data elements within a clinical record (e.g. medications, laboratory data). In some implementations, terms from within narrative documents are also used as sources for links to reference information5. Surprisingly, though, one prior study reported that these links were not frequently used as sources for reference searches, accounting for only 21% of all contexts of infobutton use (9% radiology, 7% clinical notes, 3% pathology reports, 2% discharge summaries)6.
Background
The North Shore-Long Island Jewish Health System (NSLIJ) is an integrated delivery network located in the New York Metropolitan area. It consists of 14 acute care hospitals, more than 7000 employed and voluntary physicians, ambulatory care practices, long term care facilities, and home care agencies.
NSLIJ is currently in the second year of a multiyear rollout of an inpatient electronic medical record (Sunrise Clinical Manager™ (SCM 4.5 XA™), Eclipsys Corporation, Atlanta, GA). At the time of this writing, five hospitals in the health system are utilizing SCM to access clinical results, reports and documents. These include laboratory results, radiology reports and images, pathology reports, cardiology reports and images, and transcribed documents (operative notes and discharge summaries). At two hospitals, Emergency Department triage and disposition documents are being entered into SCM. At one hospital, transcribed admission history and physicals and consult notes are available in the system.
The current state of implementation presented two challenges to the implementation of embedded links to reference information. First, because we were using a commercial product, we did not have the local programmatic ability to embed buttons or to convert onscreen text to hyperlinks. Second, other than laboratory results, the bulk of clinical information currently available was contained within text documents, many of which are unstructured narratives not constrained by or mapped to a standard clinical vocabulary.
Sunrise Clinical Manager’s user interface is designed to reflect a chart metaphor, with different types of information available in different sections. Sections are accessed by clicking virtual tabs displayed horizontally across the upper part of the screen. Sunrise Clinical Manager allows addition of custom tabs to the application. Clicking a custom tab launches locally-developed code which then executes and is displayed within the Sunrise application. The local code can access user and patient context information by means of a series of exposed Sunrise objects and interfaces referred to as ObjectsPlus. This technology was utilized to allow clinicians to invoke ClinRefLink by simply clicking a custom tab while reviewing patient data within SCM.
ClinRefLink Description
Functional Description
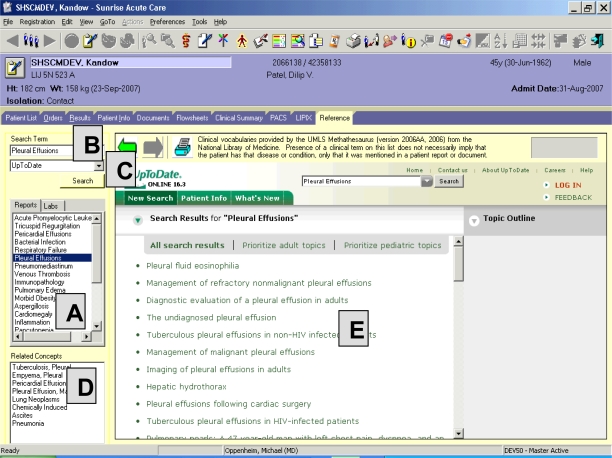
A representative screen print of the ClinRefLink application is shown in figure 1. When the clinician clicks the ‘References’ tab, ClinRefLink processes all results (discrete and textual) and documents in the patient’s record (described below). Clinical terms identified in the patient’s record are presented to the clinician in the box labeled ‘A’ which contains two tabs, one for items identified from narrative documents (labeled ‘Reports’), one for items identified from discrete laboratory values (labeled ‘Labs’). Selecting a term from a list within ‘A’ places the term in the ‘Search Term’ box (‘B’), and also displays MeSH co-occurring concepts7 for the selected term in the ‘Related Concepts’ box (‘D’). The clinician has the option to select an item from the ‘Related Concepts’ box and replace the initial search term with the newly selected term. Clicking the search button executes the actual search in the reference product displayed in ‘C’. The reference material is displayed in the main area of the screen (‘E’). At any time, the clinician can select a different reference product by clicking the dropdown arrow in ‘C’, then clicking the ‘Search’ button.
Figure 1:
ClinRefLink Screen Print
Clinicians can manually type into the ‘Search Term’ box and perform a search. This allows the clinician to either edit a term selected from the ClinRefLink list of terms (e.g. add the word ‘congenital’ prior to a selected term ‘hypothyroidism’ to search for ‘congenital hypothyroidism’), or to enter a completely free text search.
Text Reports & Documents
Terms are identified from within textual reports and documents using a three-phased process.
Preprocessing: Text is preprocessed to remove punctuation and other extraneous characters. The text is then tokenized into 3-, 2-, and 1-word strings.
Term Identification: Tokens (in singular and plural forms) are compared against a table of clinical terms derived from the Unified Medical Language System (UMLS)7. This table includes all terms of whose semantic types reflect disease entities or clinical findings. Tokens found in the table of clinical terms in more than two source vocabularies are entered into the term list.
Cleanup: The term list is analyzed to remove one of any duplicate pair (two different strings with a common Concept Unique Identifier in UMLS, e.g. acute myelogenous leukemia and acute myeloid leukemia) or terms which are substrings of other terms (e.g. myelogenous leukemia vs. acute myelogenous leukemia) and therefore less specific.
By design no attempt is made to identify terms with negators (i.e. things stated to be absent, e.g. ‘No evidence of pneumonia’) as has been described elsewhere8,9. This is because the purpose of the extraction is not to assign a diagnosis to a patient but rather to identify terms of potential interest to a clinician reading a report. A term appearing in a narrative, even if stated as being absent, may be of interest to a clinician who is unfamiliar with the specific clinical entity mentioned in the report or document. Disclaimers are displayed onscreen reminding the clinicians that the presence of a term on the list does not necessarily indicate that the patient has that disease or condition.
Laboratory Data
Laboratory data for the currently selected patient is processed to provide the clinician with search terms most likely to provide useful information. Abnormal laboratory results are stored in the clinical data repository with indicators of the direction of abnormality (above the high end of the reference range or below the low end of the reference range). A translation table was created to translate the test name / abnormality combination to a clinical descriptor where appropriate. For example, a serum sodium measurement with an elevated indicator would be translated so that the clinician would be presented with the search term ‘hypernatremia’. Abnormal laboratory tests for which there is no single descriptive term are presented to the clinician with the test name preceded by either ‘increased’ or ‘decreased’ (e.g. ‘increased alkaline phosphatase’).
Microbiology data is also processed to present the clinician with the most specific search terms. If the body site from which a culture was obtained maps to a single clinical disease entity (e.g. specimen from CSF = meningitis, blood = bacteremia), then the clinician is presented with a search term which includes the organism name and the clinical disease entity (e.g. ‘Streptococcus pneumoniae meningitis’, ‘Staphylococcus aureus bacteremia’). If the culture source is ambiguous (e.g. wound cultures, tissue cultures, and etc.) then the organism name alone is presented as the search term. Again, because the positive culture may not be reflective of disease (e.g. contaminants, blood cultures drawn through indwelling catheters), disclaimers are displayed onscreen reminding the clinicians that the presence of a disease on the list does not necessarily indicate that the patient has that disease or condition
At the time of this writing, prior to implementation of Computerized Prescriber Order Entry, medications are not yet in the clinical information system. Thus, a list of active patient meds cannot yet be provided to the clinicians to support medication-related information needs.
Available Reference Products
ClinRefLink supports searches in UpToDate, Access Medicine, Access Surgery, MDConsult, Nursing Consult, Micromedex Infobutton, the National Guideline Clearinghouse, Cochrane Library, and the UK’s National Institute for Health and Clinical Excellence. These resources are generally summary or text-based resources as opposed to journal article resources, as summary resources are likely to be more useful for inquiries made at the point of care4. The proprietary products listed are licensed for the Health System by the health science libraries.
Results
In the first 30 days after implementation of ClinRefLink, a total of 1018 searches were performed by clinicians. A ‘search’ is defined as the clicking of the ‘Search’ button to obtain reference information. Invoking ClinRefLink but not performing a search was not counted in this total. A search of the same term in two different reference products was counted as two searches.
272 distinct clinicians performed the 1018 searches. 100 clinicians performed 3 or more searches, while the remaining 172 clinicians performed 2 or 1. 8 clinicians performed 20 or more searches, with 2 of them performing more than 50 searches. This wide disparity in use is consistent with other published observations3. The breakdown of clinician types is shown in table 1.
Table 1.
| Role | Searches |
|---|---|
| Resident | 400 |
| Attending | 234 |
| RN | 122 |
| Physician Assistant | 88 |
| Fellow | 77 |
| Other | 63 |
| Nurse Practitioner | 14 |
| Medical Student | 12 |
| Technician | 5 |
| Pharmacist | 2 |
| Unit Receptionist | 1 |
As described earlier, clinicians have the option to use a term identified from the patient’s record, or to either manually edit the term or type a completely different search term. Clinicians selected a term from the presented list of terms 43% of time, and manually edited or typed the search term 57% of the time. Of the 434 searches using terms offered by ClinRefLink, 103 searches were done using Co-occurring concepts of a selected term rather than the primary term.
Attending physicians searched terms identified by ClinRefLink in 101/234 searches (43%) while residents searched system-identified terms in 130/400 searches (33%) (p<0.01, Fisher’s Exact test). RNs searched using a system-identified term in 67/122 searches (55%), and Medical Students searched a system-identified term in 12/12 searches (100%).
For searches done using terms identified by ClinRefLink, the sources of those terms are shown in table 2.
Table 2.
(Reports = text results; Documents = transcribed documents or documents entered directly into Sunrise)
| Source | Count |
|---|---|
| REPORT | 247 |
| DOCUMENT | 85 |
| LAB | 58 |
| DOCUMENT & REPORT | 28 |
| MICROBIOLOGY | 20 |
Discussion
In contrast to classic infobuttons which provide clinicians with links to online reference material in close proximity to the items of interest, ClinRefLink requires clinicians to go to a specific area within the application, and then presents an aggregate of terms about which the clinician may wish to search. While this could potentially be a barrier to use and adoption, our usage (1018 searches) in the first 30 days since implementation is, by extrapolation, very similar to other reported usage patterns. Maviglia et al. reported 7,972 uses of KnowledgeLink over a one year study period3, Cimino reported 30,374 uses of Infobutton Manager over a 5.7 year period2, and Del Fiol reported 53,127 E-Resources Manager sessions over 4 years4.
While these early usage statistics are encouraging, it should be noted that usage of ClinRefLink has not reached steady state (see table 4). Thus, it is not clear whether some of the utilization reflects clinicians looking at and evaluating the new application out of curiosity, not to truly support clinical activities. Our early data demonstrate some clinical clinicians with extremely frequent use and others who have only utilized the functionality once or twice. This is consistent with observations from other implementations3.
Table 4.
Our usage statistics show a fairly significant proportion of searches being performed utilizing terms extracted from text / narrative documents, in contrast to earlier published work6. Once again, given the relative novelty of this functionality in our environment, the usage patterns may reflect clinician curiosity and evaluation. Continued observation of usage will be necessary to determine the real value of text-and narrative-derived terms in supporting clinician’s information needs. Additionally, a user survey will be performed once clinicians have had significant time to explore and use ClinRefLink.
Another interesting observation is the frequency with which ClinRefLink is being used to perform searches using manually entered search terms (57% of all searches). While our electronic library10 is easily accessible on computer desktops, clinicians seem to appreciate the ease of simply clicking a tab within the clinical information system rather than launching a separate browser and accessing the library website. It is not known how many manual terms were actually present in reports or documents but not offered as a search term because of the limitation to only certain UMLS semantic types or requirement that the term appear in more than one vocabulary. This analysis should be done once clinicians are experienced in using ClinRefLink.
Our statistics show an overwhelming predominance of use of one particular reference product. At this point, it is not known whether this is because of a specific product preference or because it is the default selection on the list of products. Future work will focus on evaluating the impact of changes to the sequence of the products presented.
Conclusion
ClinRefLink has been successfully implemented as an add-on module within a commercial clinical information system. While not truly providing in-context access to reference information, our early usage statistics suggest that clinicians find value in the ability to access terms extracted and presented from various parts of a patient’s record. Additionally, our utilization patterns suggest that extraction of clinical terms from text and narrative documents is a useful strategy for supporting clinician’s information needs. Further monitoring of utilization over time and direct user feedback is needed to understand if these patterns of usage will be sustained with longer-term clinician use and experience with this product.
Table 3.
| Reference | Count |
|---|---|
| UpToDate | 619 |
| Access Medicine | 125 |
| MDConsult | 110 |
| MicroMedex InfoButton | 40 |
| Access Surgery | 38 |
| NursingConsult | 36 |
| Cochrane Library | 30 |
| National Institute for Health and Clinical Excellence | 13 |
| National Guideline Clearinghouse | 7 |
References
- 1.www.infobuttons.org
- 2.Cimino JJ. Use, Usability, Usefulness, and Impact of an Infobutton Manager. AMIA Annual Symposium Proceedings. 2006:151–155. [PMC free article] [PubMed] [Google Scholar]
- 3.Maviglia SM, Yoon CS, Bates DW, Kuperman G. Knowledge Link: Impact of Context-Sensitive Information Retrieval on Clinician’s Information Needs. J Am Med Inform Assoc. 2006;13:67–73. doi: 10.1197/jamia.M1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Fiol G, Rocha RA, Clayton PD. Infobuttons at Intermountain Healthcare. AMIA Annual Symposium Proceedings. 2006:180–184. [PMC free article] [PubMed] [Google Scholar]
- 5.www.dbmi.columbia.edu/cimino/Presentations/2004-CancerCenter-InfobuttonsandCancerCenterProtocols.ppt
- 6.Cimino JJ, Li J, Graham M, et al. Use of Online Resources While Using a Clinical Information System. AMIA Annual Symposium Proceedings. 2003:175–179. [PMC free article] [PubMed] [Google Scholar]
- 7.Unified Medical Language System, National Library of Medicine; version 2006AA. wwwnlm.nih.gov/research/umls; Level 0 and SNOMED CT vocabularies were used
- 8.Meystre S, Haug PJ.Evaluation of Medical Problem Extraction from Electronic Clinical Documents Using MetaMap Transfer (MMTx)2005116823–828. [PubMed] [Google Scholar]
- 9.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting Information from Textual Documents in the Electronic Health Record: A Review of Recent Research. Yearb Med Inform. 2008:128–144. [PubMed] [Google Scholar]
- 10.NSLIJ Electronic Medical Information Library (EMIL) was developed under NLM Grant # 5G08LM008090-3