Abstract
Figures are frequently used in biomedical articles to support research findings; however, they are often difficult to comprehend based on their legends alone and information from the full-text articles is required to fully understand them. Previously, we found that the information associated with a single figure is distributed throughout the full-text article the figure appears in. Here, we develop and evaluate a figure summarization system – FigSum, which aggregates this scattered information to improve figure comprehension. For each figure in an article, FigSum generates a structured text summary comprising one sentence from each of the four rhetorical categories – Introduction, Methods, Results and Discussion (IMRaD). The IMRaD category of sentences is predicted by an automated machine learning classifier. Our evaluation shows that FigSum captures 53% of the sentences in the gold standard summaries annotated by biomedical scientists and achieves an average ROUGE-1 score of 0.70, which is higher than a baseline system.
1. Introduction
Biomedical journal articles frequently incorporate figures as evidence of discovery (1). Figures are frequently used by scientists to validate research findings and to formulate novel research hypotheses. Therefore, figures serve as important evidence for scientific communication and peer review. Despite the importance of figures, their potential has not been completely recognized. Recently, however, there has been growing interest in the extraction and use of information available in biomedical figures (2).
Although associated texts are necessary for understanding the content of a figure (3), our study has shown that this content is disseminated throughout the article (4). We speculate that a text summary aggregating this scattered content may help a reader comprehend the figure’s meaning. Hence, our goal in this study is to find a way to automatically extract the sentences from a full-text article that best describes a figure in the text and use these sentences to generate a short summary of the figure. Our task can be viewed as a topic-specific or targeted summarization task (2).
2. Related Work in Biomedical Text Summarization
Summarization tasks can be divided into two approaches - extractive and abstractive. In an extractive approach, the task is to generate a summary by selecting the most informative and relevant sentences from the article. On the other hand, in an abstractive approach, the task is to understand the concepts of the article and then generate a summary by generating text based on these concepts.
Both approaches have been used to summarize biomedical articles. Ling et al. (5) generated a structured summary for genes by extracting sentences from the literature. Their approach first retrieved articles relevant to the queried gene and then used a probabilistic language modeling approach to extract relevant sentences. This approach outperformed general purpose summarization approaches.
In their summarization system, Reeve et al. (6) used the Unified Medical Language System (UMLS) to link semantically-related concepts within biomedical text. These concept chains were used to identify candidate sentences for extraction. A summary was produced by using those sentences extracted with the strongest chains. The system was evaluated by comparing the extracted summaries with the abstracts from the articles. The system’s precision and recall were 0.90 and 0.92, respectively.
Similarly, Fiszman et al. (7) developed an abstractive approach that relies on identifying the semantic categories of terms in articles using SemRep (8) and the relationships between these categories. Interestingly, instead of producing a textual summary, this approach displays summaries in graphical form, with nodes being the terms and edges being the relationships between those nodes.
3. Algorithm
Our goal is to provide a text summary describing the content of figures in biomedical articles. For this, we decided to select four sentences for each summary: one sentence providing a background for the figure, one sentence describing the methods used to obtain the figure, one sentence describing the outcome and one sentence showing the significance of the figure. This is based on our user survey study (unpublished), which finds that biologists consider a summary comprising one sentence per category to be adequate for comprehending figure content.
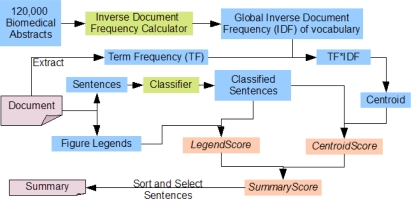
For each article, we first classified all full-text sentences into Introduction, Methods, Results and Discussion (IMRaD) categories using the classifier described in (9). Briefly, a multinomial Naïve Bayes classifier was trained on manually annotated sentences to predict IMRaD categories. We designed an algorithm, FigSum, that selects sentences that are similar to the legend of the figure being summarized and are also related to the central theme of the article, as we believe that such sentences can convey the content of a figure. Hence, for each sentence, we calculated a CentroidScore, which indicates the closeness of the sentence to the central theme of the article, and a LegendScore, which indicates the closeness of the sentence to the legend of the figure being summarized. CentroidScore and LegendScore were then combined to obtain a SummaryScore. Summaries were generated by selecting the sentences with the highest SummaryScore. Calculation of the CentroidScore and LegendScore is described in the following subsections, and the algorithm’s schematic is shown in Figure 1.
Figure 1.
Schematic representation of our summarization algorithm, FigSum.
3.1. Calculation of CentroidScore
For each sentence in the article, we calculated a CentroidScore to identify sentences that were central to the theme of the article. This score was based on the idea described in (10). We downloaded Pubmed Central’s open access subset, which contained 120,000 biomedical articles when it was downloaded. We first calculated the global IDF (Inverse Document Frequency) of every word appearing in the abstract of these 120,000 articles. IDF was calculated using the following formula –
All abstracts were normalized by lowercasing all words, and removing stop words (e.g., ‘the’, ‘and’, ‘for’), punctuation and numbers. All words were stemmed using the Porter stemmer (11). We noticed that words appearing with a frequency of less than five were mostly artifacts or misspelled words (example ‘australiaa’ and ‘resultsshow’); hence we ignored words with a frequency less than five.
For each article, we calculated the frequency of each word (term frequency) and multiplied it with the global IDF value of that word to obtain the TF*IDF (Term Frequency * Inverse Document Frequency) score. Words were sorted by this score to obtain the top 20 words believed to be central to the article. The TF*IDF values of these 20 words were divided by the number of sentences in the article to obtain a unique centroid for every article.
We then calculated the CentroidScore for each sentence as a measure of its centrality to the article. For this, word similarity between the normalized sentence and the centroid of the article was calculated and stored as the CentroidScore of the sentence.
3.2. Calculation of LegendScore
LegendScore was calculated as the similarity between the legend of the figure being summarized and the sentence. Paragraphs from the full text that directly referred to the figure were appended to the legend of that figure. Both the appended legend and sentence were normalized by lowercasing all words and removing stop words, punctuation and numbers. An ISF (Inverse Sentence Frequency) value was calculated for every word in the article using the following formula -
The frequency of every word in a sentence and figure legend, TF, was multiplied with that word’s ISF to get a TF*ISF value vector for every sentence and figure legend. The LegendScore for every sentence was calculated as the cosine similarity of the TF*ISF vector of that sentence and the figure legend.
3.3. Automatic Summary Generation
The SummaryScore of every sentence was calculated using the following formula -
where wc and wl are the weights for CentroidScore and LegendScore, respectively. We tried various combinations of wc and wl such that they add up to 1 and found that the best performance was obtained with wc set at 0.1 and wl set at 0.9. Based on the SummaryScore and the classifier tag of the sentences, the best Introduction sentence, Methods sentence, Results sentence and Discussion sentence were selected to form the figure’s summary. We refer to this summary as AutomaticSummary. A sample summary is shown in Figure 2.
Figure 2.
A summary generated by our algorithm for Figure 1 in an article (12). The four summary sentences are the sentences tagged as Introduction, Methods, Results and Discussion, in that order.
3.4. A Baseline System
A baseline system, BaseSum, was developed based on the position of the sentences relative to the first sentence referring to the figure (13). All sentences in each article were arranged as a list in the order that they appeared in the article. Based on the predicted IMRaD category of the first sentence referring to the figure, BaseSum selected the remaining three sentences by moving up or down the sentence list. Since IMRaD categories are linear, i.e. Introduction, Methods, Results and Discussion appear in that order, the direction in which the system scans to select sentences depends on the predicted category of the first referring sentence. For example, if the first referring sentence was predicted as Results, then the system would move up the sentence list to find the first predicted Introduction and Methods sentences and move down the sentence list to find the first predicted Discussion sentence. Four sentences were thus selected to form the BaselineSummary.
4. Generation of Evaluation Data
We asked four annotators to generate a summary for each figure in an article. Each annotator had an advanced degree (MS and above) in biomedical science, and they were asked to select three to four sentences best describing the background of the figure, the method used to obtain the figure, the outcome based on that figure, and the conclusion based on the figure. Annotators were free to choose the same sentence for two different categories. Hence, for every figure, we obtained a 12- to 16-sentence long summary, which we call the AnnotatorSummary.
In all, seven articles, comprising a total of 44 figures, were annotated. Six of these articles were randomly selected from the GENIA corpus (14) and were annotated by three annotators (one person annotated three articles, one annotated two articles, and one annotated a single article; there was no article overlap), and one article was randomly selected from PubMed Central’s Open Access subset and was annotated by the author of that article. The average number of unique sentences in AnnotatorSummary for each figure and the total number of figures and sentences in the full text of each article appear in Table 1. We used these figure annotations to evaluate FigSum.
Table 1.
The number of unique annotated sentences per figure for each article, the number of sentences per article, and the number of figures per article.
| Article | Average no. of unique sentences per figure in AnnotatorSummary | No. of senteninces article | No. of figures in article |
|---|---|---|---|
| 1 | 10.2 | 160 | 5 |
| 2 | 10.8 | 140 | 5 |
| 3 | 11.78 | 281 | 9 |
| 4 | 10.5 | 172 | 4 |
| 5 | 6.33 | 137 | 9 |
| 6 | 8.4 | 87 | 5 |
| 7 | 8.0 | 173 | 7 |
5. Evaluation based on Precision
Here, we report the precision of the summary generated by FigSum (AutomaticSummary) and BaseSum (BaselineSummary) against the annotator generated summary (AnnotatorSummary). For each sentence in AutomaticSummary and BaselineSummary, we checked to see if the sentence was a part of AnnotatorSummary as well. If it was, we denoted the sentence as a True Positive (TP), and if not, we denoted the sentence as a False Positive (FP). Based on the number of TP sentences and FP sentences, the precision was calculated by using the formula: Precision = TP / (TP + FP). The precision of AutomaticSummary and BaselineSummary with respect to these seven articles is shown in Table 2.
Table 2.
Precision of the Automatic Summary and the Random Summary. Ar: Article, Oa: Overall, TP: True Positives, FP: False Positives, Prec.: Precision
| AutomaticSummary | BaselineSummary | |||||
|---|---|---|---|---|---|---|
| Ar | TP | FP | Prec. | TP | FP | Prec. |
| 1 | 11 | 9 | 0.55 | 8 | 12 | 0.40 |
| 2 | 11 | 9 | 0.55 | 11 | 9 | 0.55 |
| 3 | 16 | 20 | 0.44 | 6 | 30 | 0.17 |
| 4 | 8 | 8 | 0.50 | 7 | 9 | 0.44 |
| 5 | 23 | 13 | 0.64 | 15 | 21 | 0.42 |
| 6 | 12 | 8 | 0.60 | 11 | 9 | 0.55 |
| 7 | 13 | 15 | 0.46 | 13 | 15 | 0.46 |
| Oa | 94 | 82 | 0.53 | 71 | 105 | 0.40 |
6. Evaluation based on the ROUGE Score
In addition to precision, we used the ROUGE score to obtain evaluation metrics, as ROUGE scores are widely used for evaluating text summarization tasks (15). ROUGE score evaluates test summaries by comparing it with a human-generated gold standard summary based on the n-gram overlap between the test summary and the gold standard. An n-gram is a subsequence of n words in a text. ROUGE scores range from 0 to 1, and a higher score indicates that the test summary is closer to the gold standard summary.
We calculated the ROUGE scores using the parameters established by the Document Understanding Conference 2007 (16). For every sentence in AutomaticSummary and BaselineSummary, we calculated ROUGE scores against every annotated sentence and retained the best scores. The average of the best ROUGE scores over every figure and sentence in the three summaries were then calculated. We report the ROUGE-1, ROUGE-2 and ROUGE-SU4 scores in Table 3. ROUGE-1 compares summaries based on the co-occurrence of unigrams (single words), ROUGE-2 compares summaries based on the co-occurrence of bigrams (two consecutive words), and ROUGE-SU4 compares summaries based on the co-occurrence of skip bigrams with a maximum gap length of four (15).
Table 3.
The average ROUGE-1, ROUGE-2 and ROUGE-SU4 scores for the AutomaticSummary and BaselineSummary. R-1: ROUGE-1, R-2: ROUGE-2, R-SU4: ROUGE-SU4, Std Dev: Standard Deviation
| AutomaticSummary | BaselineSummary | |||||
|---|---|---|---|---|---|---|
| Article | R-1 | R-2 | R-SU4 | R-1 | R-2 | R-SU4 |
| 1 | 0.75 | 0.62 | 0.63 | 0.62 | 0.46 | 0.48 |
| 2 | 0.73 | 0.64 | 0.63 | 0.69 | 0.61 | 0.60 |
| 3 | 0.59 | 0.48 | 0.49 | 0.45 | 0.23 | 0.26 |
| 4 | 0.71 | 0.61 | 0.60 | 0.59 | 0.48 | 0.48 |
| 5 | 0.76 | 0.68 | 0.69 | 0.58 | 0.46 | 0.47 |
| 6 | 0.71 | 0.65 | 0.64 | 0.64 | 0.57 | 0.58 |
| 7 | 0.65 | 0.55 | 0.54 | 0.65 | 0.52 | 0.53 |
| Average | 0.70 | 0.60 | 0.60 | 0.60 | 0.48 | 0.49 |
| Std Dev | 0.06 | 0.07 | 0.07 | 0.08 | 0.12 | 0.11 |
7. Discussion
Here, we have described our algorithm, FigSum, to automatically generate extractive summaries for figures in biomedical journal articles. A baseline summary was generated by selecting sentences near the first sentence that refers to the figure. On comparing with expert generated summaries, we found that FigSum performs better than the baseline system.
We found that the precision of AutomaticSummary fell in a range between 0.44 and 0.64. This could be due to variation in the quality of summary generated by the annotators. As noted earlier, human-generated summaries often display such variation (17; 18). Also, article length seemed to have an effect on the precision of AutomaticSummary. The longest article (Article 3) contained 281 sentences, and it had the worst precision score (0.44). On the other hand, the shortest article (Article 6) was 87 sentences long and had the second best precision score (0.64).
We calculated the ROUGE-1, ROUGE-2 and ROUGE-SU4 scores according to the guidelines of the Document Understanding Conference 2007. These scores reflected that FigSum performed better than BaseSum. It was interesting to note that for Article 2, although both AutomaticSummary and BaselineSummary achieved a precision of 0.55, the ROUGE scores for the AutomaticSummary were better than the ROUGE scores for BaselineSummary. This indicates that sentences chosen by FigSum are closer to the human-generated gold standard summary than the sentences chosen by BaseSum.
We also observed that the best performance was obtained when the contribution of the CentroidScore in the SummaryScore was only 10%. In fact, when the CentroidScore formed 0% of the SummaryScore the overall precision remained unaltered but the overall ROUGE score declined. The impact of the CentroidScore on the selection of summary sentences will be studied further in the future.
There are, however, certain limitations to our study. The current results are based on only 44 biomedical figures. Although this is a small number of figures, the results still indicate that our approach yields summaries that are closely related to the information deemed important by experts for explaining the contents of these figures. Another limitation is that FigSum does not take into account the semantics of the sentence. Additionally, the performance of BaseSum indicates that position of the sentence relative to a sentence referring to the figure might be useful; however, this feature was not explored in this study. To overcome these limitations, we intend to expand our study by annotating more articles to establish a better a foundation for our conclusions and use the above-mentioned features to potentially improve the performance of the system.
Our approach generates summaries by extracting sentences from articles. Hence it is not immune to the inherent problem of extractive summaries in that certain sentences do not make sense when taken out of context. In the future, we will explore methods for limiting the selection of sentences that are difficult to understand out of context.
The summaries generated by our system are available via our biomedical figure search engine – http://figuresearch.askhermes.org/.
8. Acknowledgements
We acknowledge the support of 1R21RR024933-01A1, 5R01LM009836-02, and the University of Wisconsin-Milwaukee’s RGI in 2007–2008, all to Hong Yu. We would like to thank Dr. Lamont Antieau for proofreading the manuscript.
References
- 1.Yu H, Lee M. Accessing bioscience images from abstract sentences. Advances in Neural Information Processing Systems. 2006;22(14):e547–556. doi: 10.1093/bioinformatics/btl261. [DOI] [PubMed] [Google Scholar]
- 2.Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of biomedical text mining: current progress. Brief Bioinform. 2007;8(5):358–75. doi: 10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H, Agarwal S, Johnston M, Cohen A. Are figure legends sufficient? Evaluating the contribution of associated text to biomedical figure comprehension. Journal of Biomedical Discovery and Collaboration. 2009;4(1):1. doi: 10.1186/1747-5333-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H. Towards Answering Biological Questions with Experimental Evidence: Automatically Identifying Text that Summarize Image Content in Full-Text Articles. AMIA Annu Symp Proc. 2006:2006834–838. [PMC free article] [PubMed] [Google Scholar]
- 5.Ling X, Jiang J, He X, Mei Q, Zhai C, Schatz B. Generating gene summaries from biomedical literature: A study of semi-structured summarization. Information Processing & Management. 2007;43(6):1777–1791. [Google Scholar]
- 6.Reeve L, Han H, Brooks AD. BioChain: lexical chaining methods for biomedical text summarization; Proceedings of the 2006 ACM symposium on Applied computing; Dijon, France: ACM; 2006. pp. 180–184. [Google Scholar]
- 7.Fiszman M, Rindflesch TC, Kilicoglu H. Abstraction summarization for managing the biomedical research literature; Proceedings of the HLT/NAACL 2004 Workshop on Computational Lexical Semantics; 2004. pp. 76–83. [Google Scholar]
- 8.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. Journal of Biomedical Informatics. 2003;36(6):462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal S, Yu H. Automatically Classifying Sentences in Full-Text Biomedical Articles into Introduction, Methods, Results and Discussion. In Proceedings of the AMIA Summit on Translational Bioinformatics. 2009. [PMC free article] [PubMed]
- 10.Radev DR, Jing H, Stys M, Tam D. Centroid-based summarization of multiple documents. Information Processing & Management. 2004;40(6):919–938. [Google Scholar]
- 11.van Rijsbergen CJ, Robertson SE, Porter MF. New models in probabilistic information retrieval. Cambridge, England: Computer Laboratory, University of Cambridge; 1980. [Google Scholar]
- 12.Zimmermann N, Colyer JL, Koch LE, Rothenberg ME. Analysis of the CCR3 promoter reveals a regulatory region in exon 1 that binds GATA-1. BMC Immunol. 2005:67. doi: 10.1186/1471-2172-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hovy E, Lin C. Automated Text Summarization in SUMMARIST. In ACL/EACL Workshop On Intelligent Scalable Text Summarization. 1997.
- 14.Kim J, Ohta T, Tateisi Y, Tsujii J. GENIA corpus--semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(Suppl 1):i180–2. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- 15.Lin C. ROUGE: A package for automatic evaluation of summaries Proceedings of the ACL Workshop: Text Summarization Braches Out 2004. 2004:74–81. [Google Scholar]
- 16.nist. DUC 2007: Task, Documents, and Measures. DUC 2007: Task, Documents, and Measures. 2007
- 17.Nomoto T, Matsumoto Y. Data Reliability and Its Effects on Automatic Abstracting. In Proceedings of the Fifth Workshop on Very Large Corpora. 1997.
- 18.Salton G, Singhal A, Mitra M, Buckley C. Automatic text structuring and summarization. Information Processing & Management. 1997;33(2):193–207. [Google Scholar]