Abstract
Identifying, tracking and reasoning about tumor lesions is a central task in cancer research and clinical practice that could potentially be automated. However, information about tumor lesions in imaging studies is not easily accessed by machines for automated reasoning. The Annotation and Image Markup (AIM) information model recently developed for the cancer Biomedical Informatics Grid provides a method for encoding the semantic information related to imaging findings, enabling their storage and transfer. However, it is currently not possible to apply automated reasoning methods to image information encoded in AIM. We have developed a methodology and a suite of tools for transforming AIM image annotations into OWL, and an ontology for reasoning with the resulting image annotations for tumor lesion assessment. Our methods enable automated inference of semantic information about cancer lesions in images.
Introduction
Assessment of tumor burden, the amount of cancer in the body, is a central task in cancer research and clinical practice. Tumor burden is prognostic at baseline, and it can be used to assess tumor response to treatment through serial evaluation of changes in the size of lesions that make up the tumor burden. Radiological imaging is crucial for evaluating tumor burden, because it permits physicians to identify and track cancer lesions and to evaluate their visual features. These imaging features may be descriptive, qualitative or quantitative.
The cancer clinical trial community has developed objective criteria to quantify the change in tumor burden with treatment in order to evaluate the efficacy of novel therapeutics in patient cohorts (1–3). These criteria specify the types of measurement modalities and techniques, classification of measurable and non-measurable disease, calculations for estimating the total tumor burden and change in tumor burden, and classification of tumor response to treatment. Application of these criteria to assess an individual patient’s response to treatment requires reasoning over the imaging features of tumor lesions. There are several types of specific reasoning tasks including classification and calculation. For example, response criteria specify that lesions must be classified as measurable or non-measurable disease. In the RECIST criteria (1), liver lesions with a longest diameter greater than or equal to 10 mm at baseline are classified as measurable disease and those less than 10 mm as non-measurable disease. This classification task requires knowledge of the location of the lesion and calculation of its length. A subset of the measurable disease is then classified as target lesions. Tumor burden is estimated by summing the longest dimension of the target lesions at baseline prior to the start of treatment and again at serial follow-ups after treatment has begun. Evaluation of the percent change in tumor burden from baseline yields a quantitative response rate that can then be further classified into response categories (such as complete response, partial response, stable disease, and progressive disease) given a set of thresholds.
In the current clinical research workflow for evaluating tumor burden, the radiologist identifies cancer lesions and records detailed measurements on the lesions as image annotations, summarizing the results in a textual report. The oncologist then reviews and manually extracts the information about the location and size of tumor lesions from the report and image annotations, and records the information in a flow sheet. Information in this flow sheet is then used for the tumor burden and response rate calculations. This workflow is cumbersome and error-prone (4); it could be automated if the semantics of the imaging features in the image annotations were explicit and machine-accessible; image annotations currently provide only a graphical presentation of the imaging information, but they lack semantic meaning, so machines cannot process them.
The Annotation and Image Markup (AIM) Project (5) of the National Cancer Institute’s cancer Biomedical Informatics Grid (caBIG) has recently developed an information model that describes the semantic contents in images. AIM provides an XML schema that describes the anatomic structures and visual observations in images utilizing the RadLex terminology. Information about image annotations is recorded in AIM as XML compliant with the AIM schema, enabling the consistent representation, storage, and transfer of the semantic meaning of imaging features. Tools are being developed to collect image annotations in AIM format (6).
Creating image annotations in AIM could enable the development of agents to automate the assessment of tumor burden. However, AIM does not represent annotations in a form that is directly suitable for reasoning: it provides a transfer and storage format only. There are currently no semantic reasoning methods for making inferences about cancer lesions from AIM-encoded image annotations. Our goal is to develop knowledge-based reasoning methods for automated calculation and classification of tumor response from images. In this paper we describe a methodology and suite of tools for semantic reasoning over AIM image annotations for the particular task of tumor assessment.
Methods
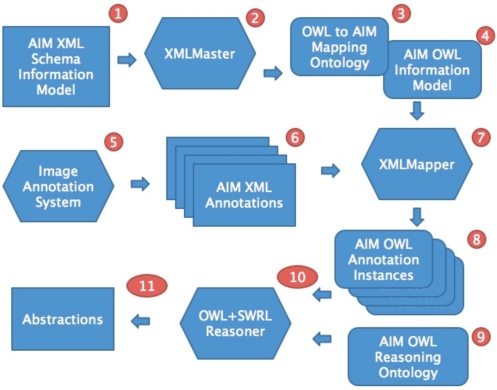
Our approach comprises three tasks: (1) produce an OWL equivalent of the AIM XML-based information model, (2) develop a mechanism to transform annotations from the AIM XML documents to instances in the OWL model, and (3) define and implement reasoning tasks that use these OWL instances. The components of these tasks are shown in Figure 1.
Figure 1:
Outline of process to take AIM image annotations from an imaging system and to perform OWL and SWRL-based reasoning with those annotations.
Transforming the AIM Information Model to OWL
To enable ontology-based reasoning, we transformed the AIM information model, which is described by XML Schema, into an ontological representation that defines a semantically equivalent information model. This model can both represent all the concepts in the AIM XML, and it can be used to store OWL instances of AIM annotations. Domain-level inferences can then be defined using this representation. This transformation was performed by creating classes and properties in the OWL information model that correspond to respective components in AIM. Our goal was to produce an OWL information model that is user-understandable and is suitable for inference.
We developed a tool called XMLMaster (7) to define this transformation. XMLMaster was written as a plugin to the popular Protégé-OWL ontology development environment (8) and provides a graphical user interface that allows users to interactively define mappings between entities in an XML document and concepts in an OWL ontology. It can be used to define mappings between an XML model and an existing OWL ontology or can generate a new OWL ontology as the target of these mappings. We used this latter mode to create an OWL AIM information model.
Transforming AIM XML Annotations to OWL Instances
The second step is to define a mechanism to transform existing AIM XML documents to their equivalent annotations encoded using the OWL information model. We developed a tool called XMLMapper to perform this task. XMLMapper uses the mappings defined by users of XMLMaster when they are specifying an XML–to-OWL transformation. These mappings are stored by XMLMaster in a mapping ontology and they contain a specification of how entities can be mapped from an XML document to instances in an OWL ontology. XMLMapper uses this mapping ontology to automatically transform XML documents to OWL ontologies. It can process streams of XML documents and populate an OWL knowledge base with the resulting transformed content.
AIM OWL Reasoning Ontology
Once AIM annotations are represented in OWL, we are in a position to develop ontology-based reasoning mechanisms to work with these annotations. For example, a reasoning task could be to classify all liver lesions greater than a particular size in a set of image annotations. This task would require structural information from referenced anatomical terminologies in addition to mass calculations on image data stored in annotations to generate the necessary inferences. A wide array of reasoning tasks of this type can be defined to support rich inferences over image annotations. As described below, the reasoning tasks in our particular use case obtain semantic information about cancer lesions in images.
We used OWL and its associated rule language SWRL (9) to define a reasoning ontology necessary to make inferences about imaging findings. SWRL allows users to write rules that can be expressed in terms of OWL concepts to provide more powerful deductive reasoning capabilities than OWL alone. Semantically, SWRL is built on the same description logic foundation as OWL and provides similar strong formal guarantees when performing inference. We used the Protégé-OWL (8) ontology authoring environment and its associated SWRLTab plugin (10) for developing the reasoning ontology. To support ontology querying we used a language that we have developed called SQWRL (11). SQWRL (Semantic Query-Enhanced Web Rule Language) is a SWRL-based query language that can be used to query OWL ontologies. SQWRL provides SQL-like operations to format knowledge retrieved from an OWL ontology.
The reasoning ontology (Figure 2) and SWRL rules (Figure 3) were developed to enable our task of automated reasoning about imaging findings based on image annotations. Specifically, the ontology and rules were developed to enable calculating the length of image findings and classifying the image findings as measurable and non-measurable.
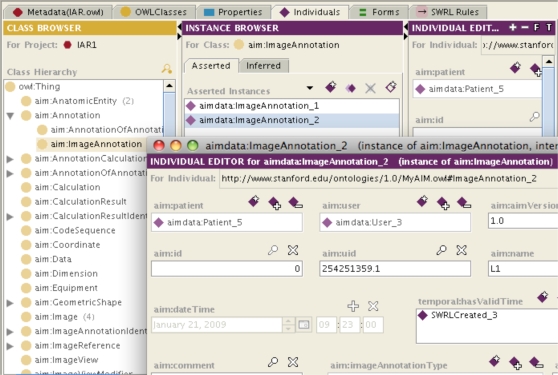
Figure 2:
A portion of the AIM ontology in the left hand panel. Two instances of AIM image annotations are shown in the middle panel, and details of the properties of one instance are shown in the window in the lower right.
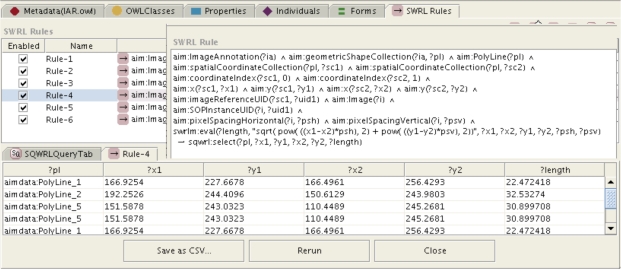
Figure 3:
Example SQWRL query to calculate length of line from (x,y) coordinates and pixel spacing information, with results shown in the panel below. The length is shown in the last column in the lower panel. An equivalent SWRL rule is used to assert these lengths into an OWL knowledge base for further reasoning.
Reasoning about Cancer Lesions
We evaluated our system in our use case related to reasoning about cancer lesions for estimating tumor burden. For this use case, we annotated 116 images from 10 cancer patients who had serial imaging studies. Lesions were annotated in AIM format. The AIM files were input into our system for processing to generate inferences about tumor burden. The results of these inferences were evaluated for face validity by an oncologist.
Results
We successfully transformed the AIM information model into OWL. Figure 2 shows a portion of the AIM ontology, two instances of AIM image annotations and a subset of the properties of one of the image annotations. Thus, the semantic image information from AIM is accessible for computer reasoning.
The image annotations we acquired for our use case were processed by our system to perform automated reasoning about the image findings. Our SWRL rules executed two subtasks: 1) calculation of the length of each image finding from pixel coordinates, and 2) classification of image findings as measurable and non-measurable using a combination of semantic information about the location and type of finding, and its calculated length.
For the first subtask, the length of each finding was calculated from the pixel (x, y) coordinates for a line, and information about the height and width of the pixels in the image using the Pythagorean theorem. Figure 3 shows an example of a SWRL rule and its output for calculating the length in this way.
For the second subtask of classification of findings as being measurable or non-measurable, several new classes were added to the OWL Reasoner and several rules were invoked. New OWL classes include concepts such as Long Axis, Short Axis, Pathologic Finding, Measurable Disease and Non-measurable Disease. Rules for this subtask include: 1) classification of the longest diameter from a pair of length annotations for a lesion; 2) classification of findings as pathologic or non-pathologic based on the imaging observation in the image annotation; these observations consist of coded terms in the AIM terminology, including mass, nodule, lesion, effusion, or children of those concepts; 3) temporal classification of the valid time of lesions as the relative time points of baseline or follow-up assessments based on their temporal relationship to the start of therapy; and 4) classification of image findings on baseline images as measurable if their observation is a mass or nodule greater than or equal to 10 mm in longest diameter, otherwise all other baseline pathologic findings classified as non-measurable.
The inferences from our system were reviewed by an oncologist who confirmed that they were correct based on the raw image annotation information. In qualitative terms, the oncologist believed our system will streamline the process of evaluating tumor burden.
Discussion
Estimating tumor burden is an important clinical and research task in oncology. Currently, this is a manual process that involves evaluation of multiple imaging features of tumor lesions including the lesion’s location, type (mass or effusion), and size. This evaluation results in further classification of imaging findings according to the rules of response criteria in order to estimate tumor burden. These evaluations, being pre-determined and labor-intensive, are opportune for automation if the image information is machine-accessible.
Recent standardization efforts have produced computer-interpretable image annotations that can be used in software systems to automate some of these reasoning tasks. The AIM information model, which was developed by caBIG, is one of the primary results of these activities. In this paper, we have outlined an approach to perform ontology-based reasoning with AIM image annotations. Our approach will ultimately be used to automate the task of estimating tumor burden. We implemented the reasoning services for two of the subtasks needed to estimate tumor burden: calculating the length of imaging findings and classifying them as measurable or non-measurable. We are currently implementing the remaining two subtasks needed to estimate tumor burden: classification of measurable disease into a subset of target lesions, and calculation of the sum of longest diameters of the target lesions. These four reasoning subtasks will provide an automated estimate of the tumor burden according to the RECIST (2) criteria on imaging studies at baseline and follow-up. This information will enable oncologists to calculate and classify the response of patients on cancer treatment.
A limitation of our method is that it requires maintenance of an ontology that corresponds to the AIM information model. The AIM information model is not complex, so this is not a prohibitively expensive task. Our method also requires construction and maintenance of SWRL rules. Since the response criteria do not change frequently, but are applied often, we believe creating this infrastructure is worthwhile.
A benefit of our approach is that it establishes an automated workflow, taking AIM XML image annotations produced by an imaging system, transforming them to OWL, and reasoning with those annotations to generate inferences necessary for the domain task. In addition, our work is extensible by extending our ontology or rules. As more tools for creating AIM annotations are developed, our approach will be able to process those image annotations, since our infrastructure complies with the AIM standard. Furthermore, as the reasoning tasks over image annotations continue to be extended, we expect our rules and ontology to more broadly support the task of automated assessing tumor response to treatment.
We believe our approach can be generalized to other types of image-based reasoning use cases beyond the cancer use case described through extension of the reasoning ontology and rules. In addition, while our discussion is specific to the AIM information model, this approach and the associated tools can be used to take any XML-based information model, generate its OWL equivalent, and then reason over it to produce high-level abstractions.
References
- 1.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2008;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 3.Cheson BD, Pfistner B, Juweid ME, Gascoyne RD, Specht L, Horning SJ, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25:579–86. doi: 10.1200/JCO.2006.09.2403. [DOI] [PubMed] [Google Scholar]
- 4.Levy MA, Rubin DL. Tool support to enable evaluation of the clinical response to treatment. AMIA Annu Symp Proc. 2008:399–403. [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin DL, Mongkolwat P, Kleper V, Supekar K, Channin D. Medical imaging on the Semantic Web: Annotation and image markup; AAAI Spring Symposium Series, Semantic Scientific Knowledge Integration; 2008. [Google Scholar]
- 6.Rubin DL, Rodriguez C, Shah P, Beaulieu C. iPad: Semantic Annotation and Markup of Radiological Images. AMIA Annu Symp Proc. 2008:626–30. [PMC free article] [PubMed] [Google Scholar]
- 7.http://protege.cim3.net/cgi-bin/wiki.pl?XMLMasterPlugin
- 8.Knublauch H, Fergerson RW, Noy NF, Musen MA. The Protege OWL plugin: An open development environment for semantic web applications. Lecture Notes in Computer Science. 2004:229–43. [Google Scholar]
- 9.Horrocks I, Patel-Schneider PF, Boley H, Tabet S, Grosof B, Dean M.SWRL: A semantic web rule language combining OWL and RuleML. W3C Member Submission. 200421
- 10.O Connor M, Knublauch H, Tu S, Grosof B, Dean M, Grosso W, Musen M. Supporting rule system interoperability on the semantic web with SWRL. Lecture notes in computer science. 2005;3729:974. [Google Scholar]
- 11.http://protege.cim3.net/cgi-bin/wiki.pl?SQWRL