Abstract
Our aim was to identify asthmatic patients as cases, and healthy patients as controls, for genome-wide association studies (GWAS), using readily available data from electronic medical records. For GWAS, high specificity is required to accurately identify genotype-phenotype correlations. We developed two algorithms using a combination of diagnoses, medications, and smoking history. By applying stringent criteria for source and specificity of the data we achieved a 95% positive predictive value and 96% negative predictive value for identification of asthma cases and controls compared against clinician review. We achieved a high specificity but at the loss of approximately 24% of the initial number of potential asthma cases we found. However, by standardizing and applying our algorithm across multiple sites, the high number of cases needed for a GWAS could be achieved.
Introduction
The NUgene Project(1) is a biobank of DNA samples coupled to electronic medical record (EMR) data from participating patients at Northwestern affiliated medical centers. This research initiative is a partnership between Northwestern’s Feinberg School of Medicine (FSM), Northwestern Memorial Hospital (NMH), and Northwestern Medical Faculty Foundation (NMFF). Participants’ DNA samples are combined with self-reported questionnaire data, completed at enrollment, and longitudinal health data from participant EMRs. Participants consent to use of their coded DNA samples and data by researchers to examine the role genes play in the development, progression, and treatment of common diseases.
Leveraging our campus-wide NUgene resource, Northwestern has joined the national Electronic Medical Records and Genomics (eMERGE)(2) network, a consortium formed to investigate how EMR linked DNA biorepositories can be leveraged for genomics and informatics science (http://www.gwas.net). One of the main goals of eMERGE is to assess whether EMRs provide suitable data to identify individuals with specific phenotypes for downstream GWAS.
We proposed asthma as one of the phenotypes for a GWAS with eMERGE for several reasons. First, asthma is the most prevalent chronic disease in the U.S.(3), affecting an estimated 7.2%(4) of the adult U.S. population and accounting for approximately $12.7 billion in health care costs(5). Second, asthma has also been the subject of several genetics studies, which provides the basis for a comparison between the results of genome wide association studies performed on EMR based phenotypes with those resulting from more traditionally derived case-control studies. Lastly, asthma is an example of a disease with a range of diagnostic criteria, often not easily captured in an EMR; therefore, we selected asthma to test the feasibility of using an automated algorithm given these limitations.
To develop our algorithms, we utilized EMR data contained within the Northwestern Medicine Enterprise Data Warehouse (EDW)(6). The EDW is an integrated repository of clinical data and biomedical research data sources from FSM, NMFF, and NMH. The core data sources include Cerner Millennium (PowerChart, RadNet, etc.), EpicCare® Ambulatory EMR, GE Centricity (outpatient billing and scheduling for NMFF), and PRIMES (inpatient billing). Other systems in the EDW include numerous smaller specialized clinical and research databases. Most EDW data is synchronized nightly with its source systems.
Methods
Asthma is diagnosed by either a 12% reversibility in FEV1 (Forced Expiratory Volume in 1 second) (≥200ml) from a spirometric test, after administration of a short-acting bronchodilator, or by hyperreactivity to methacholine. Neither is routinely assessed in the clinical setting. To develop a more pragmatic approach using commonly captured EMR data, we used diagnostic codes, medications, and discrete smoking history data from the EMR, supplemented with data from our self-report health questionnaire.
As the combination of ICD-9 (International Classification of Diseases, Ninth Revision) diagnoses and medications from the EMR has been shown to successfully identify asthmatics(7), and ICD-9 codes are a standard that is easily shared among sites, we extracted all patients with an ICD-9 code of 493.xx, as well as all patients on asthma medications (Table 1) at any time. To ensure that we were only capturing medications used for asthma, we included medications with a route of administration used for asthma (ex. inhaled), or with a trade name that implied a route used for asthma (Table 1).
Table 1.
Asthma Medications: route and trade names are listed only for those medications where we needed to use them
| Generic | Trade Name(s) |
|---|---|
| Relievers | |
| - Short-acting bronchodilators (SABA): [route: Inhalers, Nebulizers & oral] | |
| Albuterol | Ventolin, Proventil, ProAir, Accuneb |
| Pirbuterol | Maxair |
| Levalbuterol | Xopenex |
| Terbutaline | Brethine |
| Old Controllers | |
| - Methylxanthines: [route: PO (oral), IV, injections] | |
| Theophylline or aminophylline | Slo-Bid, Theo, Theodur, Theolair, Uniphyl |
| - Mast cell stabilizers: [route: Inhalers & Nebulizers] | |
| Cromolyn | Intal |
| Nedocromil | Tilade |
| Controllers | |
| - Inhaled corticosteroids (ICS): [route: Inhalers & Nebulizers] | |
| Beclomethasone | QVar |
| Budesonide | Pulmicort |
| Fluticasone | Flovent |
| Flunisolide | Aerobid |
| Triamcinolone | Azmacort |
| Mometasone | Asmanex |
| - Long-acting bronchodilators (LABA): | |
| Salmeterol | Serevent |
| Formoterol | Foradil |
| Arformoterol | Brovana |
| - ICS + LABA Combinations | |
| - Leukotriene Antagonists (LTAs): | |
| Montelukast, Zafirlukast, Zileuton | |
| - Oral Corticosteroids: | |
| Prednisone | Orasone |
| Prednisolone | Medrol |
| Dexamethasone | Decadron/Deltasone |
| - Anti IgE: Omalizumab | |
We tried two different algorithms. Initially, we extracted all diagnoses in the EMR, including billing diagnoses. We required an asthma diagnosis and a single use of an asthma medication, which could be on the same date as the diagnosis.
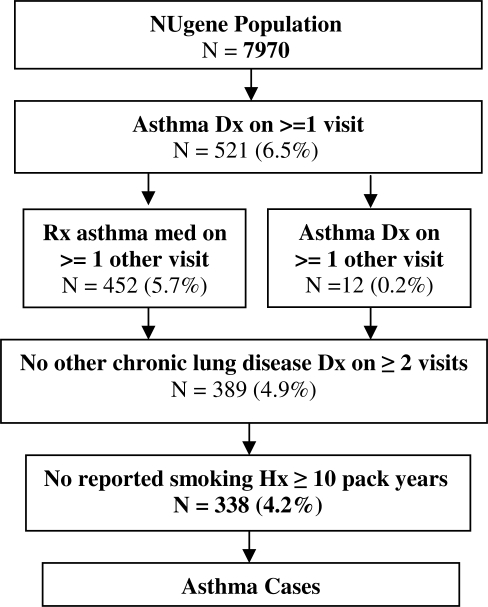
In our final algorithm, depicted in Figure 1, we used only the clinician entered diagnoses from outpatient encounters and problem lists. In addition to the initial diagnosis of asthma, we required that the patient have an asthma diagnosis or medication prescription on at least one additional date. We further required that patients receiving a prescription for an oral corticosteroid also have a prescription for a long or short acting beta agonist, or an inhaled corticosteroid.
Figure 1.
Algorithm for the Identification of Subjects with Asthma.
For both algorithms, we excluded patients with other potentially overlapping or chronic lung diseases; specifically, those diagnosed more than once (one time acute episodes were deemed acceptable) with the diseases listed in Table 2. We excluded these subjects as some of these diseases either use the same medications, or can mask or mimic the symptoms of asthma. We also excluded patients with a smoking history greater than or equal to 10 pack-years (packs/day multiplied by years used). We calculated pack-years as both number of packs/day and years smoked are recorded separately in discrete fields in our EMR and our self-administered questionnaire. We used the discrete values in the EMR in combination with the self-reported data, and if recorded in both places, we used the maximum of the 2 values, in order to incorporate the most recent estimate of years smoked.
Table 2.
ICD-9 codes for other lung diseases.
| Respiratory disease to exclude | ICD9 codes |
|---|---|
| Cystic fibrosis | 277.xx |
| Chronic pulmonary heart disease | 416.xx |
| Vocal cord dysfunction | 478.3x |
| Bronchitis, Emphysema | 490.xx–492.xx |
| Bronchiectasis, Allergic alveolitis, Chronic airway obstruction | 494.xx–496.xx |
| Pneumoconiosis | 500.xx–508.xx |
| Other respiratory disease | 510.xx–519.xx |
| Respiratory distress syndrome | 769.xx |
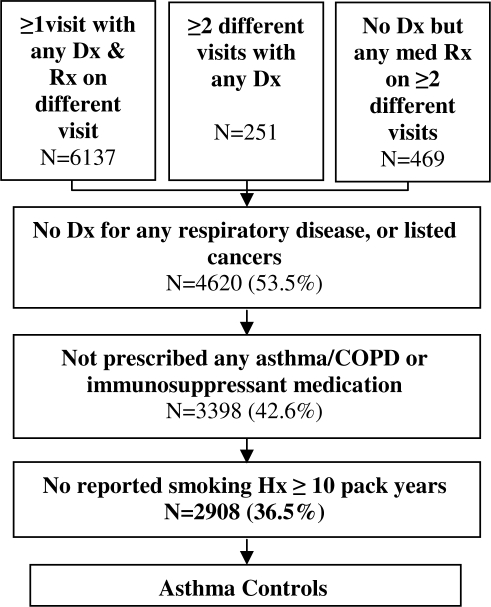
We similarly developed two control algorithms. Initially, we excluded patients with an asthma diagnosis (ICD-9 codes 493.xx), or any of the chronic conditions in Table 2. We also excluded patients with any asthma/COPD medication (Table 1). Lastly, as with cases, we excluded those patients with a smoking history ≥ 10 pack years.
For our final control algorithm we added a number of requirements. We required that controls have a minimal amount of information in the EMR equivalent to the data required for cases. Specifically, we required that they have diagnoses and/or prescriptions on at least two different dates. We additionally excluded patients with diagnoses of lung, bronchial, tracheal, or pleural cancers (ICD-9 codes 162.xx–163.xx); or hemapoeitic cancers (ICD-9 codes 200.xx–208.xx). Patients with these cancers were excluded for 2 reasons: 1) they often take prednisone as part of chemotherapy; and 2) to be consistent with the algorithm to identify patients with a healthy respiratory system, thus avoiding selection bias. For medications, we added immunosuppressants to our exclusion list as these can mask asthma symptoms. Figure 2 depicts the final algorithm for choosing potential controls.
Figure 2.
Algorithm for the Identification of Asthma Controls.
To validate the results of our algorithms, two clinicians (PA, AK) conducted a blinded review of 100 charts identified as cases and controls for both algorithms. We also reviewed charts of patients excluded as either cases or controls in order to completely assess specificity and sensitivity. Using clinician chart review as a gold standard, we generated a positive predictive value (PPV) and negative predictive value (NPV) for both of these iterations of the automated algorithms. Statistical analysis was performed using R(8), specifically the epiR(9) package, which provides functions for calculating statistics and confidence intervals (CI) for 2x2 tables.
Results
Out of 7,790 patients in our NUgene population with an EMR we found 338 cases of asthma (4.2%), and 2,908 potential controls, using our final algorithms described above and depicted in Figures 1 and 2. The final number of cases is significantly lower than our earlier algorithm, where we found 445 cases (6%). But, from blinded physician chart review, the final PPV was 95% (CI: 86.4 – 99.3), only 2 cases were found to be misclassified; and our NPV was 96% (CI: 89.2 – 99.4), only 2 controls were misclassified. This compared favorably with the 70% PPV (CI: 56.7 – 81.3) of the less stringent algorithm allowing use of billing codes and fewer inclusion criteria. Clearly non-overlapping 95% confidence intervals indicate a statistically significant improvement in PPV for the final algorithm. We determined the PPV and NPV for both algorithms as shown in Tables 3.1 and 3.2. The breakdown of the number of cases and controls at each step of the final case and control algorithms are also depicted in Figures 1 and 2 respectively.
Table 3.1.
Initial algorithm PPV & NPV.
| Predictive Value | Chart Review | |||
|---|---|---|---|---|
| Case | Not Case | Total | ||
| EMR | Case | 35 | 15 | 50 |
| Control | 0 | 50 | 50 | |
| Total | 35 | 65 | 100 | |
PPV =35/(35+15)=0.70; NPV = 50/(0+50)=1.0
Table 3.2.
Final algorithm PPV & NPV.
| Predictive Value | Chart Review | |||
|---|---|---|---|---|
| Case | Not Case | Total | ||
| EMR | Case | 42 | 2 | 44 |
| Control | 2 | 54 | 46 | |
| Total | 44 | 56 | 100 | |
PPV = 42/(42+2)=0.95; NPV = 54/(2+54)=0.96
Discussion
Published efforts to identify asthmatics in the EMR took similar approaches to ours. One study(10) used similar criteria such as use of asthma medications; number and type of encounters; and asthma diagnoses, from multiple databases, over a span of 2 years in order to identify more prevalent cases of asthma, achieving an overall PPV of 89%. As our EDW contains data from multiple databases that spans 10 years or more in some cases, with the average patient having 4 years of data, we chose to take advantage of this and search over the span of our entire EMR for our patients. Searching over a longer span of time, using a larger number of medications, and excluding other lung diseases and heavy smokers helped to improve our algorithm’s performance.
Through an iterative process, we developed an algorithm with a 95% PPV. Stricter algorithms requiring that a patient have both a diagnosis and be taking a medication ruled out too many potential cases, especially those with milder asthma that do not require medication. Less strict algorithms had a much lower PPV: for example, the initial case algorithm described above allowed all diagnoses and medications on the same date, resulting in a PPV of only 70%. Chart review indicated that such an approach included cases with an initial diagnosis of asthma that was later ruled out.
The advantage of our algorithm over manual chart review is enormous. Since it took up to 2 minutes to review the medical records of one patient, it would have taken 8 hours/day for 33 days to manually survey and apply our algorithm to the medical records of all 7,970 NUgene subjects. Once developed, the current electronic algorithm takes less than 1 minute to run across all subjects, and is readily repeatable at little cost.
We also identified and addressed a number of specific challenges. Identifying controls was a particularly difficult task. We needed to prevent contamination of our control group with cases, i.e., we needed to ensure our control group truly did not have asthma or similar diseases, as lack of a diagnosis, prescribed medications, or other data in the EMR does not necessarily imply that the patient does not have a given disease. Also, as a tertiary care center in a major metropolitan area, our EMR had sparse data for some patients due to infrequent visits only with specialists. To address these challenges, we required controls have a minimal amount of data.
In addition, many asthma medications are used for other conditions, such as other obstructive diseases (E.g. COPD (Chronic Obstructive Pulmonary Disease)). Prednisone may be used for a broad range of diseases, and therefore may not useful for finding cases of asthma, although it can be used cautiously as a marker of asthma exacerbations or severity of disease. To ensure medications prescribed in the EMR are truly used for asthma, we realized we could not use only generic ingredients for some medications (example: fluticasone propionate in inhaler form is the asthma medication Flovent, and in a nasal spray is Flonase for allergic rhinitis). The diagnosis associated with the ordered medication or its route of administration were logical data to enhance generic names, but often unavailable, as these are optional fields in our EMR (i.e., route is implied by the name of the drug in some cases, and a diagnosis is not required to order a medication). Lack of standardization across EMRs also posed a challenge. Our EDW contains medications from both inpatient and outpatient systems which use different medication vocabularies: Multum and Medispan. These two vocabularies were linked together with RxNORM(11) using the generic substance as the common link. Since pharmaceutical class cannot be determined from generic substance alone, for the medications with different uses, we used trade name where possible or generic name in combination with route or class if available.
Lastly, despite the fact that we had discrete fields for smoking history, it was still difficult to extract and not necessarily complete. For instance, within the discrete fields in the EMR, smoking history was filled out, completely or partially, in only 60% of EMR records. Not only was the data somewhat sparse, in some charts in the EMR when the discrete smoking fields were filled, the data was often not strictly numeric (ex. “+/− 5”, “>5”), requiring pattern matching to strip out these non-numerical characters.
Smoking history can also be in the EMR in the text of a note from a patient encounter. One study(12) used natural language processing (NLP) to extract information from text notes for known asthmatics, achieving 90% accuracy in extracting smoking status, and also achieving 82% accuracy in extracting principal diagnosis. Using NLP to elicit smoking history, more detailed diagnoses, medication usages, and even asthma exacerbations may increase our number of cases and allow us to characterize the cases in terms of asthma severity in the future. As we are in the exploratory stages of using NLP to extract such data from free text notes, we did not have this data to supplement our final algorithm.
We developed a similar algorithm (using diagnoses and medications, as well as lab values) for type 2 diabetes that has been successfully implemented at another eMERGE institution, despite that fact that their EMR requires NLP for extraction of at least some discrete data. As a result, we are currently pooling our DNA samples across sites to conduct a GWAS of type 2 diabetes. We plan on similarly applying the asthma algorithm. Additional research is needed to determine if standards for document sharing (such as the Continuity of Care Document, http://www.hl7.org) can further expedite cross-institutional sharing of phenotypic data.
Conclusion
In conclusion, we describe a practical approach to the identification of asthma cases and controls for GWAS using data captured in routine clinical care in commercial EMRs. To achieve the high specificity required for GWAS, we applied stringent criteria (ex. no billing diagnoses), tough temporal criteria, and dove deep in the data to find the flaws. Although overall number of cases decreases with the increased specificity needed for GWAS, we believe standardizing this algorithm across diverse EMRs holds great potential to identify the largest number of cases and controls needed for well powered GWAS.
Acknowledgments
Funding for this study is provided by the National Human Genome Research Institute (NHGRI), U01-HG-004609; and an Ernest S. Bazley Grant.
References
- 1.The NUgene Project Available from: http://www.nugene.org
- 2.Wolf WA, Chisholm RL, Chute CG, Jarvik G, Larson E, Masys DR, et al. The eMERGE Network: A national consortium of electronic health record-linked biobanks furthering largescale genetic research [#2129]. Available from: http://www.ashg.org/2008meeting/abstracts/fulltext/ The American Society of Human Genetics; November 12, 2008; Philadelphia, PA.
- 3.Mannino DM, Homa DM, Akinbami LJ, Moorman JE, Gwynn C, Redd SC. Surveillance for asthma--United States, 1980–1999. MMWR Surveill Summ. 2002 Mar 29;51(1):1–13. [PubMed] [Google Scholar]
- 4.Moorman JE, Rudd RA, Johnson CA, King M, Minor P, Bailey C, et al. National surveillance for asthma--United States, 1980–2004. MMWR Surveill Summ. 2007 Oct 19;56(8):1–54. [PubMed] [Google Scholar]
- 5.Weiss KB, Sullivan SD. The health economics of asthma and rhinitis. I. Assessing the economic impact. J Allergy Clin Immunol. 2001 Jan;107(1):3–8. doi: 10.1067/mai.2001.112262. [DOI] [PubMed] [Google Scholar]
- 6.The Northwestern Medicine Enterprise Data Warehouse. Available from: http://edw.northwestern.edu/About.aspx
- 7.Donahue JG, Weiss ST, Goetsch MA, Livingston JM, Greineder DK, Platt R. Assessment of asthma using automated and full-text medical records. J Asthma. 1997;34(4):273–81. doi: 10.3109/02770909709067217. [DOI] [PubMed] [Google Scholar]
- 8.R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 9.Stevenson M, Nunes T, Sanchez J, Thornton R. epiR: Functions for analysing epidemiological data. R package version 0.9-11 ed2008
- 10.Vollmer WM, O'Connor EA, Heumann M, Frazier EA, Breen V, Villnave J, et al. Searching multiple clinical information systems for longer time periods found more prevalent cases of asthma. J Clin Epidemiol. 2004 Apr;57(4):392–7. doi: 10.1016/j.jclinepi.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 11.RxNORM. Available from: http://www.nlm.nih.gov/research/umls/rxnorm/
- 12.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]