Abstract
Creating mappings between concepts in different ontologies is a critical step in facilitating data integration. In recent years, researchers have developed many elaborate algorithms that use graph structure, background knowledge, machine learning and other techniques to generate mappings between ontologies. We compared the performance of these advanced algorithms on creating mappings for biomedical ontologies with the performance of a simple mapping algorithm that relies on lexical matching. Our evaluation has shown that (1) most of the advanced algorithms are either not publicly available or do not scale to the size of biomedical ontologies today, and (2) for many biomedical ontologies, simple lexical matching methods outperform most of the advanced algorithms in both precision and recall. Our results have practical implications for biomedical researchers who need to create alignments for their ontologies.
Algorithms For Ontology Mapping
Ontologies in biomedicine facilitate information integration, data exchange, search and query of heterogeneous biomedical data, and other critical knowledge-intensive tasks.1 The biomedical ontologies used today often have overlapping content. Creating mappings among ontologies by identifying concepts with similar meanings is a critical step in integrating data and applications that use different ontologies. With these mappings, we can, for example, link resources annotated with terms in one ontology to resources annotated with related terms in another ontology. By doing so, we may discover new relations among the resources themselves (e.g., linking drugs and diseases).
As part of our work for the National Center for Biomedical Ontology (NCBO), we have developed BioPortal—an online repository of biomedical ontologies.* BioPortal provides access not only to ontologies, but also to mappings between them.2 Other NCBO tools use the mappings for annotating resources with terms from different ontologies and for linking these resources to each other. Having high quality mappings for BioPortal ontologies is therefore essential for the other tools.
The Unified Medical Language System (UMLS) provides a large set of carefully constructed mappings between medical terminologies. However, UMLS provides these mappings only for a limited set of ontologies and terminologies. At the same time, researchers actively continue to extend existing ontologies and to develop new ones. In fact, most of the 130 ontologies in BioPortal are not in UMLS.
Because identifying mappings among large ontologies manually is an enormous task, developing algorithms that automatically find candidate mappings is a very active area of research.3 In recent years, researchers in computer science have developed a large number of sophisticated algorithms that use a wide variety of methods, such as graph analysis, machine learning, and use of domain-specific and background knowledge. The Ontology Alignment Evaluation Initiative (OAEI) is an annual competition, in which developers of mapping algorithms can compare the performance of their tools to others on the same set of ontologies.4 Participants compare their performance on different sets of ontologies, which vary in the domains they cover (e.g., anatomy, food) and in their structure (e.g., expressive ontologies, thesauri, or directory listings). The results of the evaluation demonstrate that the accuracy of these algorithms increases every year, with the best algorithms reaching the accuracy in the range of 80–85% for the ontologies in the tests (Figure 1). Participation in OAEI has become a de facto requirement for any researcher who claims to have a mapping algorithm that advances the state of the art.
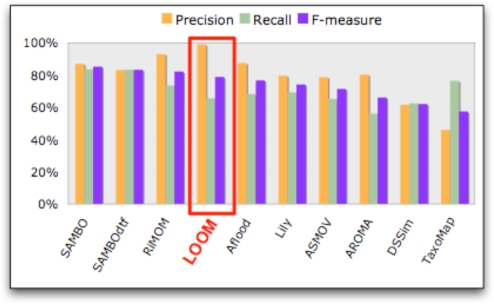
Figure 1.
Performance of LOOM and OAEI tools in the Anatomy track (Experiment 1). The graph shows the results for OAEI 2008 from the competition report4 along with LOOM results on the same set, ordered by the F-measure. The tools produced mappings for the Mouse adult gross anatomy ontology and the human anatomy part of the NCI Thesaurus. A reference alignment was used as the basis for precision and recall.
In order to generate mappings between biomedical ontologies in BioPortal, we decided to evaluate the tools that performed best in the OAEI competition and to use the best of the available tools. We also implemented a simple ontology-mapping algorithm, Lexical OWL Ontology Matcher (LOOM), to use as a baseline for comparison.
Our evaluation has demonstrated that (1) most of the advanced algorithms are either not publicly available or do not scale to the size of biomedical ontologies today, and (2) for many biomedical ontologies, simple lexical matching methods outperform most of the advanced algorithms in both precision and recall.
Methods
Implementing the LOOM algorithm
In order to have a baseline for comparison, we have developed LOOM, a simple lexical algorithm for producing mappings. LOOM takes two ontologies represented in OWL,5 a Semantic Web ontology language, and produces pairs of related concepts from the two ontologies. In order to identify the correspondences, LOOM compares preferred names and synonyms of the concepts in both ontologies. It identifies two concepts from different ontologies as similar, if and only if their preferred names or synonyms are equivalent based on a modified string-comparison function. Our string-comparison function first removes all delimiters from both strings (e.g., spaces, underscores, parentheses, etc.). It then uses an approximate matching technique to compare the strings, allowing for a mismatch of at most one character in strings with length greater than four and no mismatches for shorter strings.
Obtaining the OAEI tools
The OAEI competition had different tracks: each track had a specific set of ontologies that the tools needed to compare. In our experiments, we used the only track that had biomedical ontologies: the anatomy track. In this track, the tools had to provide mappings between the Mouse adult gross anatomy ontology and the human-anatomy part of the National Cancer Institute (NCI) Thesaurus. We tried to access, install, and run on BioPortal ontologies all the OAEI 2008 tools that performed well on the anatomy track.
Experiment 1: Using LOOM for OAEI 2008 anatomy track
To compare LOOM performance to the performance of all the tools that participated in the OAEI competition, we used LOOM on the ontologies in the OAEI anatomy track. The OAEI organizers provided a manually created reference alignment between the two anatomy ontologies.6 Figure 1 contains performance results for the OAEI tools, with this reference alignment used as a gold standard.† To compute the precision of LOOM results on the anatomy-track ontologies, we randomly selected 300 of the mappings returned by LOOM. With the help of a domain expert, we manually evaluated the correctness of these mappings. We did not use the reference alignment provided by the OAEI organizers in computing the precision because the reference alignment did not contain 4.8% of the correct mappings returned by LOOM. To be consistent with the OAEI performance results, we evaluated the recall of the LOOM results using the OAEI reference set of correct mappings.
Experiment 2: Comparing LOOM performance to AROMA
As we discuss in the Results section, only one of the tools that participated in OAEI 2008, AROMA,7 was both available for us to use and scalable to the size of the ontologies in BioPortal. Thus, we compared the performance of LOOM and AROMA on several ontologies in BioPortal (Table 2). We chose both pairs of ontologies where we expected to have high overlap, based on their domain of coverage, and pairs of ontologies where we expected low overlap. To compute the precision of LOOM and AROMA on these ontology pairs, we randomly selected 100 mappings for each ontology pair and each algorithm. With the help of a domain expert, we evaluated the correctness of each of the selected mappings by looking at the definitions of the concepts in the ontologies. Furthermore, we assessed the quality of the mappings that AROMA found, but that LOOM missed. To perform this assessment, once again we randomly selected 100 such mappings for each of the ontology pairs and evaluated the selected mappings manually. Because we did not have a complete set of mappings to use as a reference, we did not evaluate recall in experiment 2.
Table 2.
Comparison of LOOM and AROMA performance (Experiment 2).There was wide variability in the number of matches found by LOOM and AROMA. LOOM’s precision exceeded AROMA’s precision for three out of four experiments. The table also presents the number of mappings and the precision of the mappings found by AROMA, but not LOOM, and vice versa.
| Ontologies used | Number of results found | Precision | Overlap, # of results | Found by AROMA, but not LOOM | Found by LOOM, but not AROMA | ||||
|---|---|---|---|---|---|---|---|---|---|
| AROMA | LOOM | AROMA | LOOM | # of results | Precision | # of results | Precision | ||
| Mouse adult gross anatomy–Zebrafish anatomy and development | 1,080 | 235 | 25% | 99% | 135 | 945 | 4% | 100 | 98% |
| Cell type–Fungal gross anatomy | 34 | 12 | 24% | 83% | 8 | 26 | 0% | 4 | 50% |
| Cell cycle ontology (S. Pombe)–Cell cycle ontology (A. Thaliana) | 1,571 | 15,178 | 94% | 78% | 1,533 | 38 | 0% | 13,645 | 76% |
| Cell type–Cell cycle ontology (S.Pombe) | 538 | 4 | 0% | 50% | 0 | 538 | 0% | 4 | 50% |
Results
Getting the OAEI tools
We attempted to access, install, and evaluate all the tools from the OAEI 2008 anatomy track (Figure 1). For the tools that were not available for download, we contacted their authors requesting access to the tools. Of all OAEI tools, only RiMOM, Lily, and AROMA were publicly available or made available upon request. RiMOM was configured to run only on a benchmark data set. The available demo of Lily crashed due to extensive memory use when we ran it on two relatively small ontologies (fewer than 3,000 classes) on a computer with 2GB of RAM. Thus, of all the tools, we could use only the AROMA algorithm to produce mappings for the ontologies in our sample. AROMA uses statistical analysis to compare the vocabulary used to describe terms in the ontologies.
Experiment 1: LOOM performance for the OAEI 2008 anatomy track
Figure 1 compares precision, recall, and F-measure for LOOM to the tools that were used in the OAEI 2008 Anatomy track. LOOM identified 1136 mappings, with 99% precision. Its recall was 65%. The F-measure was 79%. Thus, only three of the OAEI tools—SAMBO, SAMBOdtf, and RiMOM—outperform LOOM with respect to F-measure. No OAEI algorithm had better precision than LOOM. Though we might expect the recall of our algorithm to be low since other algorithms take advantage of methods other than lexical matching, LOOM’s recall was within 10% of all but the top two algorithms.
Table 1 shows the difference in results between LOOM and other tools. Specifically, column 4 shows the number of mappings found by each algorithm that LOOM missed. The percentage of the missed mappings that were correct varies widely, ranging from 15% to 70%, with a median of 53% (standard deviation 20%). In other words, about half of the mappings that LOOM missed and other tools found were correct.
Table 1.
Difference between mappings found by LOOM and other algorithms in the OAEI 2008 anatomy track (Experiment 1).The first column has the name of the algorithm; the second column shows the number of mappings the algorithm found. The third column is the number of mappings found by both the OAEI algorithm and LOOM. The fourth column is the percentage of overlap between LOOM and the algorithm (column 3 / column 2). The fifth column is the number of mappings found by an OAEI algorithm that LOOM did not find (column 2 – column 3). The last column is the percentage of mappings among the ones that LOOM missed that were correct.
| Tool | # of results found | Overlap with LOOM | Mappings missed by LOOM | ||
|---|---|---|---|---|---|
| # of results | % of results | # of results | Precision | ||
| loom | 1,136 | N/A | N/A | N/A | N/A |
| aroma | 964 | 732 | 76% | 232 | 64% |
| asmov | 1,262 | 890 | 71% | 372 | 32% |
| rimom | 1,205 | 1,084 | 90% | 121 | 70% |
| lily | 1,325 | 878 | 66% | 447 | 50% |
| dssim | 1,545 | 890 | 58% | 655 | 14% |
| sambo | 1,465 | 1,065 | 73% | 400 | 60% |
| sambodtf | 1,527 | 1,065 | 70% | 462 | 53% |
| taxomap | 2,533 | 1,053 | 42% | 1480 | 15% |
| aflood | 1,187 | 950 | 80% | 237 | 56% |
Experiment 2: LOOM versus AROMA on BioPortal ontologies
Table 2 compares the results of running both LOOM and AROMA on four pairs of BioPortal ontologies. There was significant variability in the number of mappings found by the two tools. For instance, for the S.Pombe cell cycle and A.Thaliana cell cycle ontologies, LOOM found almost ten times as many mappings as AROMA. Moreover, LOOM’s precision on this pair of ontologies was 78%. On the Mouse adult gross anatomy–Zebrafish anatomy and development pair, AROMA found many more mappings than LOOM. However, the precision of the mappings that AROMA found and LOOM did not was only 4% (i.e., almost all of these “extra” mappings found by AROMA were incorrect). Similarly, AROMA found 538 mappings to LOOM’s 4 mappings on the Cell type–S.Pombe cell cycle pair of ontologies, all of which were incorrect. The two ontologies have very little overlap.
The last two columns in Table 2 show the mappings found by LOOM, but not AROMA. For three out of four pairs, the precision for these mappings is essentially the same as the overall LOOM precision. In other words, the mappings found by LOOM but missed by AROMA were similar in accuracy as LOOM mappings overall.
Discussion
We found that the majority of ontology alignment tools are not readily available, and those that are available either (1) do not outperform simple lexical matching methods, or (2) do not scale well to the size of biomedical ontologies, or (3) lack sufficient documentation for proper use. Thus, researchers who want to use some of the best tools must be prepared to spend significant resources to get the tools and get them to run.
We limited our search to the tools that participated in the OAEI competition for several reasons. First, most developers of ontology-mapping tools participate in the competition to position their tool among other tools. Second, the OAEI provides a rigorous comparison and evaluation framework and make all the data public. Third, using the tools and the test ontologies from the competition enabled us to compare our own algorithm to other available tools.
Our results on the anatomy test set from OAEI show that the performance of our simple algorithm is comparable to the performance of the best available advanced mapping algorithms. There are several possible explanations for this result. First, many of the advanced algorithms rely on their analysis of the structure of the ontology, and biomedical ontologies—while usually large in size—have relatively little structure, with only a few relationships. Second, biomedical ontologies often have rich terminological information, with many synonyms specified for each concept. Therefore, just using synonyms as a source for mappings provides good results. Third, there is probably less variability in the language used to name biomedical concepts compared to other domains. All these factors make lexical techniques effective.
Our results from the evaluation of LOOM against AROMA support the hypothesis that lexical matching is an efficient method for generating mappings for biomedical ontologies. The low precision of AROMA on mappings missed by LOOM demonstrates that AROMA was not very useful for finding mappings that we were unable to find simply with lexical matching techniques. Additionally, the precision of mappings found by LOOM and missed by AROMA shows that LOOM can add, with high accuracy, a significant number of useful mappings for ontologies with a high degree of overlap.
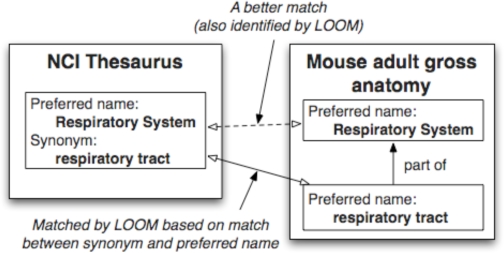
It is also instructive to analyze the mappings that LOOM missed or identified incorrectly. First, if a pair of concepts has identical preferred names or synonyms, LOOM always returns them as matches. However, there are cases where names or synonyms of classes match exactly but the classes are not equivalent. Consider the example in Figure 2. The concept “Respiratory_System” in NCI Thesaurus has synonym “respiratory tract”. The Mouse adult gross anatomy ontology has a concept named “respiratory tract”. However, “Respiratory_System” in NCI Thesaurus is not equivalent to “respiratory tract” in Mouse adult gross anatomy ontology because there is another class, “respiratory system”, in Mouse adult gross anatomy that is a better match for “Respiratory_System.”
Figure 2.
Example of incorrect mapping produced by lexical matching. The synonym of a class in NCI Thesaurus matches the preferred name for a class in Mouse adult gross anatomy. However, this match is misleading because there is a more precise match for the class Respiratory system.
Second, LOOM allows for one-character mismatches. While in many ontologies this feature significantly increases recall without affecting precision, in some cases, one-character differences are critical. For instance, the only case where LOOM did not have near-perfect precision was for the Cell Cycle ontologies, where many names differ by one character and this difference is significant (e.g., “ATP binding” and “ADP binding,” “DNA helicase activity” and “RNA helicase activity,” etc.).
More research is needed to understand better why such simple techniques as the ones that LOOM uses perform so well on biomedical ontologies and why advanced algorithms perform relatively poorly. One possible explanation is that developers of biomedical ontologies use a relatively controlled language. In fact, researchers performing term extraction from biomedical annotations observed a similar phenomenon to ours: very simple information-extraction methods, such as simple string matching works surprisingly well in identifying ontology concepts in text.8, 9 At the same time, biomedical ontologies have relatively limited structural information, so algorithms that rely heavily on analyzing the structure do not have an advantage.
In related work, Johnson and colleagues10 analyzed a number of lexical methods in finding relations among terms in biomedical ontologies. They have found that methods have a wide variety of correctness, depending on the ontologies that are being compared. However, their methods used more advanced techniques than LOOM did. For example, even when using exact string matching, they broke up preferred names and synonyms into separate words and matched them. This method performed very well on some ontologies but had low precision on others. By using only complete terms, LOOM ensures high precision, and our experiments show that it does not lose much in recall either.
Other researchers have investigated mapping techniques that are specific to biomedical ontologies. For instance, Zhang and Bodenreider focused on anatomy ontologies and compared the performance of different mapping techniques, such as lexical mapping and the use of external reference sources.11 We have intentionally chosen a spectrum of ontologies for our Experiment 2—with ontologies from both medicine and biology—to evaluate lexical matching on different types of biomedical ontologies.
In general, we need to understand better what characteristics of ontologies may predict which algorithms will perform well and which will not in comparing the ontologies.
Conclusions and Future Plans
We have demonstrated that simple lexical matching can be very effective in creating mappings between biomedical ontologies. Their performance is comparable to the performance of the more advanced algorithms. LOOM is publicly available at http://bioontology.org/tools.html.
Now that we have shown that LOOM performs quite well on biomedical ontologies, we plan to run LOOM on all pairs of ontologies in BioPortal to produce pair-wise mappings between them. Users and developers will be able to access the mappings via Web services and to download sets of mappings that they need. We will use these mappings in annotating biomedical resources and in other tools and services that NCBO provides.
Acknowledgments
This work was supported by the National Center for Biomedical Ontology, under roadmap-initiative grant U54 HG004028 from the National Institutes of Health. We are very grateful to Nigam Shah for his help as a domain expert and other advice on the paper.
Footnotes
Precision measures what fraction of the mappings returned by the algorithm was in the reference alignment (i.e., how many were correct). Recall measures what fraction of the mappings from the reference alignment was returned by the algorithm. F-measure is a combined measure of precision and recall (harmonic mean).
References
- 1.Rubin DL, Shah NH, Noy NF.Biomedical ontologies: a functional perspective Brief Bioinform2008 January120089175–90. [DOI] [PubMed] [Google Scholar]
- 2.Noy NF, Griffith N, Musen MA. Collecting Community-Based Mappings in an Ontology Repository; 7th Int. Semantic Web Conf. (ISWC 2008); Karlsruhe, Germany. 2008. [Google Scholar]
- 3.Euzenat J, Shvaiko P.Ontology matchingBerlin; New York: Springer; 2007 [Google Scholar]
- 4.Caracciolo C, et al. Results of the Ontology Alignment Evaluation Initiative 2008; 3d Int. Workshop on Ontology Matching (OM-2008) at ISWC 2008; Karlsruhe, Germany. 2008. [Google Scholar]
- 5.Bechhofer S, et al. OWL Web Ontology Language ReferenceW3C Recommendation: W3C; 2004
- 6.Bodenreider O, Hayamizu TF, Ringwald M, De Coronado S, Zhang S. Of mice and men: aligning mouse and human anatomies. AMIA Annual Symposium. 2005:61–5. [PMC free article] [PubMed] [Google Scholar]
- 7.David J, Guillet F, Briand H. Association rule ontology matching approach. Int. J. on Semantic Web and Information Systems. 2007;3(2):27–49. [Google Scholar]
- 8.Shah N, et al. Ontology-driven indexing of public datasets for translational bioinformatics. BMC Bioinformatics. 2009;10:1471–2105. doi: 10.1186/1471-2105-10-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai M. An Efficient Solution for Mapping Free Text to Ontology Terms. AMIA Summit on Translational Bioinformatics; San Francisco, CA: 2008. [Google Scholar]
- 10.Johnson H, et al. Evaluation of lexical methods for detecting relationships between concepts from multiple ontologies. Pac Symp Biocomput. 2006:28–39. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Bodenreider O. Alignment of multiple ontologies of anatomy: Deriving indirect mappings from direct mappings to a reference. AMIA Annual Symposium. 2005:864–8. [PMC free article] [PubMed] [Google Scholar]