Abstract
Infections with Methicillin-Resistant Staphylococcus aureus (MRSA) account for almost 20,000 deaths per year. Early identification of patients with MRSA infection or colonization aids in stopping spread. We compared automated identification of MRSA using HL7 lab result messages to current manual infection control practices at a local hospital during July–September 2008. We used data from infection control providers (ICPs), the microbiology lab, and a Regional Healthcare Information Exchange to assess the accuracy of manual and automated methods. Three hundred seventy MRSA cases were identified from July–September 2008. Manual identification recognized 314 (sensitivity 84.9%, positive predictive value 99.4%) MRSA cases and automated detection from HL7 messages identified 341 (sensitivity 92.2%, positive predictive value 98.8%). Automated processing of HL7 lab report messages is a more sensitive method of capturing MRSA cases than current standard infection control practice, with minimal loss of specificity.
Introduction
Infections with Methicillin-Resistant Staphylococcus aureus (MRSA) pose an increasing public health concern. Infections with MRSA have steadily increased nationwide since the 1960s.(1; 2) The public health burden of MRSA is significant; leading to increased morbidity and mortality as well as higher healthcare costs.(3)
Despite advances in laboratory methods for diagnosing MRSA, capturing and maintaining accurate data about MRSA colonization or infection remains a challenge. Current practice relies heavily on infection control providers (ICPs) who review microbiology data on a daily basis and manually maintain a list of all known MRSA cases. Admissions are screened against this list and infection control precautions implemented when appropriate. The use of manual review requires significant personnel time and limits list maintenance to hours when ICPs are on duty.
We hypothesized that automated identification of MRSA patients using HL7 laboratory result messages will generate a more accurate and timely list of MRSA patients than current manual infection control practices. Automated recognition of MRSA infection or colonization enables accurate and early point of care clinical decision support(4–6) and creates a platform to support epidemiologic studies needed to understand and prevent spread.
Methods
We compared manual MRSA case identification by ICPs (ICP method) and automated identification using HL7 laboratory report messages (HL7 method) during July–September 2008 at a local healthcare facility. We linked cases from three data sets using the medical record number.
Manual Identification of MRSA by ICPs
ICPs identify MRSA cases by reviewing daily microbiology reports and keeping a list of all known cases. Since May 2007, ICPs maintain this list using standardized web collection forms created as part of a citywide electronic infection control network built on the infrastructure of the Indiana Network for Patient Care (INPC), a Regional Healthcare Information Exchange.(7) Using the INPC database, we extracted a list of unique ICP-identified MRSA cases from a local hospital during July–September 2008.
Automated Identification of MRSA Using HL7 Laboratory Report Messages
We collected HL7 laboratory report messages from July–September 2008 from the INPC message queues. We used natural language processing (NLP) software to identify MRSA positive HL7 lab report messages as previously described by Friedlin et al.(8) We extracted all positive MRSA messages associated with a specific facility and collapsed the data into one positive record for each unique medical record number.
Identification of MRSA from Microbiology Data
We collected MRSA culture data from the local hospital’s microbiology laboratory by extracting culture results directly from a bioMerieux Vitek Bacterial Analyzer (Durham, NC). The Vitek Bacterial Analyzer provides organism identification and antibiotic susceptibility for each culture processed. Lab technicians reviewed this information and cultures identified as oxacillin resistant S. aureus were manually renamed from S. aureus to MRSA.
The hospital does not use alternative organism identification methods, such as genetic analysis.
We queried the analyzer for all cultures with an organism identified as MRSA or S. aureus. The data were extracted, formatted using PERL, and imported into a relational database table. To ensure that all oxacillin resistant S. aureus had been recognized as MRSA, we queried this table for organisms with oxacillin resistance and found no instances where oxacillin resistant S. aureus was not recognized by the organism name MRSA. We extracted unique medical record numbers from this table for any patients with positive MRSA cultures during July–September 2008. Because this process involved manual coding, a small number of laboratory data entry errors were expected.
Definition of a Gold Standard
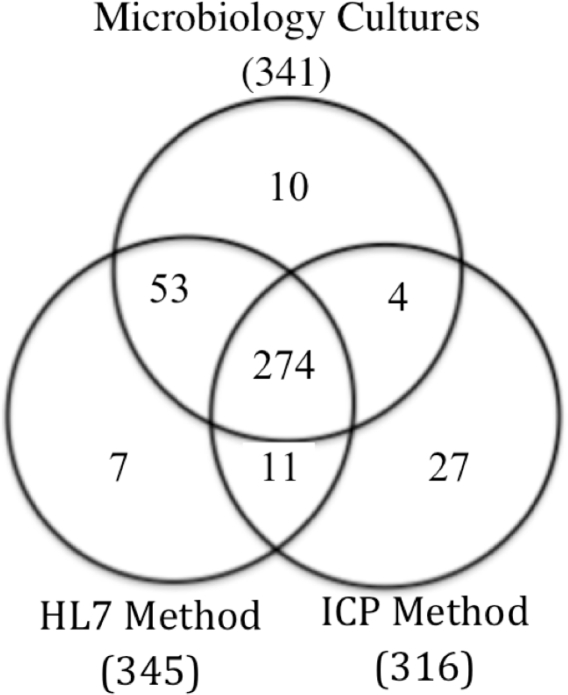
In order to fairly compare the accuracy of manual and automated methods, we determined the true MRSA status of each case. True positive MRSA status required (1) verification of a positive MRSA culture in chart documentation or microbiology data, and (2) recognition as positive by the HL7 method, ICP method, or both. Figure 1 displays relationships between the three data sets after linking MRSA cases using medical record numbers.
Figure 1.
Identifying cases for targeted chart review.
We performed chart reviews for all patients identified as MRSA positive without verifying cultures in the microbiology data. We also reviewed patient records for ten cases found only in the microbiology data. Cases involving data input errors were reconciled using alternative patient identifiers. After correction of data input errors, cases meeting our criteria for true positive MRSA status were placed into a gold standard list.
Of 341 cases in the microbiology data, 331 met our criteria as true positive cases and were added to the gold standard list. Ten cases in the microbiology data were not recognized by either method and involved laboratory data input errors of the medical record number. After correcting the errors, nine of the cases were recognized by both the HL7 and ICP methods and one by the HL7 method.
Of the 27 unique ICP-identified cases, two (2/27) had no evidence of a positive MRSA culture in the patient record (false positive cases) and 25 (25/27) had positive MRSA cultures in June. Of the seven unique HL7-identified cases, four (4/7) did not have a positive MRSA culture in the patient record (false positive cases) and two (2/7) had positive MRSA cultures in the patient record but not in microbiology data. One (1/7) case reconciled with a previously identified lab data entry error.
There were 11 medical record numbers concurrently identified by the HL7 and ICP methods with no culture verification in microbiology data. Nine (9/11) of these cases were associated with previously reconciled lab data entry errors and two (2/11) had culture verification in the patient record.
After reconciling these cases and verifying positive MRSA cultures, all cases meeting our true positive criteria were compiled into a gold standard list. This list was used as the basis for comparison to evaluate the sensitivity of HL7 and ICP methods of MRSA recognition.
Statistical Analysis
We created Venn diagrams to visualize the logical relationships between our data sets (R statistical software, Vienna, Austria). Sensitivity, specificity, positive predictive value, and negative predictive value were calculated using standard methodology and sensitivities compared using McNemar’s test (α=0.05) with continuity correction.
Results
A total of 370 true positive cases of MRSA were identified in this study. Microbiology data verified 331 cases occurring between July–September 2008. Forty-five cases were identified by the ICP or HL7 methods with no associated positive MRSA cultures in microbiology data from this time range. A trained clinician (DS) reviewed these charts finding 39 culture proven MRSA cases not represented in the microbiology data during July–September and six cases with no proof of a positive MRSA culture anywhere in the patient record.
Of the 39 cases not in the microbiology data, 25 were identified by the ICP method and found to have positive MRSA cultures performed by the microbiology lab in June; one month prior to our study period. The remaining 14 cases were composed of ten lab data entry errors and 4 cases without verifying cultures in microbiology data. After reconciling the ten lab data entry errors, nine cases were recognized by both the HL7 and ICP methods and one case was recognized by the HL7 method. The four cases without culture evidence in the microbiology data did have culture verification elsewhere in the patient record.
Six false positive cases were found during the chart review. Four cases related to the NLP software misinterpreting a negation in HL7 lab messages and two related to ICP data input mistakes.
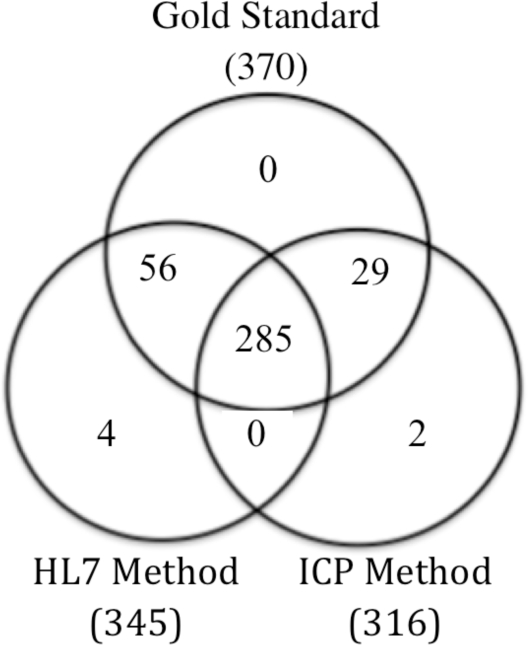
The Venn diagram (Figure 2) shows how ICP and HL7 methods performed against each other and against the gold standard. The automated HL7 method of MRSA identification had a sensitivity of 92.2% compared to 84.9% for the manual ICP method (Table 1). The two methods combined captured 100% of MRSA cases.
Figure 2.
Comparison of HL7 and ICP methods using gold standard as basis of comparison.
Table 1.
Comparison of ICP and HL7 methods using the gold standard as a basis of comparison.
| Identification Method | *SN(%) | PPV (%) |
|---|---|---|
|
ICP (TP=314, FN=56, FP=2) |
84.9 | 99.4 |
|
HL7 (TP=341,FN=29, FP=4) |
92.2 | 98.8 |
SN = sensitivity, PPV = positive predictive value. TP=True Positive, FP=False Positive, FN=False Negative.
p-value = 0.005
Discussion
Capturing MRSA cases by processing HL7 lab messages with NLP is superior to manual infection control practices. The HL7 method recognized more cases and had a significantly higher sensitivity (92.2% vs. 84.9%, p = 0.005) than the ICP method. Our study identified a number of issues related to manual maintenance of MRSA cases and provides focus for future work on automating the process.
Manual processes are prone to failure at multiple points.(9) For this reason, we reviewed charts for MRSA cases not found in microbiology data and ten unique to microbiology data. A modest number of errors (2.7%, 10/370) occurred when lab personnel input medical record numbers into the microbiology analyzer, but ICPs committed surprisingly few data input errors (0.5%, 2/370).
In order to compare ICP and HL7 methods using the most comprehensive and accurate set of MRSA cases, we created a gold standard from cases verified by either microbiology data or through chart review. We created this gold standard to compensate for data entry errors in the lab (10), delay in ICP data input (25), and cases not in the microbiology data but verified in the patient record (4).
Our initial culture verification step, using microbiology lab data, provided insight into the accuracy of this data set. There were 39 cases originally identified as false positives by ICP and HL7 methods, which upon chart review were found to be culture proven cases. All but four of these cases could be reconciled to microbiology data. These four required chart review to verify culture evidence of MRSA. Two of these cases were found by both the HL7 and ICP methods and two solely by the HL7 method. They did not involve duplicate medical record numbers and could not be found in the microbiology data. The most likely explanation for absence of these cases in the microbiology data is accidental deletion from the bacterial analyzer.
A chart review of the false positive cases from the ICP method revealed 25 ICP observations associated with positive MRSA cultures in June; one month prior to the study period. These cases were included in the gold standard data set in order to fairly assess ICP accuracy, without penalizing for delayed data input. These cases represent true positive MRSA patient identification by the ICP method, but were initially recognized as false positives because the corresponding culture was performed one month prior to our study period.
Lag time in ICP data entry is a significant issue and we found an average of eight days (range 0–172, standard deviation=17.7) elapsed before ICPs entered a patient's MRSA status into the infection control network. Given that patients often receive care at multiple facilities,(10) this delay leaves ample time for an encounter at a neighboring facility and increases the potential spread of MRSA. Due to workflow constraints, ICPs tended to enter MRSA cases into the infection control network in bulk; waiting until they have a number of cases and entering them all at the same time. A logical technique given the workload of ICPs; however, the objective of the infection control network is to provide citywide MRSA status and delays in data entry increase the risk of transmission to surrounding facilities.
In a separate analysis, we extended the search for ICP-identified MRSA cases by looking at the months following our study period. We wanted to assess how many MRSA patients with culture evidence in September had ICP observations recorded in October; one month after the study period We found 6 cases of positive MRSA cultures in September associated with ICP observations in October. Including these cases in our comparison did not significantly affect the sensitivity (HL7 sensitivity 92.2%, ICP sensitivity 86.5%, p-value = 0.04) of the ICP method.
The ICP method was prone to false negative errors while the HL7 method was prone to false positive errors, which equated with a higher sensitivity of the automated process. Both error types pose differing risk; false negatives lead to delay or absence of an appropriate infection control response at receiving healthcare institutions. False positives may result in patients erroneously placed into contact precautions, leading to increased risk for an adverse event.(11) Discussions with our infection control providers and infectious disease consultants suggest that a hybrid model incorporating the efficiencies of automated identification with the irreplaceable expert input of ICPs likely achieves the ideal balance.
Limitations
Linking patient data using medical record numbers poses a source of error. A full review assessing all medical record numbers for duplicates was not performed. A small number of medical record numbers in the microbiology data were truncated and could not be used in our comparison. For future comparisons, we will perform a chart review of all study cases and use a combination of patient identifiers in order to more accurately link patient data.
Additionally, a more extensive chart review may identify cases where ICPs made a decision based on MRSA testing from a neighboring facility. We did not identify any such instances in this study but the possibility should be addressed in future work.
Our data were collected from one facility and these results may not be applicable to other institutions. A larger comparison of these methods involving multiple institutions would provide stronger evidence.
Conclusion
The increasing prevalence of MRSA has serious consequences leading to preventable morbidity and mortality. Accurate and timely MRSA case identification enables interventions to prevent transmission in both the hospital and ambulatory settings. Overburdened ICPs may lack dedicated time to maintain an institutional list of MRSA cases. Augmenting ICP detection of MRSA with automated methods provides more accurate and timely point of care clinical decision support, allowing ICPs to focus on the greater task of preventing infections.
In this study, we provide evidence that an automated method of MRSA identification is a more sensitive and timely means of identifying and tracking MRSA cases than ICP identification alone. Understanding the epidemiology of MRSA transmission is paramount to implementing prevention/intervention protocols and an automated process can provide an accurate real time platform for this purpose.
Future Work
We aim to incorporate automated MRSA identification into our existing infection control network as part of our goal to provide accurate and timely infection control data at the point of care and across multiple institutions.
Acknowledgments
This work was performed at the Regenstrief Institute, Indianapolis, IN, and was supported in part by grant T15 LM07117 from the National Library of Medicine.
This project was funded in part by contract number 290-04-0015 from the Agency for Healthcare Research and Quality, U.S. Department of Health and Human Services. The opinions expressed in this document are those of the authors and do not reflect the official position of AHRQ or the U.S. Department of Health and Human Services.
Other contributors: J. Marc Overhage, MD, PhD (Regenstrief Institute and Indiana University School of Medicine), Pat Lineback MT(ASCP) (Hospital Microbiology Lab), Marc Rosenman, MD (Regenstrief Institute, Inc.), Dan Thompson (Regenstrief Institute, Inc.).
References
- 1.Styers D, Sheehan DJ, Hogan P, Sahm DF. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Annals of Clinical Microbiology and Antimicrobials. 2006;5(1):2. doi: 10.1186/1476-0711-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA-J. Am. Med. Assoc. 2007;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 3.Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infection Control and Hospital Epidemiology. 2005 Feb 1;26(2):166–174. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- 4.Evans RS, Larsen RA, Burke JP, Gardner RM, Meier FA, Jacobson JA, et al. Computer surveillance of hospital-acquired infections and antibiotic use. JAMA. 1986 Aug 22;256(8):1007–1011. [PubMed] [Google Scholar]
- 5.Evans RS, Wallace CJ, Lloyd JF, Taylor CW, Abouzelof RH, Sumner S, et al. Rapid identification of hospitalized patients at high risk for MRSA carriage. J Am Med Inform Assoc. 2008 Jul 1;15(4):506–512. doi: 10.1197/jamia.M2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kho AN, Dexter PR, Warvel JS, Belsito AW, Commiskey M, Wilson SJ, et al. An effective computerized reminder for contact isolation of patients colonized or infected with resistant organisms. International Journal of Medical Informatics. 2008;77(3):194–198. doi: 10.1016/j.ijmedinf.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald CJ, Overhage JM, Barnes M, Schadow G, Blevins L, Dexter PR, et al. The Indiana Network for Patient Care: a working local health information infrastructure. Health Aff. 2005 Sep 1;24(5):1214–1220. doi: 10.1377/hlthaff.24.5.1214. [DOI] [PubMed] [Google Scholar]
- 8.Friedlin J, Grannis S, Overhage JM. Using natural language processing to improve accuracy of automated notifiable disease reporting. AMIA Annu Symp Proc. 2008:207–11. [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald C. Protocol-based computer reminders, the quality of care and the non-perfectability of man. N Engl J Med. 1976 Dec 9;295(24):1351–1355. doi: 10.1056/NEJM197612092952405. [DOI] [PubMed] [Google Scholar]
- 10.Kho AN, Lemmon L, Commiskey M, Wilson SJ, McDonald CJ. Use of a regional health information exchange to detect crossover of patients with MRSA between urban hospitals. J Am Med Inform Assoc. 2008;15(2):212–216. doi: 10.1197/jamia.M2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stelfox HT, Bates DW, Redelmeier DA. Safety of patients isolated for infection control. JAMA. 2003 Oct 8;290(14):1899–1905. doi: 10.1001/jama.290.14.1899. [DOI] [PubMed] [Google Scholar]