Abstract
Objective:
To explore the feasibility of real-time mortality risk assessment for ICU patients.
Design/Methods:
This study used retrospective analysis of mixed medical/surgical intensive care patients in a university hospital. Logistic regression was applied to 7048 development patients with several hundred candidate variables. Final models were selected by backward elimination on top cross-validated variables and validated on 3018 separate patients.
Results:
The real-time model demonstrated strong discrimination ability (Day 3 AUC=0.878). All models had circumstances where calibration was poor (Hosmer-Lemeshow goodness of fit test p < 0.1). The final models included variables known to be associated with mortality, but also more computationally intensive variables absent in other severity scores.
Conclusion:
Real-time mortality prediction offers similar discrimination ability to daily models. Moreover, the discrimination of our real-time model performed favorably to a customized SAPS II (Day 3 AUC=0.878 vs AUC=0.849, p < 0.05) but generally had worse calibration.
Introduction
Estimation of risk for intensive care unit (ICU) patients has drawn considerable interest from the medical community in recent decades. Most researchers have focused on providing simplistic “severity of illness” scores, such as APACHE [1], MPM [2] or SAPS [3]. While most models have been designed for risk estimation at 24 hours after ICU admission, they have been applied to subsequent days [4, 5]. However, if risk estimates are to be used for individual care decisions, more frequent—even real-time—estimates may be helpful. Acute deterioration due to the onset of septic shock, for example, could easily occur between daily scores. Moreover, while others have taken common severity scores and augmented them with additional information [6, 7], the scores emphasize simple, common ICU observations. However, in an era of digital information, computers should be able to assist the caregivers in interpreting complex data patterns.
In this paper we explore models for predicting ICU mortality. We create three types of models: (1) a stationary acuity score, SDAS, that uses daily summary data; (2) daily acuity scores, DASn, that use daily summary data for individual days n ∈ {1, 2, 3, 4, 5}; and (3) a real-time acuity score, RAS, that uses all observations from all days. We compare the models against each other and against a customized SAPS II score for ICU days 1 through 5. If real-time ICU risk models are feasible, they might eventually make their way to the bedside and help caregivers interpret a wealth of ICU data.
Methods
Study data were retrospectively extracted from the MIMIC II database [8]. The MIMIC II database contains medical ICU, critical care unit and surgical ICU data collected from the Beth Israel Deaconess Medical Center, Boston, USA between 2001 and 2007. The data were collected and analyzed with institutional approval by the local IRB.
Data inclusion/exclusion criteria
We performed retrospective analysis on 10066 intensive care patients. Our selection criteria required patients to have at least one valid observation for the following: (a) heart rate, (b) Glasgow Coma Scale (GCS), (c) hematocrit, and (d) BUN. Patients were excluded based on any of the following, as they indicated significantly different risk profiles: (a) an ICD9 code indicating Chronic Renal Failure; (b) received neurological service (NSICU); or (c) received trauma service (TSICU).
Additional limitations were placed on individual patients. If a patient had multiple hospital visits, only the first ICU visit was included. Patient data were also limited to the first 7 ICU days and episodes where the patients received full treatment (i.e., full code). Periods after dialysis was started were also excluded.
Study Outcomes
Our study outcome was mortality in the ICU or within 30 days of ICU discharge. If a patient was discharged from the hospital alive (censored), survival was assumed.
Data preparation
A large set of candidate variables was considered for our models. Variables directly extracted from MIMIC II included real-valued nurse-charted observations (e.g., vital signs, lab values, etc.), categorical nurse-charted observations (e.g., ventricular fibrillation, ICU Service type, etc.), intravenous medications, input/output variables, and demographic variables.
From the variables directly extracted from MIMIC II, a number of meta variables and derived variables were added. These included binary indicator variables that marked the presence or absence of non-uniformly available measurements such as central venous pressure. Other derived variables involved simple calculations such as pulse pressure. In addition, several variables generalized more specific variables, such as indicators for type of medication (e.g., Neo-Synephrine and Levophed both map to sympathomimetic agent). A number of variables also sought to capture the temporal dynamics of the values, such as the 28-hour slope for blood pressure (from linear best-fit line) or the cumulative time spent on vasopressor medications. Other computationally intensive variables were also included, such as the range (maximum-minimum) up to the current point in a patient’s stay, the deviation of an observation from the evolving patient baseline (using prior information up to the current time), or the ratio of the blood pressure value while on vasopressors to the blood pressure value while not on pressors.
Additional variables were included based on literature suggesting their usefulness in predicting mortality. Rivera-Fernández et al. suggested several types of events that help augment typical severity scores [7]. Similarly, Silva et al. suggest similar events that they used with artificial neural networks to predict mortality [6]. Examples of variables based on these studies include the number of minutes that the systolic blood pressure (SBP) is continuously out of range (OOR) within a two hour window, or the number of SpO2 observations that fall below a critical threshold.
Several demographic variables that might help determine the risk level for patients were included. The variables shown in Table 1 (with the exception of ICU length of stay and SAPS II) and the ICU service type were included, along with three chronic disease variables that were extracted from ICD9 codes: Metastatic Carcinoma, Hematologic Malignancy, and AIDS.
Table 1:
Demographics.
| Age (y) | Male | Female | Hosp (d) | ICU (d) | SAPS II* |
|---|---|---|---|---|---|
| 65 ± 16 | 59.7% | 40.3% | 1.8 ± 3.6 | 2.8 ± 2.1 | 36 ± 17 |
Hosp=days in hospital prior to ICU admission; ICU=days in ICU;
no type of admission
Observation frequencies varied greatly between variables. Some variables, such as chemistry labs, were typically measured daily, while other variables, such as blood pressure, were updated at least once per hour. To limit sparseness, it was assumed that variables were observed when necessary, and old observations were held until updated (with a variable-dependent upper limit typically set to 28 hours). Some variables, such as INR, were assumed normal when absent. A detailed discussion of the data and the data preparation can be found in [9].
Before multivariate modeling, we ranked the variables according to their performance using univariate logistic regression Wald Z scores (the coefficient estimate divided by the estimated standard error of the coefficient). We discarded variables with p-values less than 0.05 (i.e., Z2 < 3.841). Furthermore, we eliminated variables with strong collinearity by keeping the best univariate variable among variables with Spearman rank correlation coefficients greater than 0.8.
Model Selection
We used logistic regression to create our predictive models. In previous work, we explored the use of survival models, but found that with limited followup information survival analysis methods provided no clear benefit over logistic regression at predicting ICU mortality [10]. Others have made similar conclusions by noting that ICU patients who expire in the ICU often experience prolonged ICU stays without benefit [11].
The data used for our models are listed in Table 2. For the aggregate data, the large number of variables resulted from four functions used for daily summaries: min, max, mean, and standard deviation (sd).
Table 2:
Data for SDAS and DASn (Aggregated 24h), and for RAS (Real-Time)
| Patients | Obs | Vars | |
|---|---|---|---|
| Aggregated 24h (Agg) Data | 10066 | 32480 | 1752 |
| Agg Development Partition | 7048 | 22888 | 349 |
| Agg Validation Partition | 3018 | 9592 | 349 |
| Real-Time (RT) Data | 10066 | 1044982 | 438 |
| RT Development Partition | 7048 | 736218 | 200 |
| RT Validation Partition | 3018 | 308764 | 200 |
For each model, we performed five-fold cross validation using backward elimination with Akaike’s Information Criterion (AIC) on each fold of the development data. By iteratively increasing the AIC threshold, plots showing the sensitivity of the model to the number of variables were analyzed for overfitting. Selecting the best variables (best 25 for SDAS, best 20 for DASn, and best 60 for RAS) from the top 4 cross validation models (least overfit), we fit the final model using the entire development data and performed backward elimination once more. A final refinement step involved manually removing similar variables (such as multiple output measurements that share influence in the model) and manually adding AIDS, Metastatic Carcinoma, and Hematologic Malignancy if their contribution was significant at the p = 0.1 level. Further details regarding the methodology can be found in [9].
Model Validation and Comparison
Using the 30% held-out validation data, we validated each model’s performance on unseen data. This was done by examining the discrimination performance as measured by the ROC curve area (AUC) and the model calibration as measured by the Hosmer-Lemeshow goodness of fit (Ĥ). For the Ĥ statistic, deciles of risk were used and the result was compared to the χ2 distribution with 8 d.f. for training data and 10 d.f. for validation data. A significant difference (e.g., p < 0.1) indicates poor model calibration (i.e., a significant difference between the observed and the expected mortalities in the risk deciles).
Models were compared against each other using subsets of the development data that had predictions available from each model for a given day. We also looked at the performance on patients that remained in the unit for at least five days and had valid predictions from each model for each day.
SAPS II
For comparison, we calculated pseudo-SAPS II scores for the MIMIC-II patients. Currently MIMIC II does not contain the SAPS II “type of admission” field. We omitted this contribution to the score, but added the cardiac surgery recovery unit service (svCSRU) and the medical ICU service (svMICU) indicators to our SAPS II logistic equation as proxies. To avoid confusion, we refer to our SAPS II approximation as SAPSIIa.
Results
The characteristics of our seven trained models, along with the SAPS II logistic models, are provided in Table 3. Models 1 and 2 show the top 22 variables (ranked by Wald Z score) for SDASand RAS, respectively. Positive Z scores indicate positive correlation with mortality. Transformations applied to a variable follow in parenthesis. The “dev” transformation measures the variable’s difference from the population’s mean value. A number of predictive variables represent summaries of an observation over time, such as the time SpO2 was out of range (OOR) during the past 2 hours. Only one long-term slope was included (Platelets over 28 hours), but it had a significant contribution to all models except DAS1 and DAS2. All models were heavily influenced by the Glasgow Coma Scale (GCS) and patient Age. In comparing Models 1 and 2, it is important to note that the significance of infrequently observed RASinputs are likely inflated due to the sample-and-hold approach taken for low-frequency observations.
Table 3:
Acuity Model Characteristics (on training data)
| Model (day) | Train Obs | Missing | d.f. | AUC | R2 |
|---|---|---|---|---|---|
| RAS (1–5) | 528850 | 207368 | 43 | 0.885 | 0.436 |
| SDAS (1–5) | 20130 | 2758 | 35 | 0.898 | 0.456 |
| DAS1 (1) | 6364 | 684 | 22 | 0.900 | 0.447 |
| DAS2 (2) | 5179 | 397 | 24 | 0.910 | 0.463 |
| DAS3 (3) | 3526 | 182 | 26 | 0.904 | 0.463 |
| DAS4 (4) | 2351 | 116 | 20 | 0.892 | 0.467 |
| DAS5 (5) | 1690 | 60 | 15 | 0.883 | 0.450 |
| SAPSII (1) | 6008 | 504 | 20 | 0.796 | 0.238 |
| SAPSII (2) | 5247 | 164 | 20 | 0.857 | 0.328 |
| SAPSII (3) | 3512 | 97 | 20 | 0.845 | 0.313 |
| SAPSII (4) | 2321 | 72 | 20 | 0.842 | 0.326 |
| SAPSII (5) | 1620 | 62 | 20 | 0.830 | 0.321 |
Validation on Held-Out Data
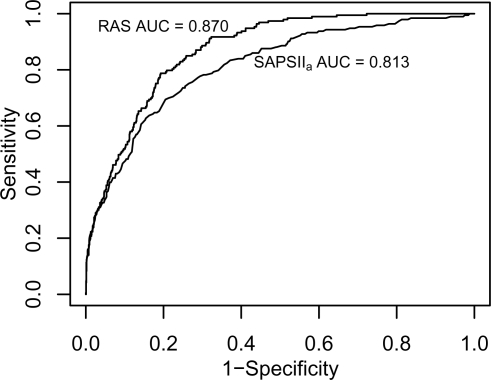
Figure 1 shows the ROC curves for RASand SAPSIIa on day one. Table 4 shows the AUC for each day on the subset of patients with valid predictions from all models for that day. One important consideration when comparing models was the summary function used to compare multiple RAS predictions against single predictions from daily models. We used the mean prediction over each day.
Figure 1:
ROC Curves for RAS and SAPSIIa on Day 1
Table 4:
AUC by day on matched validation patients
| Day | RAS | SDAS | DASn | SAPSIIa | n |
|---|---|---|---|---|---|
| 1 | 0.875 | 0.876 | 0.879 | 0.809 | 1954 |
| 2 | 0.880 | 0.885 | 0.881 | 0.839 | 1849 |
| 3 | 0.878 | 0.887 | 0.876 | 0.849 | 1245 |
| 4 | 0.871 | 0.878 | 0.871 | 0.826 | 836 |
| 5 | 0.853 | 0.863 | 0.858 | 0.805 | 596 |
Model 1.
SDAS (showing top 22 of 35 inputs)
| Obs | d.f. | P | C | R2 | Brier | |
|---|---|---|---|---|---|---|
| 20130 | 35 | 0 | 0.898 | 0.456 | 0.074 | |
| Wald Z | P | |||||
| Max GCS (squared) | −12.85 | 0 | ||||
| Mean INR (inv) | −12.85 | 0 | ||||
| Pacemaker | −7.92 | 0 | ||||
| CSRU Service | −7.31 | 0 | ||||
| Min Platelets Slope 28 hr | −6.66 | 0 | ||||
| Mean Riker SAS | −6.66 | 0 | ||||
| Mean Hourly Urine Out (sqrt) | −6.18 | 0 | ||||
| ... | ... | |||||
| Mean PaO2 : FiO2 (if Ventilated) | 6.16 | 0 | ||||
| Mean Na (dev) | 6.45 | 0 | ||||
| Min Mg (squared) | 6.46 | 0 | ||||
| Max Shock Index | 6.49 | 0 | ||||
| Mean Platelets (inv) | 6.76 | 0 | ||||
| Prior Hospital Time (sqrt) | 7.05 | 0 | ||||
| ICU Day (squared) | 7.17 | 0 | ||||
| Jaundiced Skin | 7.18 | 0 | ||||
| Mean CO2 (inv) | 7.23 | 0 | ||||
| Max Lasix (log dev) | 7.33 | 0 | ||||
| Mean Beta-Blocking Agnt (log dev) | 7.44 | 0 | ||||
| Min Sympathomimetic Agent | 9.27 | 0 | ||||
| Mean SpO2 OOR past 30 m (sqrt) | 9.83 | 0 | ||||
| Min BUN : Creatinine (sqrt) | 12.05 | 0 | ||||
| Age (squared) | 17.40 | 0 | ||||
Model 2.
RAS (showing top 22 of 43 inputs)
| Obs | d.f. | P | C | R2 | Brier | |
|---|---|---|---|---|---|---|
| 528850 | 43 | 0 | 0.885 | 0.436 | 0.084 | |
| Wald Z | P | |||||
| GCS (squared) | −76.68 | 0 | ||||
| Intercept | −61.51 | 0 | ||||
| CSRU Service | −57.19 | 0 | ||||
| Pacemaker | −45.04 | 0 | ||||
| All Output (log) | −38.39 | 0 | ||||
| Pressors Start Day 1 | −35.32 | 0 | ||||
| ... | ... | |||||
| Lasix per Kg (log dev) | 29.99 | 0 | ||||
| Hourly Urine OOR past 2 hr | 30.28 | 0 | ||||
| Antiarrhythmic Agent | 30.35 | 0 | ||||
| Na (dev) | 30.66 | 0 | ||||
| Hematoligic Malignancy | 30.87 | 0 | ||||
| Beta-Blocking Agent | 30.96 | 0 | ||||
| SpO2 OOR past 2 hr (sqrt) | 32.54 | 0 | ||||
| PaO2 : FiO2 (if Ventilated) | 36.49 | 0 | ||||
| Prior Hospital Time (sqrt) | 38.44 | 0 | ||||
| Minutes in ICU | 40.57 | 0 | ||||
| Jaundiced Skin | 42.60 | 0 | ||||
| Std Pressor Sum (sqrt) | 49.11 | 0 | ||||
| Creatinine (log) | 50.31 | 0 | ||||
| INR (log) | 62.11 | 0 | ||||
| BUN : Creatinine (sqrt) | 69.45 | 0 | ||||
| Age (squared) | 91.28 | 0 | ||||
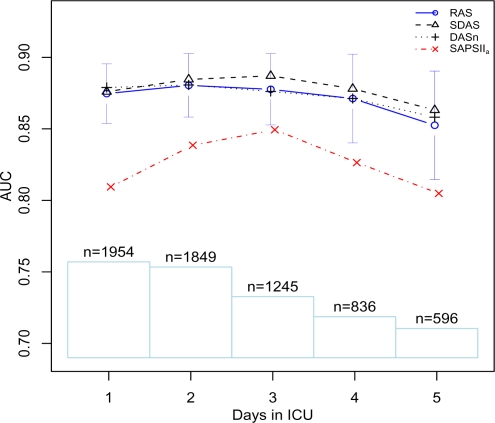
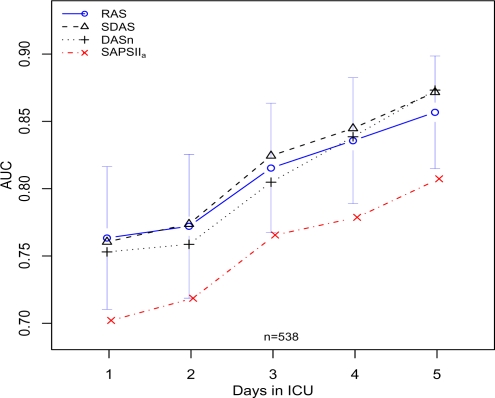
Figure 2 graphically depicts the change in AUC over time for matched patients. Similarly, Figure 3 shows the performance on the subset of patients who are in the ICU for at least 5 days and have predictions for each day from all models.
Figure 2:
AUC versus number of days in the ICU. Confidence intervals (95%) are shown for RAS AUC values along with a histogram showing the number of patients used by the models on each day.
Figure 3:
AUC versus number of days in the ICU for patients that stay at least 5 days and have predictions available from all models for the first 5 days. Confidence intervals (95%) are shown for RAS AUC values.
Table 5 lists the Ĥ statistic p-values. As suggested by [12], we combined adjacent deciles to make all expected frequencies at least 4 (the Ĥ statistic relies on large expected frequencies). For each row merger, the degrees of freedom for the χ2 comparison was reduced by 1.
Table 5:
Hosmer-Lemeshow calibration statistic Ĥ, matched validation patients
| Day | RAS p (d.f) | SDAS p (d.f) | DASn p (d.f) | SAPSIIap (d.f) | n |
|---|---|---|---|---|---|
| 1 | 0.002 (7) | 0.045 (7) | 0.001 (7) | 0.130 (9) | 1954 |
| 2 | 0.006 (6) | 0.018 (6) | 0.008 (6) | 0.439 (7) | 1849 |
| 3 | 0.063 (6) | 0.542 (6) | 0.087 (6) | 0.765 (7) | 1245 |
| 4 | 0.097 (6) | 0.110 (6) | 0.064 (6) | 0.884 (7) | 836 |
| 5 | 0.143 (6) | 0.183 (5) | 0.248 (5) | 0.275 (7) | 596 |
Discussion
Our model performed well at discriminating mortality on the separate validation data. In terms of AUC, each model outperformed SAPSIIa across all days (p < 0.05 for each comparison). The svCSRU and svMICU inputs greatly improved SAPSIIa discrimination, increasing day 1 AUC from 0.680 to 0.809 and the Ĥ p-value from 0.0001 to 0.130. With this correction, the performance of SAPSIIa aligns nicely with SAPS II numbers found in literature, especially in terms of calibration (e.g., see [13]). By using a subset of patients with predictions available from all models, the results were slightly biased towards the most input-constrained model, SDAS, which retained nearly all of its validation patients. When each model was validated on all validation patients with valid predictions, only marginal improvement against SDAS was observed, except for Day 1 SAPSIIa, which performed worse (AUC=0.781, and Ĥ p-value = 0.085). Further exploration of the calibration, e.g., using cross validation, is needed to better understand the sensitivity of the Ĥ statistic within our results.
For days 3, 4, and 5, the models demonstrate reasonable calibration (i.e., all p-values are > 0.05). SAPSIIa, however, is the only model with strong calibration on days 1 and 2. The predictions from both models have a large positive skew (most patients survive). RAS, however, has a much longer tail with its 10th decile covering a probability of 0.690 (versus 0.562 for SAPSIIa). The SAPSIIa logistic regression equation includes a logarithm of the the SAPS II score, along with the SAPS II score, that helps its calibration performance.
On the training data the goodness of fit for the model was much better in general, but still lacking for RASand SDASon day 1. Furthermore, SAPSIIa calibration on day 1 of the training data was weak (p < 0.0001, 9 d.f.). This was surprising as SAPS II was developed for use over the first 24 hours of an ICU stay and performed well on our validation data. In the original SAPS II model Le Gall et al. report p-values of 0.883 and 0.101 on their development and validation data, respectively [3].
Figure 2 shows that the models performed best on days 2 and 3. For RAS and SDAS this observation can be partly explained by the large number of observations available on days 2 and 3 biasing the model against earlier and later days. DASn has little improvement between days 1 and 3.
Early mortality discrimination for patients that stay in the ICU at least five days is difficult. Figure 3 shows that all of the models improve for these patients over time. Patients with long stays likely fall in the difficult middle risk group between the patients who recover or expire within the first few days.
Limitations and Future Work
A number of important limitations exist in this work: (1) The results reflect only one hospital population. Validation on external data from a separate institution is necessary before fully generalizing the conclusions. (2) As done with other severity of illness scores, missing observations could be considered normal in order to improve the model’s coverage at the potential cost to model performance [13]. This was done with negligible change in performance with two variables, INR and PaO2:FiO2, which were missing more frequently than other variables (but were still present enough for inclusion). (3) The models that we developed also include therapies which make them dependent on caregiver practice. The influence of therapies, however, was found to be small—possibly as a result of variation in practice between caregivers. (4) Future work is underway to explore the performance of our real-time acuity models in the context of secondary outcomes such as septic shock or weaning of vasopressors.
Conclusions
Real-time acuity scores can offer similar discrimination performance (risk ranking) to daily acuity scores with superior performance over customized SAPS II. Calibration performance (adequacy of risk estimates) was also similar between real-time models and daily models. Furthermore, automatic variable selection from several hundred candidates returned variables known to be correlated with mortality (e.g., GCS) but also a variety of other variables, including computationally intensive inputs (e.g., the time that SpO2 is out of range) and interventions (e.g., vasopressor medications). Many of these additional inputs are significant contributors to mortality prediction models.
Acknowledgments
This work was supported in part by the National Library of Medicine (NLM) Medical Informatics Traineeship (LM 07092) and the US National Institute of Biomedical Imaging and Bioengineering (NIBIB) under Grant Number R01 EB001659. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the NLM or the NIH.
References
- [1].Knaus WA, Wagner DP, Draper EA, Zimmerman JE, Bergner M, Bastos PG, et al. The APACHE III prognostic system. Risk prediction of hospital mortality for critically ill hospitalized adults. Chest. 1991 Dec;100(6):1619–1636. doi: 10.1378/chest.100.6.1619. [DOI] [PubMed] [Google Scholar]
- [2].Lemeshow S, Klar J, Teres D, Avrunin JS, Gehlbach SH, Rapoport J, et al. Mortality probability models for patients in the intensive care unit for 48 or 72 hours: a prospective, multicenter study. Crit Care Med. 1994 Sep;22(9):1351–1358. doi: 10.1097/00003246-199409000-00003. [DOI] [PubMed] [Google Scholar]
- [3].Le Gall JR, Lemeshow S, Saulnier F. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270(24):2957–2963. doi: 10.1001/jama.270.24.2957. [DOI] [PubMed] [Google Scholar]
- [4].Wagner DP, Knaus WA, Harrell FE, Zimmerman JE, Watts C. Daily prognostic estimates for critically ill adults in intensive care units: results from a prospective, multicenter, inception cohort analysis. Crit Care Med. 1994 Sep;22(9):1359–1372. doi: 10.1097/00003246-199409000-00004. [DOI] [PubMed] [Google Scholar]
- [5].Timsit JF, Fosse JP, Troch G, Lassence AD, Alberti C, Garrouste-Orgeas M, et al. Accuracy of a composite score using daily SAPS II and LOD scores for predicting hospital mortality in ICU patients hospitalized for more than 72 h. Intensive Care Med. 2001 Jun;27(6):1012–1021. doi: 10.1007/s001340100961. [DOI] [PubMed] [Google Scholar]
- [6].Silva A, Cortez P, Santos MF, Gomes L, Neves J. Mortality Assessment in Intensive Care Units via Adverse Events Using Artificial Neural Networks. Artificial Intelligence in Medicine. 2005;36:3. doi: 10.1016/j.artmed.2005.07.006. [DOI] [PubMed] [Google Scholar]
- [7].Rivera-Fernndez R, Nap R, Vzquez-Mata G, Miranda DR. Analysis of physiologic alterations in intensive care unit patients and their relationship with mortality. J Crit Care. 2007 Jun;22(2):120–128. doi: 10.1016/j.jcrc.2006.09.005. [DOI] [PubMed] [Google Scholar]
- [8].Saeed M, Lieu C, Raber G, Mark RG. MIMIC II: a massive temporal ICU patient database to support research in intelligent patient monitoring. Comput Cardiol. 2002;29:641–644. [PubMed] [Google Scholar]
- [9].Hug C.Detecting Hazardous Patient Episodes using Predictive Modeling Massachusetts Institute of Technology; 2009. In preparation. Available from: http://dspace.mit.edu/ [Google Scholar]
- [10].Hug CW. Predicting the Risk and Trajectory of Intensive Care Patients using Survival Models. Massachusetts Institute of Technology; 2006. [Google Scholar]
- [11].Schoenfeld D.Survival methods, including those using competing risk analysis, are not appropriate for intensive care unit outcome studies Crit Care 2006February101103Available from: http://dx.doi.org/10.1186/cc3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hosmer DW, Lemeshow S. Applied Logistic Regression. 2nd ed. New York: Wiley; 2000. [Google Scholar]
- [13].Ohno-Machado L, Resnic FS, Matheny ME. Prognosis in critical care. Annu Rev Biomed Eng. 2006;8:567–599. doi: 10.1146/annurev.bioeng.8.061505.095842. [DOI] [PubMed] [Google Scholar]