Abstract
The world's largest Q fever outbreak is ongoing in The Netherlands with around 3,000 confirmed cases since the first half of 2007. Increased awareness has resulted in early referral of patients for diagnostics. An important drawback to serological diagnosis of acute Q fever is the lag phase in antibody response. Therefore, we evaluated the performance of a real-time PCR for detection of Coxiella burnetii DNA using serum samples from patients with acute Q fever. PCR, targeting IS1111, was retrospectively performed on acute-phase and follow-up convalescent-phase serum samples from 65 patients with acute Q fever as diagnosed by immunofluorescence assay. The results obtained by PCR were related to disease stage as defined by subsequent appearance of phase II IgM, phase II IgG, phase I IgM, and phase I IgG (IgM-II, IgG-II, IgM-I, and IgG-I, respectively) antibodies and time since onset of disease. In addition, we analyzed seronegative acute-phase serum samples from patients with inconclusive Q fever serology, because no convalescent-phase serum samples were available. PCR was scored positive in 49/50 (98%) seronegative sera, 9/10 (90%) sera with isolated IgM-II antibodies, 3/13 (23%) sera with IgM-II/IgG-II antibodies, 2/41 (5%) sera with IgM-II/IgG-II/IgM-I antibodies, 0/15 (0%) sera with IgM-II/IgG-II/IgM-I/IgG-I antibodies, and 0/1 (0%) serum sample with IgM-II/IgG-II/IgG-I antibodies. The latest time point after onset of disease in which C. burnetii DNA could be detected was at day 17. In patients with inconclusive Q fever serology, PCR was positive in 5/50 (10%) cases. We conclude that real-time PCR with serum samples is indispensable for early diagnosis of acute Q fever. C. burnetii DNA becomes undetectable in serum as the serological response develops.
Q fever, an infection caused by the bacterium Coxiella burnetii, results in a self-limiting disease in 40 to 50% of infected cases. Pneumonia is the predominant presenting symptom in acute Q fever, although fever and hepatitis are also frequently observed (9, 10). Failure to diagnose acute Q fever and delay in treatment may lead to prolonged morbidity and increased hospital admission rates (4, 7, 11, 14).
During three consecutive years, large Q fever epidemics occurred in an area in the south of The Netherlands where the disease was formerly not prevalent (11). In 2007 there were a total of 191 confirmed cases reported, in 2008 a total of 998, and in 2009 more than 2,000 confirmed cases were reported, which ranks the outbreak as the largest Q fever epidemic recorded to date. The affected area has a large density of dairy goats, of which a number have tested positive for Q fever. Next to the differences in sizes of the epidemics, the interval between onset of disease and date of diagnosis decreased from a median of 77 days in 2007 to 29 days in 2008 and 17 days in 2009 (12). Moreover, the hospital admission rates were reduced from 40% in 2007 to 20% in 2008 (11). Both observations are most likely due to increased awareness among physicians in the affected area resulting in early submission of clinical samples to the laboratory, subsequent earlier diagnosis, and probably fewer undiagnosed cases. The majority of diagnostic samples from both epidemics were submitted to our laboratory, which lies in the center of the epidemic area and serves a catchment area of roughly 500,000 persons in a semirural district supporting two hospitals and surrounding general practitioners.
The gold standard for serological diagnosis of an infectious disease is either a seroconversion or a 4-fold rise in antibody titer. The reference test for serological diagnosis of Q fever is the immunofluorescence assay (IFA) (8). Antibodies are expressed against phase II antigens during the acute infection and against phase I antigens in the established infection. For both antigens, IgM antibody production precedes IgG production, and thus three phases can be distinguished in acute Q fever: a seronegative phase followed by IgM/IgG phase II seroconversion during the acute infection and subsequent IgM/IgG phase I seroconversion in the established infection. However, an important drawback to serological diagnosis of acute Q fever is the lag phase in antibody response of 7 to 15 days after onset of clinical symptoms (8).
Apart from serology, C. burnetii-specific PCR of serum samples can be an additional tool to diagnose Q fever in the early acute phase, but conflicting sensitivities have been reported (3, 13). Here, we evaluated the performance of an in-house-developed real-time PCR assay for detection of C. burnetii DNA in serum samples from patients with acute Q fever.
MATERIALS AND METHODS
Patients.
First, we retrospectively analyzed 130 paired sera from 65 adult patients (age, ≥18 years) with acute Q fever as diagnosed by IFA for phase II IgM, phase II IgG, phase I IgM, and phase I IgG (IgM-II, IgG-II, IgM-I, and IgG-I, respectively) antibodies. We selected 65 acute-phase serum samples from patients that presented with absent antibodies (n = 50), isolated IgM-II antibodies (n = 10), or IgM-II/IgG-II antibodies (n = 5) to C. burnetii. Antibody distribution in the 65 follow-up convalescent-phase serum samples yielded IgM-II/IgG-II antibodies (n = 8), IgM-II/IgG-II/IgM-I antibodies (n = 41), IgM-II/IgG-II/IgM-I/IgG-I antibodies (n = 15), and IgM-II/IgG-II/IgG-I antibodies (n = 1). In 6-month follow-up serum samples (not submitted to PCR analysis), all patients had progressed to an antibody profile with both phase II and phase I antibodies. Forty-five patients were referred for acute Q fever diagnostics by their general physicians, while hospital physicians referred the remaining 20 patients. Previously, we showed in this epidemic that pneumonia, besides fever, is highly prevalent in both outpatient and inpatient populations (1). None of the patients developed clinical signs or a serological response suggestive of chronic Q fever during follow-up. All sera selected for PCR analysis were obtained between March and September 2008. Date of onset of disease, when available, was retrieved from the hospital information system.
Next, we retrospectively analyzed 50 sera from 50 adult patients with a respiratory tract infection of unknown etiology referred to our laboratory in June 2008, when the Q fever outbreak was at its peak (212 confirmed cases with date of onset of disease in June 2008 reported to the regional Municipal Health Service GGD Hart voor Brabant). We specifically selected acute-phase serum samples from patients in whom Q fever serology was not conclusive, because the acute-phase serum sample was seronegative while a convalescent-phase serum sample was either not received or not referred for Q fever serology. In 10/50 acute-phase sera there was a dubious or positive antibody titer against Mycoplasma pneumoniae (Fujirebio Inc., Tokyo, Japan). These 10 sera were not excluded from PCR analysis, as false-positive M. pneumoniae serology has been reported during acute Q fever (5).
Finally, we retrospectively analyzed 10 sera from 10 adult patients with a nonspecific IgM-II antibody titer against C. burnetii. We specifically selected acute-phase sera from patients that presented with either solitary dubious (n = 8) or solitary positive (n = 2) IgM-II antibodies that did not progress to a serological profile with either IgG-II, IgM-I, or IgG-I antibodies in follow-up convalescent-phase serum samples.
Individual patient consent was not obtained, because all sera used in this study were drawn for routine serological analysis. The Internal Review Board of the Jeroen Bosch Hospital approves anonymous use of discarded blood for scientific purposes. All patients that donate blood are informed of this possibility with right of refusal. All sera had been stored at −20°C until the day of analysis.
DNA isolation.
Volumes of 200 or 500 μl of serum and 10 μl of phocine herpes virus (PhHV), which served as internal control, were added to 2 ml of lysis buffer. DNA was extracted using the NucliSens EasyMAG extraction system (bioMérieux, Boxtel, The Netherlands) according to the protocol provided by the manufacturer.
C. burnetii real-time PCR.
Oligonucleotides to detect IS1111 of C. burnetii were designed using Primer Express version 2.0.0 software. To amplify a 70-bp fragment, forward primer AAA ACG GAT AAA AAG AGT CTG TGG TT, reverse primer CCA CAC AAG CGC GAT TCA T, and probe 6-carboxyfluorescein (FAM)-AAA GCA CTC ATT GAG CGC CGC G-6-carboxytetramethylrhodamine were used. For detection of the PhHV internal control, forward primer GGG CGA ATC ACA GAT TGA ATC, reverse primer GCG GTT CCA AAC GTA CCA A, and probe 6-FAM-TTT TTA TGT GTC CGC CAC CAT CTG GAT C-TAMRA were used (Sigma-Genosys Ltd., Haverhill, United Kingdom) (15). Twenty-five microliters of amplification mixture contained 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2 (prepared from 10× PCR buffer delivered with Platinum Taq polymerase), 0.75 U of Platinum Taq polymerase (Invitrogen BV, Breda, The Netherlands), 4% glycerol (molecular biology grade; CalBiochem, VWR International BV, Amsterdam, The Netherlands), 200 μM each deoxynucleoside triphosphate (Invitrogen BV), 0.5 μl Rox reference dye (Invitrogen BV), and 10 μl of DNA isolate. Primer and probe concentrations were 900 nM forward primer, 900 nM reverse primer, and 200 nM probe. Real-time PCR was performed using an ABI Prism 7500 SDS apparatus (Applied Biosystems, Nieuwerkerk aan de IJssel, The Netherlands). PCR conditions were standard: 30 s at 95°C, followed by 45 cycles of 3 s at 94°C and 30 s at 60°C. To avoid contamination of PCR with amplicons from previous reactions, the preparation of the PCR mixes, the isolation of DNA, and the amplification by PCR were carried out in three separate rooms designated for these activities. In all runs we included DNA isolation controls (NC) and no-template controls (NTC) to monitor the presence of contaminants in isolation and/or PCR reagents. In 395 NC plus NTC controls we once detected a signal with a cycle threshold (CT) of 36.22 in an isolation control well that was located next to a relatively high-positivity sample (CT, 28.58), while all other 394 controls tested negative.
To investigate the clinical specificity of the C. burnetii real-time PCR, we analyzed a serum panel consisting of five sera obtained simultaneously with blood cultures which had yielded Streptococcus pneumoniae, five sera that were positive with PCR for Legionella pneumophila, three sera obtained in the acute phase of a Chlamydophila psittaci infection as shown by seroconversion, three sera obtained in the acute phase of a Mycoplasma pneumoniae infection as shown by seroconversion or a 4-fold rise in antibody titer, three sera obtained simultaneously with throat swabs that were positive with PCR for influenza A virus, one serum obtained in the acute phase of an influenza B virus infection as shown by seroconversion, two sera obtained simultaneously with respiratory samples which had yielded Mycobacterium tuberculosis either by PCR or culture, and three sera obtained simultaneously with respiratory samples which had yielded ordinary bacterial isolates (Staphylococcus aureus, Haemophilus influenzae, or Pseudomonas aeruginosa).
C. burnetii IFA.
Serologic diagnosis of Q fever was made by IFA (Focus Diagnostics, Cypress, CA) that measured IgM and IgG antibodies against both phase I and phase II antigens, according to the manufacturer's instructions. Titers of ≥1:32 were considered positive. In general, the first antibody to appear in acute Q fever patients is IgM-II, followed by more or less simultaneous IgG-II and IgM-I responses and subsequent appearance of IgG-I antibodies, allowing for distinction of time-dependent serologic profiles (2).
RESULTS
C. burnetii real-time PCR.
To detect C. burnetii, a new quantitative real-time PCR was designed that targets the insertion element IS1111. The presence of this element in multiple copies (7 to 120 copies) of the C. burnetii genome ensures sensitive detection of the bacterium (6). Tenfold dilutions of DNA from the C. burnetii Nine Mile strain showed linear detection from 3 to 3 × 106 genome equivalents (GEq) per reaction mixture (CT = −3.3951 × log10GEq + 42,224; correlation coefficient [R2], 0.9987). The limit of detection was 4 GEq per reaction mixture (CT, 33.5 ± 1.2, mean ± standard deviation [SD]; n = 20).
To investigate whether the volume of serum used for DNA isolation influences the chance of C. burnetii DNA detection, we compared input volumes of 200 μl and 500 μl of acute-phase sera from 10 patients with serologically proven Q fever infection. With 200 μl of serum, 8 out of 10 samples were positive in duplicate and 2 out of 10 samples yielded one positive and one negative result. Using 500 μl of serum, all 10 samples were positive in duplicate. The mean CT of the eight samples that were double positive with both volumes was 33.4 ± 3.7 (200 μl [mean ± SD]) and 31.7 ± 2.1 (500 μl). Although the sample size was small, increasing the input volume from 200 μl to 500 μl yielded lower CT values and thus increased the chance of C. burnetii DNA detection. Therefore, for further analysis, DNA was isolated from 500 μl of serum and PCR was performed in duplicate. A CT value of <45.0 was considered positive. When the test yielded both a positive and a negative result in the duplicate run, the PCR was considered positive.
Based on results in silico, the sequence of the amplicon showed no homology to GenBank, EMBL, DDBJ, or PDB sequences other than those from C. burnetii (BLAST search at http://blast.ncbi.nlm.nih.gov/Blast.cgi). To further investigate the specificity of the test, 25 serum samples from patients suffering from respiratory tract infections caused by pathogens other than C. burnetii were tested in the IS1111 PCR. None of these serum samples (0/25) tested positive in the C. burnetii real-time PCR.
Patients.
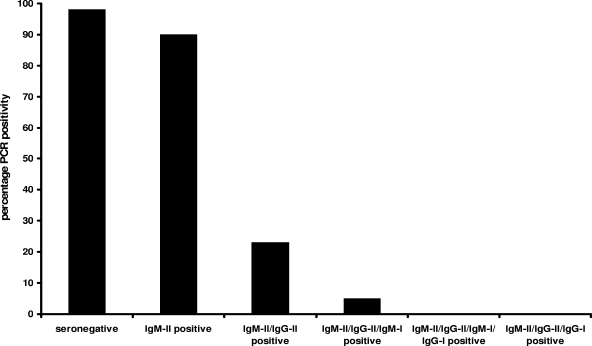
C. burnetii real-time PCR was retrospectively performed on 65 acute-phase and 65 follow-up convalescent-phase serum samples from 65 patients with acute Q fever as diagnosed by IFA. PCR was positive in 49/50 (98%) seronegative sera, 9/10 (90%) sera with isolated IgM-II antibodies, 3/13 (23%) sera with IgM-II/IgG-II antibodies, 2/41 (5%) sera with IgM-II/IgG-II/IgM-I antibodies, 0/15 (0%) sera with IgM-II/IgG-II/IgM-I/IgG-I antibodies, and 0/1 (0%) serum sample with IgM-II/IgG-II/IgG-I antibodies (Fig. 1). CT values in PCR-positive sera varied from 27.5 to 43.5. Of the 63 serum samples with a positive PCR, 6 acute-phase and 3 convalescent-phase serum samples yielded only one positive result in the duplicate run.
FIG. 1.
Coxiella burnetii real-time PCR positivity percentages in acute-phase and follow-up convalescent-phase serum samples from 65 patients with acute Q fever. Sera were grouped according to antibody profiles as determined by immunofluorescence assay. PCR was positive in 49/50 (98%) seronegative sera, 9/10 (90%) sera with isolated IgM-II antibodies, 3/13 (23%) sera with IgM-II/IgG-II antibodies, 2/41 (5%) sera with IgM-II/IgG-II/IgM-I antibodies, 0/15 (0%) sera with IgM-II/IgG-II/IgM-I/IgG-I antibodies, and 0/1 (0%) serum with IgM-II/IgG-II/IgG-I antibodies.
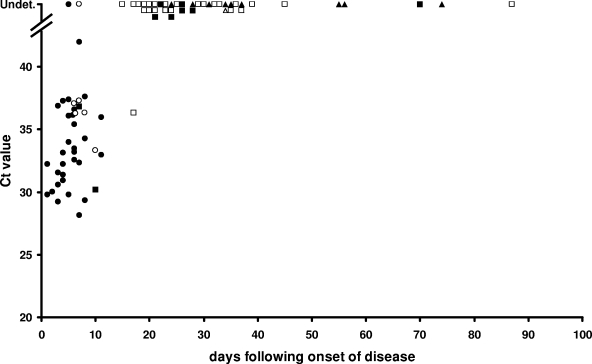
Date of onset of disease could be retrieved from the hospital information system for 40/65 (62%) patients. The latest time point at which C. burnetii DNA could still be detected was at day 17 after onset of disease. This particular serum yielded only one positive result in the duplicate run, while IFA revealed the presence of IgM-II/IgG-II/IgM-I antibodies (Fig. 2).
FIG. 2.
Relation of CT value, number of days following onset of disease, and antibody profile in 40 acute-phase and 40 follow-up convalescent-phase serum samples from 40 patients with acute Q fever for which date of onset of disease could be retrieved from the hospital information system. Symbols reflect different antibody profiles: •, seronegative sera (n = 31); ○, sera with isolated IgM-II antibodies (n = 6); ▪, sera with IgM-II/IgG-II antibodies (n = 9); □, sera with IgM-II/IgG-II/IgM-I antibodies (n = 24); ▴, sera with IgM-II/IgG-II/IgM-I/IgG-I antibodies (n = 9); ▵, serum with IgM-II/IgG-II/IgG-I antibodies (n = 1). Undet., C. burnetii DNA was undetectable.
In the 50 acute-phase sera from patients with a respiratory tract infection of unknown etiology and inconclusive Q fever serology, PCR was positive in 5/50 (10%) cases. Of these five PCR-positive sera, one serum showed a positive antibody titer against Mycoplasma pneumoniae. All 10 acute-phase sera from patients with a nonspecific IgM-II antibody titer against C. burnetii tested negative in the PCR.
DISCUSSION
Laboratory diagnosis of acute Q fever is important because of atypical clinical presentations of the disease, requirement of specific antibiotics, and follow-up of development of chronic disease. A number of reports have demonstrated the presence of C. burnetii in blood by culture and PCR during acute Q fever (3, 8, 13). Sensitivities of C. burnetii-specific PCRs on serum samples from seronegative acute Q fever patients that vary from 26% to 89% have been reported (3, 13). We detected C. burnetii DNA in serum from 98% of seronegative acute Q fever patients and in 90% of patients with isolated IgM-II, the first antibody to appear in the serologic response. Ultimately, the PCR becomes negative as the serological response develops, with subsequent appearance of IgG-II, IgM-I, and IgG-I antibodies.
Although the number of IS1111 elements in the C. burnetii strain(s) causing the Dutch outbreak is unknown, it is conceivable that high copy numbers may explain our high detection rate. Alternatively, and in our view more likely, it might be the small amplification product (70 bp) that renders it sensitive as a diagnostic assay. Since blood cells and bacteria are removed from serum by centrifugation of blood, we speculate that we detect DNA fragments rather than intact C. burnetii genomes. The 26% detection rate reported by Fournier et al. was based on amplification of a 485-bp fragment, followed by a nested amplification of a 260-bp fragment (3). With regard to the detection threshold of this nested assay, those authors deduced that they “were able to obtain positive PCR products from the 10−5 dilution of our C. burnetii suspension (equivalent of 1 DNA copy of the bacterium).” We defined the limit of detection (LOD) of our assay as the lowest concentration of genome equivalents at which 20 of the 20 replicates tested positive, and we found this limit to be 4 GEq. The relative low mean CT value of 33.5 at the LOD (4 GEq) might be due to the fact that it is based on CsCl gradient-purified high-molecular-weight DNA containing clusters of IS1111 rather than single IS1111 copies. Thus, the absolute sensitivities of the two assays are in a similar range. The 89% detection rate reported by Turra et al. was based on amplification of a 217-bp fragment (13). Another important difference contributing to the sensitivity of our method is that we extracted DNA from 500 μl of serum, while 200 μl of serum was used in both of the other studies (3, 13).
With regard to the specificity of our PCR assay, despite the increasing number of sequences available in NCBI databases, in silico no other sequences than C. burnetii-related sequences with similarity to the amplicon were retrieved by BLAST search. In addition, none of the serum specimens from 25 patients with respiratory tract infections caused by pathogens other than C. burnetii tested positive. Thus, the PCR for detection of C. burnetii appeared highly specific. We also analyzed 10 patients with isolated IgM-II antibodies in an acute-phase serum sample that did not progress to a serological profile with any of the three other antibodies in follow-up convalescent-phase serum samples. This finding is indicative of a nonspecific IgM-II antibody titer. This was confirmed in the sense that none of these 10 samples proved PCR positive, whereas 90% of samples with isolated IgM-II antibodies from patients that subsequently developed at least one of the three other antibodies were found to be PCR positive. This result further underscores the specificity of our PCR in acute Q fever.
We did not detect C. burnetii DNA in 1/50 seronegative serum samples from patients with serologically proven acute Q fever. The acute-phase serum sample that tested negative was obtained 5 days after onset of disease. A follow-up convalescent-phase serum sample with IgM-II/IgG-II antibodies was obtained at day 22. This particular patient presented in the acute phase with a C-reactive protein (CRP) level of 40 mg/liter. Recently, we showed in a group of 65 patients that acute Q fever induces a strong CRP response (169 ± 90 mg/liter [mean ± SD]), while other infection markers like procalcitonin and white blood cell count are in the normal range or only marginally increased (1). The relatively low CRP level in the one acute Q fever patient without PCR positivity might be indicative of mild infection and concomitant absence of circulating DNA. In this context, it would be interesting, if at all possible, to study the presence of C. burnetii DNA in serum from persons with asymptomatic Q fever. Also, the observation that six acute-phase serum samples yielded only one positive result in the duplicate run indicates that in some patients C. burnetii DNA is present around the lower limit of detection. The CRP level in these six patients was also relatively low (80 ± 46 mg/liter). In addition, we cannot exclude that, at least in some patients, there might be a small period of time following initial PCR positivity in which C. burnetii DNA disappears but IgM-II antibodies have not yet appeared.
We retrospectively analyzed 50 seronegative acute-phase serum samples that were obtained during the peak incidence of the 2008 epidemic but that were submitted without follow-up. In 10% of these samples, we detected C. burnetii DNA, demonstrating that, under these conditions, serology had failed to support the laboratory diagnosis of acute Q fever. It has been reported that it may take up to 15 days after onset of disease for C. burnetii antibodies to appear (8), which is in line with data from our own study, in which two acute-phase serum samples, obtained 11 days after onset of disease from patients with subsequent seroconversion in a convalescent-phase serum sample were seronegative. Another drawback of serology is that false-positive cross-reactions, as have been reported for M. pneumoniae, may divert clinicians and public health authorities from finding the true underlying causative agent of a beginning Q fever epidemic (5). Thus, submitting acute-phase serum samples for the diagnosis of Q fever without request for C. burnetii PCR may either lead to failure to diagnose Q fever or result in delayed diagnosis and treatment.
We conclude that C. burnetii PCR with serum samples should be routinely included in the diagnostic work-up of a patient with suspected acute Q fever. Early diagnosis may reduce Q fever-associated morbidity and the number of hospital admissions (12). A tool for adequate early diagnosis may assist in public health issues and containment, especially when two epidemics with more or less identical clinical symptoms, such as novel influenza A (H1N1) virus infection and Q fever, occur simultaneously.
Acknowledgments
We thank Annika Pettersson-Fernholm and Dries Budding from the VU University Medical Center in Amsterdam for providing us with the quantified Coxiella burnetii Nine Mile stock.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.de Wit, N. C. J., C. P. C. de Jager, J. C. E. Meekelenkamp, M. Schoorl, A. B. Gageldonk-Lafeber, A. C. A. P. Leenders, et al. 2009. Markers of infection in inpatients and outpatiens with acute Q-fever. Clin. Chem. Lab. Med. 47:1407-1409. [DOI] [PubMed] [Google Scholar]
- 2.Dupont, H. T., X. Thirion, and D. Raoult. 1994. Q fever serology: cutoff determination for microimmunofluorescence. Clin. Diagn. Lab. Immunol. 1:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fournier, P. E., and D. Raoult. 2003. Comparison of PCR and serology assays for early diagnosis of acute Q fever. J. Clin. Microbiol. 41:5094-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gikas, A., D. P. Kofteridis, A. Manios, J. Pediaditis, and Y. Tselentis. 2001. Newer macrolides as empiric treatment for acute Q fever infection. Antimicrob. Agents Chemother. 45:3644-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karagiannis, I., G. Morroy, A. Rietveld, A. M. Horrevorts, M. Hamans, P. Francken, et al. 2007. Q fever outbreak in the Netherlands: a preliminary report. Euro Surveill. 12:E070809.2. [DOI] [PubMed] [Google Scholar]
- 6.Klee, S. R., H. Ellerbrok, J. Tyczka, T. Franz, and B. Appel. 2006. Evaluation of a real-time PCR assay to detect Coxiella burnetii. Ann. N. Y. Acad. Sci. 1078:563-565. [DOI] [PubMed] [Google Scholar]
- 7.Marrie, T. J. 2003. Coxiella burnetii pneumonia. Eur. Respir. J. 21:713-719. [DOI] [PubMed] [Google Scholar]
- 8.Maurin, M., and D. Raoult. 1999. Q fever. Clin. Microbiol. Rev. 12:518-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker, N. R., J. H. Barralet, and A. M. Bell. 2006. Q fever. Lancet 367:679-688. [DOI] [PubMed] [Google Scholar]
- 10.Raoult, D., T. Marrie, and J. Mege. 2005. Natural history and pathophysiology of Q fever. Lancet Infect. Dis. 5:219-226. [DOI] [PubMed] [Google Scholar]
- 11.Schimmer, B., G. Morroy, F. Dijkstra, P. M. Schneeberger, G. Weers-Pothoff, A. Timen, et al. 2008. Large ongoing Q fever outbreak in the south of The Netherlands, 2008. Euro Surveill. 13:18939. [PubMed] [Google Scholar]
- 12.Schimmer, B., F. Dijkstra, P. Vellema, P. M. Schneeberger, V. Hackert, R. ter Schegget, et al. 2009. Sustained intensive transmission of Q fever in the south of the Netherlands, 2009. Euro Surveill. 14:19210. [DOI] [PubMed] [Google Scholar]
- 13.Turra, M., G. Chang, D. Whybrow, G. Higgins, and M. Qiao. 2006. Diagnosis of acute Q fever by PCR on sera during a recent outbreak in rural south Australia. Ann. N. Y. Acad. Sci. 1078:566-569. [DOI] [PubMed] [Google Scholar]
- 14.van de Veerdonk, F. L., and P. M. Schneeberger. 2006. Patient with fever and diarrhea. Clin. Infect. Dis. 42:994-995, 1051-1052. [DOI] [PubMed] [Google Scholar]
- 15.van Doornum, G. J., J. Guldemeester, A. D. Osterhaus, and H. G. Niesters. 2003. Diagnosing herpesvirus infections by real-time amplification and rapid culture. J. Clin. Microbiol. 41:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]