Abstract
There is no licensed vaccine against the intracellular pathogen Francisella tularensis. The use of conventional mouse strains to screen protective vaccine antigens may be problematic, given the differences in the major histocompatibility complex (MHC) binding properties between murine and human antigen-presenting cells. We used engineered humanized mice that lack endogenous MHC class II alleles but that express a human HLA allele (HLA-DR4 transgenic [tg] mice) to identify potential subunit vaccine candidates. Specifically, we applied a biochemical and immunological screening approach with bioinformatics to select putative F. tularensis subsp. novicida T-cell-reactive antigens using humanized HLA-DR4 tg mice. Cell wall- and membrane-associated proteins were extracted with Triton X-114 detergent and were separated by fractionation with a Rotofor apparatus and whole-gel elution. A series of proteins were identified from fractions that stimulated antigen-specific gamma interferon (IFN-γ) production, and these were further downselected by the use of bioinformatics and HLA-DR4 binding algorithms. We further examined the validity of this combinatorial approach with one of the identified proteins, a 19-kDa Francisella tularensis outer membrane protein (designated Francisella outer membrane protein B [FopB]; FTN_0119). FopB was shown to be a T-cell antigen by a specific IFN-γ recall assay with purified CD4+ T cells from F. tularensis subsp. novicida ΔiglC-primed HLA-DR4 tg mice and cells of a human B-cell line expressing HLA-DR4 (DRB1*0401) functioning as antigen-presenting cells. Intranasal immunization of HLA-DR4 tg mice with the single antigen FopB conferred significant protection against lethal pulmonary challenge with an F. tularensis subsp. holarctica live vaccine strain. These results demonstrate the value of combining functional biochemical and immunological screening with humanized HLA-DR4 tg mice to map HLA-DR4-restricted Francisella CD4+ T-cell epitopes.
Francisella tularensis is a Gram-negative bacterium and the etiological agent of the zoonotic disease tularemia. F. tularensis is classified into four subspecies, namely, F. tularensis subsp. tularensis, F. tularensis subsp. holarctica, F. tularensis subsp. mediasiatica, and F. tularensis subsp. novicida (F. novicida) on the basis of their biochemical and genetic profiles, virulence properties, and geographical origins (51). To this end, F. tularensis subsp. tularensis (type A) is the most virulent subspecies, with the inhalation of as few as 10 organisms causing disease and mortality rates of between 30 and 60% in untreated cases of pneumonic tularemia (53). The live vaccine strain (LVS) derived from F. tularensis subsp. holarctica has been used as a prophylactic vaccine against tularemia (48). Millions of individuals in the Soviet Union were immunized with live vaccine strains between 1946 and 1960 (52). However, LVS has not been licensed for use in the United States due to a lack of understanding of the genetic mutations that are responsible for attenuation of this strain, although it is used as an investigational new drug (IND) to immunize at-risk workers, primarily tularemia researchers. F. novicida, which causes disease only in immunocompromised humans but which is highly virulent for mice, has been used as a comparative model organism due to the high degree of genetic similarity with type A strains (98.1% homology between sequences common to strains U112 and SCHU S4 [45]). We recently reported that a defined vaccine strain (ΔiglB) generated in strain U112 was effective in inducing heterologous protection against various Francisella strains in a mouse model of pulmonary tularemia, suggesting the conservation of protective antigens (12).
Cell-mediated immunity has been documented to play an important role in protection against tularemia (2, 18, 19, 49, 56). The role of antibodies, via neutralization and Fc receptor-mediated clearance (43, 44) in response to infection, has also gained significant attention. Therefore, the availability of a combination of multiple Francisella antigens containing T-cell and/or B-cell epitopes would be desirable for formulating an effective multivalent vaccine against this organism. However, the use of conventional mouse strains to identify protective antigens may not be feasible, given the differences in the major histocompatibility complex (MHC) binding properties between murine and human MHCs. These constraints can be overcome with the use of engineered humanized mice, such as the HLA-DR4 transgenic (tg) mouse. This mouse was generated to express the extracellular human α1 and β1 domains of the HLA-DRA and HLA-DRB1*0401 haplotypes, which form the peptide binding sites for antigen presentation, in conjunction with the murine α2 and β2 domains (29). These chimeric molecules have been shown to exhibit the same antigen-binding specificity as HLA-DRB1*0401 and to be functional in presenting antigens to T cells (29). The frequency of the HLA-DR allele in humans is 29% in Caucasian individuals, 10% in African American individuals, and 34% in other individuals (38), underscoring the translational value of the epitopes identified in these mice for humans. We recently demonstrated the feasibility of using the HLA-DR4 tg mouse for the identification of vaccine antigens against genital Chlamydia infection (39), demonstrating the value of the use of these animals in the rational selection of vaccine candidates.
In this study, we utilized a robust biochemical membrane protein fractionation method, cytokine recall assays, and humanized HLA-DR4 tg mice to identify putative CD4+ T-cell-reactive antigens from U112. Moreover, using bioinformatics tools, we further validated one of the identified antigens (FTN_0119), designated Francisella outer membrane protein B (FopB), as a potential subunit vaccine candidate against pneumonic tularemia in HLA-DR4 tg mice.
MATERIALS AND METHODS
Bacteria.
Francisella novicida strain U112 was provided by Francis Nano (University of Victoria, Victoria, British Columbia, Canada). KKF24, an iglC mutant of U112, was constructed as reported previously (32). F. tularensis subsp. holarctica LVS (lot 703-0303-016) was obtained from Rick Lyons (University of New Mexico). Strains were grown overnight at 37°C in tryptic soy broth (TSB) supplemented with 0.025% sodium pyruvate, 0.025% sodium metabisulfite, 0.025% ferrous sulfate, and 0.1% l-cysteine. Frozen stocks were prepared and stored in TSB containing 30% glycerol. The bacterial titer of the frozen stocks and the numbers of viable bacteria in the inocula were determined by dilution plating.
Mice.
HLA-DRA-IEα/HLA-DRB1*0401-IEβ transgenic (HLA-DR4 tg) mice were generated in the C57BL/6 genetic background and backcrossed to MHC class II-deficient (IAβ−, IEα−) mice to eliminate any effect of endogenous MHC class II proteins, as described previously (29). Specific-pathogen-free HLA-DR4 tg mice were bred and housed at the University of Texas at San Antonio (UTSA) animal facility. C57BL/6 mice were obtained from the National Cancer Institute (Bethesda, MD). All animal care and experimental procedures were performed in compliance with the Institutional Animal Care and Use Committee (IACUC) guidelines at UTSA.
Biochemical membrane protein fractionation.
F. novicida U112 was grown to late logarithmic phase, and the cells were pelleted by centrifugation (8,000 × g, 10 min). The cells were resuspended in extraction buffer (50 mM Tris, pH 6.8, 150 mM NaCl) and were disrupted by two freeze-thaw cycles, followed by three cycles of sonication (30% amplitude for 30 s with 1-min intervals) on ice with a digital sonifier (Branson, Danbury, CT). This disrupted cell suspension was centrifuged (8,000 × g, 10 min), and the resulting supernatant was discarded. The pellet material was washed exhaustively with extraction buffer, and proteins were extracted for 1 h at 4°C with extraction buffer containing 4% Triton X-114 (TX-114). The TX-114-treated pellet material was centrifuged (8,000 × g, 10 min), and the supernatant was decanted. The decanted supernatant was allowed to partition at 30°C, resulting in a biphasic solution (aqueous and detergent phases). The aqueous phase served as the source of protein for this study. The proteins in the aqueous phase were concentrated by cold acetone precipitation, dissolved in 5% glycerol containing 1.75% ampholyte (pH range, 3.7 to 9.3; Bio-Rad, Hercules, CA), and separated into 20 fractions by liquid-phase isoelectric focusing with a Rotofor apparatus (Bio-Rad) under constant power of 15 W for 3 h, according to the manufacturer's instructions. The proteins were also fractionated on the basis of their molecular mass with a Bio-Rad whole-gel elution system. The proteins were dissolved in SDS sample-loading buffer (250 mM Tris buffer, pH 6.8, containing 10% SDS, 30% glycerol, 5% β-mercaptoethanol, and 0.02% bromophenol blue) without heating and were electrophoretically separated on 12% SDS-polyacrylamide preparative gels. The gel was placed on a whole-gel elution unit, and an electric current (10 V, 30 min) was applied to elute the separated proteins into 14 fractions in an elution buffer (25 mM Tris buffer, pH 8.3, containing 19.2 mM glycine and 0.01% SDS), according to the manufacturer's instructions. The endotoxin content of each fraction was determined by use of a Limulus amebocyte lysate kit (QLC-1000; Lonza, Walkersville, MD). Aliquots of each fraction were stored at −80°C until further use.
Protein identification.
Multiple proteins were present in each eluted whole-gel subfraction. Proteins were resolved on a precast Ready-gel (Bio-Rad) and were stained with Coomassie blue (Invitrogen, Carlsbad, CA); and the protein band of each fraction was excised, subjected to trypsin digestion, and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry in the Proteomics Core of the University of Tennessee Health Science Center (13). The resulting spectra were searched against those in the published F. novicida U112 genomic database (45) to identify proteins.
Bioinformatics analyses.
The PSLpred algorithm (4) was used to identify secreted and membrane-bound proteins in the list of sequenced proteins, followed by a BLASTP search against the proteins obtained from the PSLpred database to exclude human and mouse homologous proteins. The HLA-DR4Pred algorithm (5) was then used to identify proteins containing epitopes with HLA-DR4 allele binding scores greater than 1.4, which are considered to be high-binding epitopes.
Antigen-specific cytokine recall assays.
HLA-DR4 tg mice (three to four mice per group) were anesthetized with 3% isoflurane using a rodent anesthesia machine (Harvard Apparatus, Holliston, MA) (31) and were intranasally (i.n.) immunized with either 106 CFU of KKF24 (ΔiglC mutant of U112) in 25 μl of phosphate-buffered saline (PBS) or PBS alone as a control. This vaccination dose was found to be optimal, as it caused no outward signs of illness and was highly protective against subsequent pulmonary F. novicida U112 challenge (41). The spleens were collected from the vaccinated mice 14 days postimmunization and then pooled, and single cells were made and stimulated with the Rotofor apparatus and/or whole-gel elution fractionated proteins (0.25 μg/200 μl/well) for 72 h, as described previously (41). In some experiments, CD4+ T cells were enriched from nonadherent spleen cells (pooled from four mice) by using an EasySep mouse CD4+ T-cell enrichment kit (StemCell Technologies, Vancouver, British Columbia, Canada) and stimulated with recombinant proteins in the presence of homozygous HLA-DR4 (DRB1*0401) human B cells as antigen-presenting cells (APCs) (21). The cells also were cultured with the unrelated antigen hen egg lysozyme (HEL). The culture supernatants were collected after 72 h and assayed for gamma interferon (IFN-γ), interleukin-4 (IL-4), and IL-5 using optEIA kits (BD Biosciences, San Diego, CA), according to the manufacturer's recommendations.
Detection of antibody and isotype levels by ELISA.
Microtiter plates were coated overnight at 4°C with 50 ng of protein fractions in sodium bicarbonate buffer (0.1 M, pH 9.5) and blocked for an additional 1 h at room temperature with PBS containing 5% fetal bovine serum (HyClone, Logan, UT) and 0.1% Brij (Sigma-Aldrich, St. Louis, MO). Serial dilutions of sera obtained from mice vaccinated with antigens or mock vaccinated with PBS alone (day 30) were added to the wells, and the plates were incubated for 2 h at room temperature. The plates were washed and incubated for 2 h with goat anti-mouse total Ig, IgG1, and IgG2a conjugated with horseradish peroxidase (Southern Biotech, Birmingham, AL). The plates were subsequently washed and incubated with tetramethylbenzidine (TMB) substrate for 30 min, and the absorbance (630 nm) was measured with an enzyme-linked immunosorbent assay (ELISA) microplate reader (μQuant; Biotek Instruments, Winooski, VT). End-point titers were calculated by using an absorbance of 0.2 as the cutoff value. No binding of immune sera was observed when the plates were coated with the unrelated antigen HEL (50 ng/well).
Expression of recombinant FopB protein.
PCR was used to amplify FopB-encoding sequences (FTN_0119) from F. novicida U112 genomic DNA with specific primers containing an engineered NcoI site (underlined) in the sense primer (5′-GCCATGGAAAAAACTATACTAGGAGC) and an XhoI site (underlined) in the antisense primer (5′-GCTCGAGTTGCATCTTAGCTATAA CATC). FopB PCR amplicons were cloned by using the pGEM-T vector system (Promega, Madison, WI) and Escherichia coli TAM1 (Active Motif, Carlsbad, CA). Cloned plasmid pGEM-FopB was isolated from transformed E. coli TAM1 bacteria and digested with NcoI and XhoI restriction enzymes. The 0.54-kb fopB digest was inserted into the pET28a expression vector, using the same restriction sites, and the resulting plasmid (pET28-FopB) was used to transform E. coli BL21(DE3) (EMD Biosciences, Gibbstown, NJ) for protein expression. Plasmid pET28-FopB also contained a C-terminal polyhistidine (His6) sequence to facilitate protein purification. Transformed BL21 cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h, and the expressed secreted recombinant FopB (rFopB) protein was isolated from the culture medium by affinity chromatography with His.Bind resin (EMD Biosciences), according to the manufacturer's instructions. Isolated rFopB protein with an endotoxin level of less than 2 unit/μg was used in all experiments.
Intranasal rFopB vaccination and pulmonary challenge.
C57BL/6 and HLA-DR4 tg mice were anesthetized i.n. with 3% isoflurane by use of a rodent anesthesia system and were immunized i.n. on day 0 with 7.5 μg rFopB in 25 μl of sterile PBS. This was accompanied by immunization with 10 μg of CpG oligodeoxynucleotide (5′-TCCATGACGTTCCTGACGTT-3′; Operon, Huntsville, AL) on days −1, 0, and +1. Some groups of mice were immunized subcutaneously (s.c.) on day 0 with 7.5 μg of rFopB and 10 μg of CpG formulated in incomplete Freund's adjuvant (Sigma-Aldrich). Mice were boosted once on day 14 with the same amount of antigen and were challenged i.n. with approximately 3.5 × 104 CFU (∼5 50% lethal doses [LD50s]) of LVS on day 30. Mock (PBS)-vaccinated age-matched mice were used as controls. Animals were monitored daily for morbidity and mortality.
Statistical analyses.
SigmaStat software (Systat Software Inc., San Jose, CA) was used to perform all tests of significance. Statistical analyses for the survival experiments were performed by the Kaplan-Meier test. Student's t test was used to determine differences in the levels of IFN-γ production. All data are reported as means ± standard deviations from each experimental animal group and are representative of at least two independent experiments.
RESULTS
Fractionation and identification of immunogenic F. novicida membrane proteins.
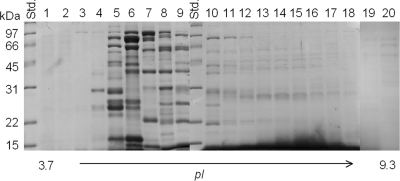
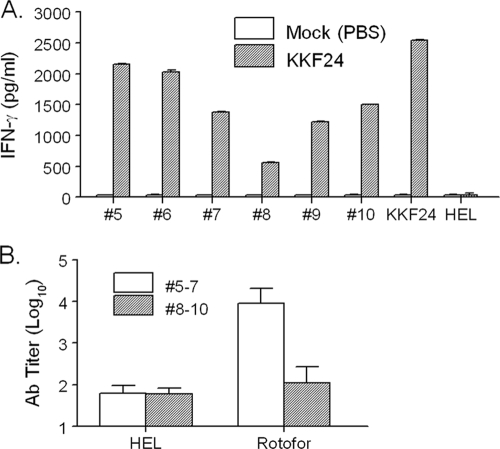
We utilized a biochemical approach to extract membrane proteins from F. novicida and examined these antigens as potential vaccine candidates against pulmonary tularemia using humanized HLA-DR4 tg mice. Specifically, membrane proteins were extracted from F. novicida U112 using the nonionic detergent TX-114. Proteins recovered from the aqueous phase of the TX-114 extraction process were subjected to isoelectric focusing (pH 3.7 to 9.3) using a Rotofor apparatus. A total of 20 fractions (referred to here as the Rotofor fractions) were obtained from this process, and the majority of the hydrophilic proteins extracted with TX-114 were observed in fractions 5 to 10, as shown in Fig. 1. The endotoxin levels in fractions 5 to 10 were found to be minimal (less than 1 EU/μg protein). To identify potential subunit vaccine candidates within these fractions, we applied a novel screening approach using humanized HLA-DR4 tg mice to assess antigen-specific cellular cytokine recall responses. To this end, the HLA-DR4 tg mice were immunized with a highly attenuated defined U112 mutant, strain KKF24 (ΔiglC) (33), and the mouse spleens were collected 14 days postvaccination. Prepared splenocytes were stimulated with selected Rotofor fractions 5 to 10 for 3 days, and the supernatants were collected for cytokine analyses. As shown in Fig. 2A, Rotofor fractions 5 to 10 induced significant (P < 0.005) IFN-γ production from KKF24-primed splenocytes but not from cells obtained from mock-vaccinated animals. As expected, UV-inactivated KKF24 induced robust IFN-γ production specifically from the KKF24-primed spleen cells (Fig. 2A). No detectable IL-4 and IL-5 responses were observed in spleen cells obtained from mice immunized with KKF24 when they were stimulated with the Rotofor fractions (data not shown), and a negligible IFN-γ response was observed when the cells were stimulated with the nonspecific antigen HEL or were incubated with medium alone (data not shown). In order to evaluate the humoral response, we pooled fractions 5 to 7 and 8 to 10, given their similar protein profiles on SDS-polyacrylamide gels. HLA-DR4 tg mice were immunized i.n. with the respective pooled fractions plus CpG, boosted on day 14, and bled on day 28; ELISA analysis for serum antibody was subsequently performed. As shown in Fig. 2B, sera from mice vaccinated with pooled fractions 5 to 7 induced significantly (P < 0.005) greater antigen-specific total antibody production than sera from animals immunized with fractions 8 to 10. Minimal binding of immune sera to the unrelated antigen HEL was observed, and as expected, sera from mock-treated mice did not react to any of the Rotofor fraction groups (data not shown). Collectively, Rotofor fractions 5 to 7 were subjected to further analysis, given the robust antigen-specific IFN-γ and antibody response.
FIG. 1.
Fractionation of TX-114-extracted F. novicida proteins. Cell wall- and membrane-associated proteins partitioned in the TX-114 aqueous phase were fractionated (pI 3.7 to 9.3) with a Rotofor apparatus. The protein profiles of the Rotofor fractions separated by SDS-PAGE were visualized by Coomassie blue staining. Std., molecular mass standard.
FIG. 2.
Analysis of cell-mediated and humoral responses induced by Rotofor fractions. (A) Rotofor fractions 5 to 10 were used to stimulate spleen cells from ΔiglC (KKF24)-primed or mock (PBS)-treated HLA-DR4 tg mice using a cytokine recall assay; (B) sera were collected at day 28 to determine the antibody profiles for the respective pooled proteins.
Subfractionation of Rotofor fractions.
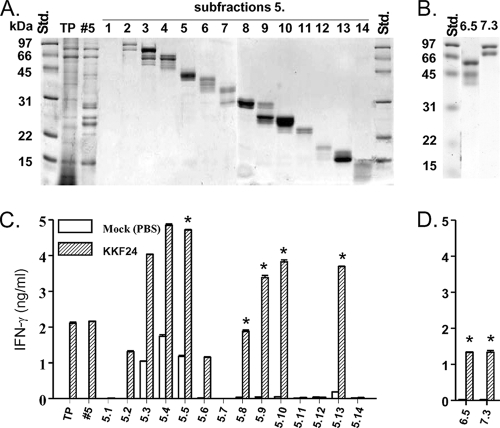
Each of the selected Rotofor fractions (fractions 5 to 7) was further subfractionated by whole-gel elution. As shown in Fig. 3A, proteins from fraction 5 were separated primarily on the basis of molecular mass; however, multiple Coomassie blue-stained bands were visible in most of the 14 whole-gel subfractions, indicating the presence of more than one protein in each individual subfraction. Rotofor fractions 6 and 7 were also subfractionated by whole-gel elution. To further identify potential subunit vaccine candidates within these fractions, we utilized a similar screening approach using humanized HLA-DR4 tg mice to assess antigen-specific cellular cytokine recall responses. These analyses revealed marked cellular cytokine recall responses with total TX-114-extracted protein (TP); Rotofor fraction 5; and subfractions 5.2, 5.3, 5.4, 5.5, 5.6, 5.8, 5.9, 5.10, and 5.13 (Fig. 3C). Additionally, antigen-specific IFN-γ responses were observed in subfractions 6.5 and 7.3 (Fig. 3D). The protein profiles of subfractions 6.5 and 7.3 are shown in Fig. 3B. Negligible IFN-γ production was observed with cells from mock-vaccinated animals or with cells from vaccinated animals stimulated with HEL or medium alone (data not shown). Therefore, on the basis of their immunogenicities and molecular masses, subfractions 5.5, 5.8, 5.9, 5.10, 5.13, 6.5, and 7.3 were selected for protein identification.
FIG. 3.
Screening of respective whole-gel elution fractions that contain potential T-cell antigens. (A) Rotofor fraction 5 was fractionated into 14 subfractions (subfractions 1 to 14), and the protein bands were visualized by Coomassie blue staining. (B) Rotofor fractions 6 and 7 were subfractionated by whole-gel elution in a fashion identical to that described for Rotofor fraction 5. Subfractions of Rotofor fraction 5 (C) and subfraction 6.5 and 7.3 (D) were used to stimulate spleen cells from ΔiglC (KKF24)-primed or mock (PBS)-treated HLA-DR4 tg mice in an IFN-γ recall assay. Total TX-114-extracted proteins (TP) were used as a positive control for the cytokine recall assay. *, fractions selected for protein identification.
Identification of putative CD4+ HLA-DR4-restricted T-cell antigens.
Each of the seven selected subfractions was subjected to electrophoresis on 12% SDS-polyacrylamide gels, and Coomassie blue-stained bands were excised, digested with trypsin, and analyzed by MALDI-TOF mass spectrometry. A range of 7 to 14 proteins from each subfraction and a total of 83 distinct proteins were identified. These proteins were filtered, using the bioinformatics strategy outlined in Fig. 4, in order to identify membrane proteins that had minimal homology to human and mouse proteins and that exhibited a high score for MHC class II binding epitopes. Specifically, the PSLpred algorithm was used to eliminate putative cytosolic proteins. The exclusion of cytosolic proteins in this study was done primarily to achieve a manageable number of antigens for characterization and secondarily because cytosolic proteins are theoretically less likely than surface proteins to serve as targets for neutralizing antibody. The BLASTP algorithm (E value = 0.001, filtered by use of the SEG program) (1) was utilized to select proteins with minimal homology to human and mouse proteins. These proteins (19 total) were subsequently subjected to analysis with the HLA-DR4Pred algorithm (5) in order to identify promiscuous HLA-DR4-restricted epitopes with binding scores greater than 1.4. Collectively, these analyses resulted in the identification of the eight proteins listed in Table 1. This list of HLA-DR4-restricted CD4+ T-cell antigens consisted of three hypothetical proteins (FTN_0320, FTN_0918, and FTN_1381) with unknown function and five proteins exhibiting significant homology to other characterized bacterial proteins, according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (30). FTN_0218 is a putative dihydropteridine reductase, which catalyzes the reduction of nitroaromatic compounds and quinines (42). FTN_0545 is a putative glycosyltransferase and may be responsible for catalyzing the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, which results in the formation of glycosidic bonds (7). FTN_0597 is a protein disulfide isomerase belonging to the DsbA family and the Com1-like subfamily. Com1 is a 27-kDa outer membrane-associated immunoreactive protein found in Coxiella burnetii strains (26). FTN_1220 is a sugar transferase involved in lipopolysaccharide synthesis (60). FTN_0119, FTN_0545, and FTN_1220 belong to the cell wall/cell membrane biogenesis category under the cluster of orthologous groups of protein classification (57).
FIG. 4.
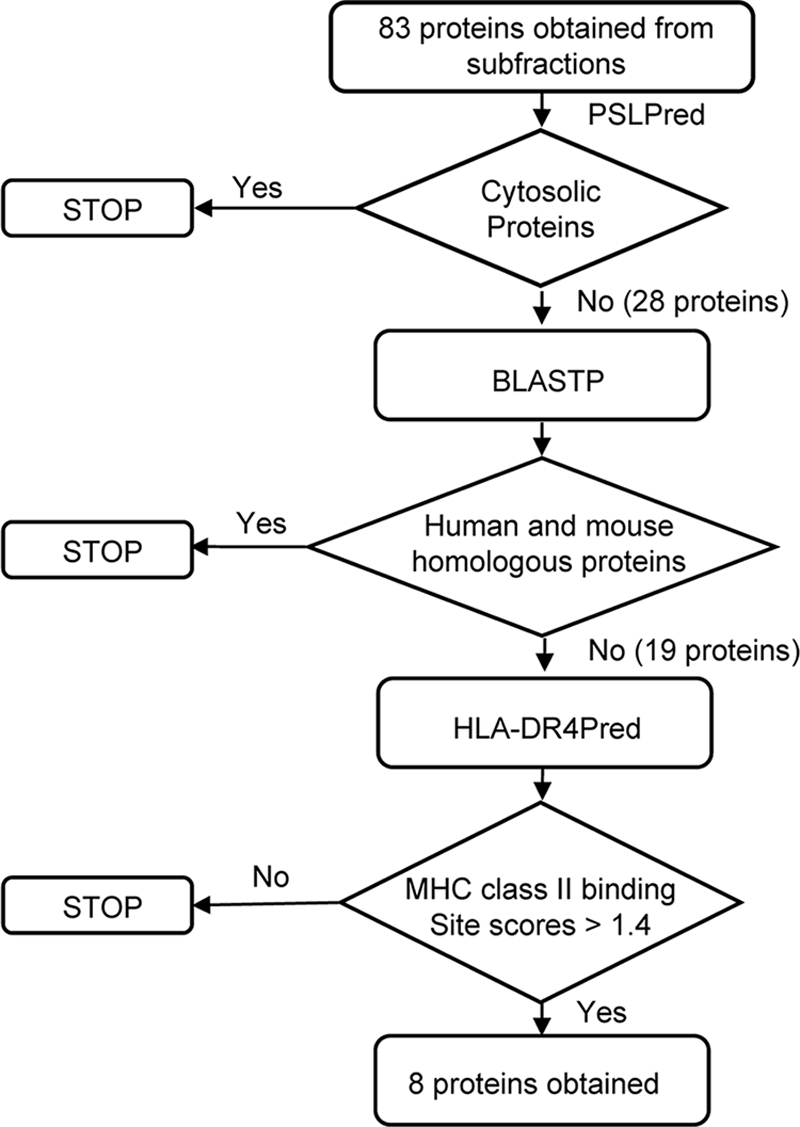
Diagrammatic representation of the sequence of in silico steps used to select putative CD4+ T-cell-reactive antigens.
TABLE 1.
Putative T-cell antigens of Francisella tularensis subsp. novicida containing potential HLA-DR4 binding peptides
| ORFa in U112 | Fraction identified | Protein function | Accession no. | Molecular mass (kDa) | ORF in SCHU S4 (% identity) |
|---|---|---|---|---|---|
| FTN_0119 | 5.10 | Conserved outer membrane protein | YP_897784 | 19.4 | FTT_1747 (100) |
| FTN_0218 | 5.8 | Dihydropteridine reductase | YP_897882 | 24.8 | FTT_0304 (99.08) |
| FTN_0320 | 5.9 | Hypothetical protein | YP_897980 | 29.3 | FTT_1589 (97.98) |
| FTN_0545 | 5.10 | Glycosyltransferase, group 2 family protein | YP_898194 | 36.1 | FTT_0454 (100) |
| FTN_0597 | 5.9 | Protein disulfide isomerase | YP_898246 | 27.9 | FTT_0507 (98.43) |
| FTN_0918 | 5.9 | Conserved hypothetical lipoprotein | YP_898560 | 23.5 | FTT_1040 (97.54) |
| FTN_1220 | 6.5 | Sugar transferase involved in lipopolysaccharide synthesis | YP_898855 | 52.9 | FTT_0790 (98.06) |
| FTN_1381 | 7.3 | Hypothetical protein | YP_899005 | 53.3 | FTT_1415 (100) |
ORF, open reading frame.
Validation of FTN_0119 as a CD4+ HLA-DR4-restricted T-cell antigen.
As a first step toward validating the list of T-cell antigens identified, we further characterized the immunogenicity of FTN_0119. This protein, an OmpH-like protein, has been annotated as an outer membrane protein with unknown function (45) and was designated FopB in this study. The full-length fopB gene, amplified from the genomic DNA of F. novicida U112 by PCR, was cloned into a pET28a expression vector. The resulting plasmid (pET28a-FopB) was used to transform E. coli BL21 for overexpression of an rFopB protein. As shown in Fig. 5, rFopB containing a C-terminal His tag was expressed only in pET28a-FopB-transformed bacteria with IPTG induction and not in uninduced or pET28a vector-transformed bacteria. It was also observed that rFopB was markedly secreted into the culture medium, affording easy purification of rFopB by nickel affinity chromatography. To validate FopB as a T-cell antigen, HLA-DR4 tg mice were immunized with 106 CFU of KKF24 or were mock immunized with PBS. Spleens were collected on day 14, single cells were prepared, and HLA-DR4-specific CD4+ T cells were enriched (>95% purity) from nonadherent cells and stimulated with rFopB for 72 h in the presence of a homozygous human HLA-DR4 (DRB1*0401)-expressing human B-cell line as antigen-presenting cells. Analyses of the culture supernatants revealed a marked induction of IFN-γ (P < 0.001) in a dose-dependent manner by the KKF24-primed HLA-DR4 CD4+ T cells stimulated with rFopB in the presence of the HLA-DR4-expressing human APCs (Fig. 6). As expected, CD4+ T cells primed with UV-inactivated KKF24 induced a greatly elevated IFN-γ response. Minimal IFN-γ responses were observed in the culture supernatants of cells cultured in the presence of the unrelated antigen HEL or medium alone.
FIG. 5.
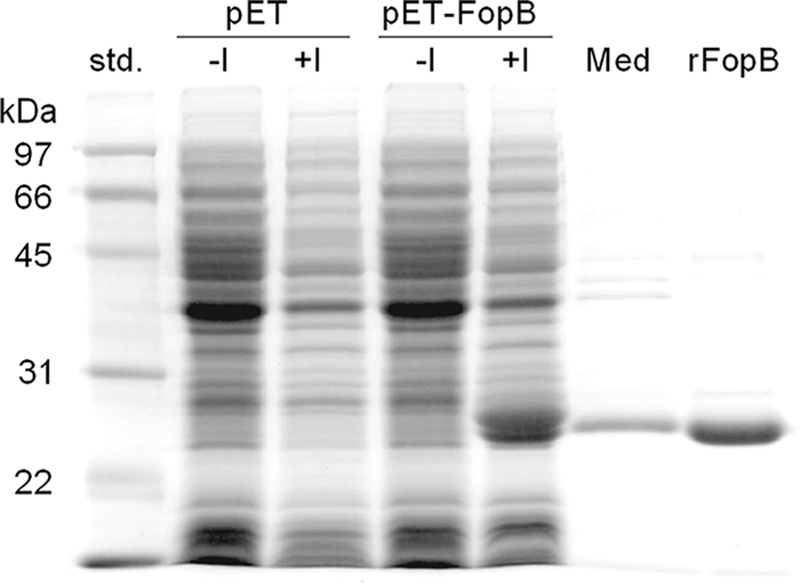
Expression and isolation of recombinant FopB protein. Total protein was prepared from pET28a-FopB-transformed E. coli BL21(DE3) cells or from the pET28a vector with or without IPTG induction and was visualized by Coomassie blue staining. Also shown are the culture medium (Med) prepared from the IPTG-induced pET28a-FopB transformant and the rFopB purified by nickel-affinity chromatography.
FIG. 6.
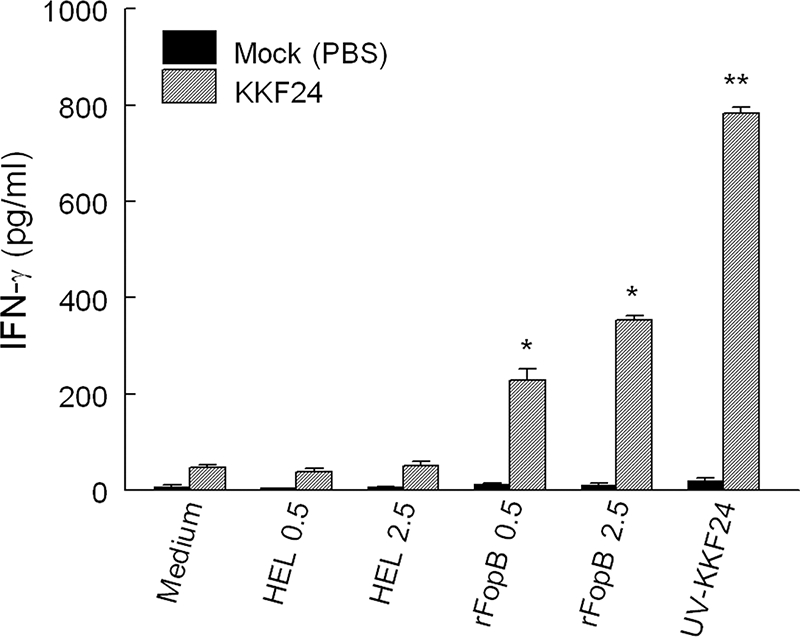
FopB is a T-cell reactive antigen. CD4+ T cells were enriched from KKF24- or mock (PBS)-vaccinated HLA-DR4 tg mice and stimulated with increasing concentrations of rFopB in the presence of human APCs expressing HLA-DR4. The level of induction of IFN-γ was measured after stimulation for 72 h. Differences in IFN-γ stimulation between KKF24- and mock-vaccinated CD4+ T cells was significant at P values of <0.001 for rFopB (*) and <0.0001 for UV-inactivated KKF24 (**).
Efficacy of intranasal vaccination with rFopB.
Given the immunogenicity of FopB in inducing robust IFN-γ responses, we evaluated the protective immunity induced following vaccination with rFopB. C57BL/6 and HLA-DR4 tg mice were vaccinated i.n. or s.c. with rFopB plus CpG, boosted once on day 14, and subsequently challenged on day 30 with LVS. The use of CpG as the adjuvant was based on findings described in our previous publications (11, 34), in which we demonstrated that the inclusion of CpG augmented protection following i.n. vaccination. As shown in Fig. 7, i. n. vaccination with rFopB conferred 50% protection (P < 0.05) against an otherwise lethal pulmonary challenge of 5 LD50s of LVS (3.5 × 104 CFU) in both C57BL/6 and HLA-DR4 tg mice compared with the protection conferred to LVS-challenged mock-vaccinated mice, all of which succumbed to the infection by day 10. In contrast, s.c. immunization with rFopB provided only 33.3% and 16.7% protection in C57BL/6 and HLA-DR4 tg mice, respectively (Fig. 7). Given the protective efficacy conferred by vaccination with this single antigen, we also directly assessed the ability of FopB to induce IFN-γ production. HLA-DR4 tg mice were vaccinated with rFopB plus CpG, and 14 days later spleen cells were analyzed for FopB- and LVS-induced cytokine recall responses. As shown in Fig. 8A, splenocytes stimulated with rFopB induced a marked IFN-γ response (P < 0.0001) in a dose-dependent manner but induced negligible IL-5 production (data not shown). Moreover, UV-inactivated LVS also induced an appreciable IFN-γ response (P < 0.01), further indicating the presence of this protein on the bacterium. Cells obtained from mock (PBS)-vaccinated animals exhibited a minimal IFN-γ response, as did cells stimulated with the unrelated antigen HEL. Vaccinated HLA-DR4 tg mice also produced significant amounts of serum antibody on day 28 (Fig. 8B). Isotype analyses indicated that both Th1-type (IgG2a) and Th2-type (IgG1) antibodies were produced in FopB-vaccinated mice. No FopB-specific antibody was detected in mock (PBS)-vaccinated mice, and none of the serum samples showed reactivity to the unrelated antigen HEL (data not shown). Collectively, i.n. immunization with FopB induced antigen-specific humoral and cell-mediated immune responses and provided partial protection against lethal pulmonary LVS challenge in C57BL/6 and humanized HLA-DR4 tg mice.
FIG. 7.
Intranasal immunization with rFopB-protected mice against pulmonary LVS challenge. C57BL/6 and HLA-DR4 tg mice (6/group) were i.n. or s.c. primed and boosted with rFopB. The mice were rested for 16 days after boosting, challenged i.n. with 35,000 CFU of LVS (∼5 LD50s), and monitored for survival. LVS-challenged mock-vaccinated mice were used as controls. The difference in the rate of survival between i.n. rFopB-immunized and mock-vaccinated mice was significant at a P value of <0.05 in both strains of mice.
FIG. 8.
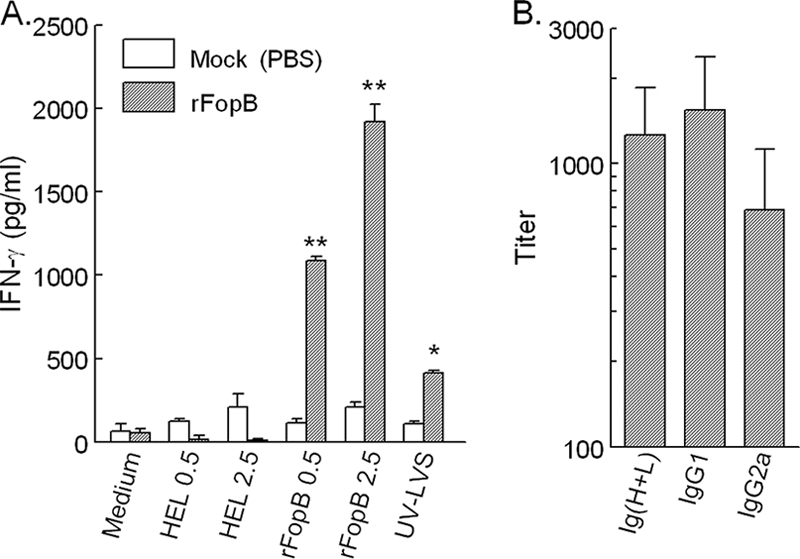
Immune response induced in HLA-DR4 tg mice by intranasal rFopB immunization. (A) Spleens were collected at 14 days postvaccination, and single cells were prepared and assayed for FopB- and LVS-induced IFN-γ production. Differences in IFN-γ stimulation between rFopB- and mock-vaccinated mice were significant at P values of <0.0001 for rFopB (**) and <0.01 for UV-LVS (*). (B) Sera were analyzed for their antibody profiles 14 days postboosting.
DISCUSSION
By using conventional infection models, peptides presented via mouse MHC class II molecules may be different from those presented via HLA molecules in humans. The HLA-DR4 haplotype is one of the major human class II MHC molecules, and the humanized HLA-DR4 tg mouse is an ideal platform for screening for F. tularensis T-cell antigens. Using a robust biochemical membrane fractionation system and HLA-DR4 tg mice, we have identified at least eight putative F. tularensis antigens highly reactive with T cells. We further validated that one of these antigens, FopB, is processed and presented by human APCs expressing HLA-DR4 (DRB1*0401) and show that vaccination with this single antigen conferred protection against pulmonary LVS challenge.
In this study, we screened antigens only from the aqueous phase of TX-114-extracted membrane-associated cellular components due to the better separation in the subsequent Rotofor fractionation step. A total of 83 proteins were initially identified from fractions that induced cellular IFN-γ responses, and these were subsequently narrowed down to eight proteins exhibiting a high binding affinity for HLA-DR4. This biochemical isolation strategy relied on the use of Triton X-114 to isolate proteins from the cell membrane (6, 46). The low cloud point (20°C) of TX-114 permits partitioning of proteins into the detergent and aqueous phases on the basis of the hydrophobicity of the protein. Although most antigen identification in previously reported studies (23, 36, 58) has been focused on detergent-phase proteins, those proteins present in the aqueous phase may have considerable value as antigens. Specifically, Mycobacterium tuberculosis cell membrane proteins partitioned from the aqueous phase have been shown to stimulate peripheral blood mononuclear cells from Mycobacterium bovis BCG-vaccinated humans to a comparable degree to detergent-phase proteins (50). Moreover, an outer membrane cell wall protein (SOWgp) of Coccidioides immitis partitioned in the TX-114 aqueous phase was found to elicit both humoral and cell-mediated immune responses in patients with coccidioidomycosis (28).
F. novicida was used in this study due to the high degree of genetic similarity and protein homology to the genes and proteins of virulent type A organisms (45). Moreover, F. novicida exhibits a high degree of virulence in mice (33, 40) and exhibits features of evasion of phagolysosomal fusion and phagosome escape within infected human-derived macrophages similar to those of F. tularensis subsp. tularensis (9, 47). F. novicida has also proven to be greatly amenable to genetic manipulation (22), in contrast to other F. tularensis subspecies, allowing the identification and study of defined attenuating mutations by several laboratories (8, 14, 15, 25, 32, 37). Our laboratory has utilized F. novicida as a comparative organism to discern mechanisms of adaptive immunity between U112, LVS, and type A strain SCHU S4 and have recently shown that i.n. vaccination with a defined F. novicida strain induces protective immunity against homotypic and heterotypic challenge (12, 40, 41, 43, 44), providing evidence for the presence of conserved protective antigens. This is further evident from the results of our current screening with HLA-DR4 tg mice, in which the proteins identified in F. novicida exhibited a high degree of identity to SCHU S4.
Of the eight T-cell-reactive antigens identified, FopB (FTN_0119) was the first protein that we further characterized for immunogenicity and evaluated as a potential subunit vaccine candidate. FopB is a 19-kDa protein and has distant homology to OmpH of Coxiella (BLASTP expectation score, 5e−08). The OmpH-like proteins are present in many bacterial species (35); however, the functions of these proteins have not been well characterized. OmpH-like proteins have been reported to confer protective immunity to Flavobacterium psychrophilum (16) and Chlamydia pneumoniae (20). Our results showed that FopB is a HLA-DR4-restricted T-cell antigen and can be presented by homologous human APCs. Conventional C57BL/6 and HLA-DR4 tg mice vaccinated i.n. with the single antigen rFopB plus CpG were shown to be partially protected (50% survival) against lethal pulmonary LVS challenge, suggesting that this antigen may be a good candidate for use in the formulation of a more effective multivalent tularemia vaccine. Moreover, intranasal immunization with rFopB plus CpG was found to induce protection greater than that conferred by the s.c. route. Intranasal vaccination has the added advantage of directly targeting mucosal surfaces and may augment the levels of local secretory antibodies and cellular immunity at the site of primary infection. Indeed, reports have shown that aerosol/i.n. vaccination with LVS results in better protection than scarification/subcutaneous vaccination against pulmonary type A challenge in mice, monkeys, and humans (17, 27, 59). Given the success seen with FopB, expression and immunovalidation of the remaining seven novel proteins containing putative HLA-DRB*0401 binding epitopes are under way. The translational value of HLA-DR4 tg mice has been shown with the identification of T-cell-reactive peptides against the outer surface protein of Borrelia burgdorferi, which tends to generate a similar immune response in patients with treatment-resistant Lyme spirochete-induced arthritis (24). Moreover, we have recently reported on the utility of the HLA-DR4 tg mice in mapping a specific HLA-DR4-restricted chlamydial epitope (39). Given that Francisella-specific antigens can be presented in an HLA-DR-restricted manner in LVS-vaccinated humans (54, 55), proteins identified with HLA-DR4 tg mice provide a powerful platform that may be used to map candidate CD4+ T-cell antigens that may be highly immunogenic in humans.
Effective antigen processing and presentation via the MHC II pathway results in the induction of antigen-specific CD4+ T cells, which facilitate the development of robust humoral immunity. To this end, we have shown (43, 44) the requirement of antigen-specific CD4+ T cells in conferring protective immunity against F. novicida and F. tularensis challenge. A similar contribution of CD4+ T cells in conferring protective immunity against pulmonary Francisella LVS challenge has been shown by others (3). Antigen-specific CD4+ T cells are a significant source of inflammatory cytokines, such as IFN-γ, that have been shown to be vital in providing protective immunity against F. tularensis by participating in the activation of phagocytic cells and the containment of bacterial dissemination. Moreover, IFN-γ is a known isotype switch factor for IgG2a production (10) and the subsequent induction of effective humoral responses.
In summary, by using a combinatorial biochemical and bioinformatics approach, we have reported on the feasibility of using humanized HLA-DR4 tg mice for the identification of putative Francisella-specific CD4+ T-cell antigens. The platform developed in this study may be used to identify potential subunit vaccine candidates that may lead to an effective multivalent vaccination strategy to induce protective immunity against F. tularensis in humans.
Acknowledgments
This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Department of Health and Human Services, under grant PO1 AI057986.
We thank Shilpa Sanapala for technical support and Yufeng Wang from the University of Texas at San Antonio for expertise in bioinformatics.
Footnotes
Published ahead of print on 16 December 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anthony, L. S., and P. A. Kongshavn. 1987. Experimental murine tularemia caused by Francisella tularensis, live vaccine strain: a model of acquired cellular resistance. Microb. Pathog. 2:3-14. [DOI] [PubMed] [Google Scholar]
- 3.Baron, S. D., R. Singh, and D. W. Metzger. 2007. Inactivated Francisella tularensis live vaccine strain protects against respiratory tularemia by intranasal vaccination in an immunoglobulin A-dependent fashion. Infect. Immun. 75:2152-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhasin, M., A. Garg, and G. P. Raghava. 2005. PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics 21:2522-2524. [DOI] [PubMed] [Google Scholar]
- 5.Bhasin, M., and G. P. Raghava. 2004. SVM based method for predicting HLA-DRB1*0401 binding peptides in an antigen sequence. Bioinformatics 20:421-423. [DOI] [PubMed] [Google Scholar]
- 6.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 7.Breton, C., and A. Imberty. 1999. Structure/function studies of glycosyltransferases. Curr. Opin Struct. Biol. 9:563-571. [DOI] [PubMed] [Google Scholar]
- 8.Brotcke, A., D. S. Weiss, C. C. Kim, P. Chain, S. Malfatti, E. Garcia, and D. M. Monack. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74:6642-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72:3204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, J. T., and W. A. Dunnick. 1993. Germline transcripts of the murine immunoglobulin gamma-2A gene-structure and induction by IFN-gamma. Int. Immunol. 5:885-891. [DOI] [PubMed] [Google Scholar]
- 11.Cong, Y., M. Jupelli, M. N. Guentzel, G. Zhong, A. K. Murthy, and B. P. Arulanandam. 2007. Intranasal immunization with chlamydial protease-like activity factor and CpG deoxynucleotides enhances protective immunity against genital Chlamydia muridarum infection. Vaccine 25:3773-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong, Y., J. J. Yu, M. N. Guentzel, M. T. Berton, J. Seshu, K. E. Klose, and B. P. Arulanandam. 2009. Vaccination with a defined Francisella tularensis subsp. novicida pathogenicity island mutant (ΔiglB) induces protective immunity against homotypic and heterotypic challenge. Vaccine 27:5554-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cummings, E. D., J. M. Brown, S. T. Sarva, R. H. Waldo, and G. M. Hilliard. 2007. High-throughput proteomics processing of proteins in polyacrylamide in a multiwell format. J. Proteome Res. 6:1603-1608. [DOI] [PubMed] [Google Scholar]
- 14.de Bruin, O. M., J. S. Ludu, and F. E. Nano. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, K., R. J. Blick, W. Liu, and E. J. Hansen. 2006. Identification of Francisella tularensis genes affected by iron limitation. Infect. Immun. 74:4224-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumetz, F., E. Duchaud, S. E. LaPatra, C. Le Marrec, S. Claverol, M. C. Urdaci, and M. Le Henaff. 2006. A protective immune response is generated in rainbow trout by an OmpH-like surface antigen (P18) of Flavobacterium psychrophilum. Appl. Environ. Microbiol. 72:4845-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eigelsbach, H. T., J. J. Tulis, E. L. Overholt, and W. R. Griffith. 1961. Aerogenic immunization of the monkey and guinea pig with live tularemia vaccine. Proc. Soc. Exp. Biol. Med. 108:732-734. [DOI] [PubMed] [Google Scholar]
- 18.Elkins, K. L., A. Cooper, S. M. Colombini, S. C. Cowley, and T. L. Kieffer. 2002. In vivo clearance of an intracellular bacterium, Francisella tularensis LVS, is dependent on the p40 subunit of interleukin-12 (IL-12) but not on IL-12 p70. Infect. Immun. 70:1936-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkins, K. L., S. C. Cowley, and C. M. Bosio. 2007. Innate and adaptive immunity to Francisella. Ann. N. Y. Acad. Sci. 1105:284-324. [DOI] [PubMed] [Google Scholar]
- 20.Finco, O., A. Bonci, M. Agnusdei, M. Scarselli, R. Petracca, N. Norais, G. Ferrari, I. Garaguso, M. Donati, V. Sambri, R. Cevenini, G. Ratti, and G. Grandi. 2005. Identification of new potential vaccine candidates against Chlamydia pneumoniae by multiple screenings. Vaccine 23:1178-1188. [DOI] [PubMed] [Google Scholar]
- 21.Forsthuber, T. G., C. L. Shive, W. Wienhold, K. de Graaf, E. G. Spack, R. Sublett, A. Melms, J. Kort, M. K. Racke, and R. Weissert. 2001. T cell epitopes of human myelin oligodendrocyte glycoprotein identified in HLA-DR4 (DRB1*0401) transgenic mice are encephalitogenic and are presented by human B cells. J. Immunol. 167:7119-7125. [DOI] [PubMed] [Google Scholar]
- 22.Frank, D. W., and T. C. Zahrt. 2007. Genetics and genetic manipulation in Francisella tularensis. Ann. N. Y. Acad. Sci. 1105:67-97. [DOI] [PubMed] [Google Scholar]
- 23.Garg, R., J. K. Srivastava, A. Pal, S. Naik, and A. Dube. 2005. Isolation of integral membrane proteins of Leishmania promastigotes and evaluation of their prophylactic potential in hamsters against experimental visceral leishmaniasis. Vaccine 23:1189-1196. [DOI] [PubMed] [Google Scholar]
- 24.Gross, D. M., T. Forsthuber, M. Tary-Lehmann, C. Etling, K. Ito, Z. A. Nagy, J. A. Field, A. C. Steere, and B. T. Huber. 1998. Identification of LFA-1 as a candidate autoantigen in treatment-resistant Lyme arthritis. Science 281:703-706. [DOI] [PubMed] [Google Scholar]
- 25.Hager, A. J., D. L. Bolton, M. R. Pelletier, M. J. Brittnacher, L. A. Gallagher, R. Kaul, S. J. Skerrett, S. I. Miller, and T. Guina. 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62:227-237. [DOI] [PubMed] [Google Scholar]
- 26.Hendrix, L. R., L. P. Mallavia, and J. E. Samuel. 1993. Cloning and sequencing of Coxiella burnetii outer membrane protein gene com1. Infect. Immun. 61:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornick, R. B., and H. T. Eigelsbach. 1966. Aerogenic immunization of man with live tularemia vaccine. Bacteriol. Rev. 30:532-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung, C. Y., N. M. Ampel, L. Christian, K. R. Seshan, and G. T. Cole. 2000. A major cell surface antigen of Coccidioides immitis which elicits both humoral and cellular immune responses. Infect. Immun. 68:584-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, K., H. J. Bian, M. Molina, J. Han, J. Magram, E. Saar, C. Belunis, D. R. Bolin, R. Arceo, R. Campbell, F. Falcioni, D. Vidovic, J. Hammer, and Z. A. Nagy. 1996. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J. Exp. Med. 183:2635-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohn, D. F., S. K. Wixson, W. J. White, and G. J. Benson. 1997. Anesthesia and analgesia in laboratory animals. Academic Press, New York, NY.
- 32.Lauriano, C. M., J. R. Barker, F. E. Nano, B. P. Arulanandam, and K. E. Klose. 2003. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229:195-202. [DOI] [PubMed] [Google Scholar]
- 33.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U. S. A. 101:4246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, W., A. K. Murthy, M. N. Guentzel, J. Seshu, T. G. Forsthuber, G. Zhong, and B. P. Arulanandam. 2008. Antigen-specific CD4+ T cells produce sufficient IFN-gamma to mediate robust protective immunity against genital Chlamydia muridarum infection. J. Immunol. 180:3375-3382. [DOI] [PubMed] [Google Scholar]
- 35.Marchler-Bauer, A., J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, A. Tasneem, N. Thanki, R. A. Yamashita, D. Zhang, N. Zhang, and S. H. Bryant. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205-D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meens, J., M. Selke, and G. F. Gerlach. 2006. Identification and immunological characterization of conserved Mycoplasma hyopneumoniae lipoproteins Mhp378 and Mhp651. Vet. Microbiol. 116:85-95. [DOI] [PubMed] [Google Scholar]
- 37.Mohapatra, N. P., A. Balagopal, S. Soni, L. S. Schlesinger, and J. S. Gunn. 2007. AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect. Immun. 75:390-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori, M., P. G. Beatty, M. Graves, K. M. Boucher, and E. L. Milford. 1997. HLA gene and haplotype frequencies in the North American population: the National Marrow Donor Program Donor Registry. Transplantation 64:1017-1027. [DOI] [PubMed] [Google Scholar]
- 39.Murthy, A. K., Y. Cong, C. Murphey, M. N. Guentzel, T. G. Forsthuber, G. Zhong, and B. P. Arulanandam. 2006. Chlamydial protease-like activity factor induces protective immunity against genital chlamydial infection in transgenic mice that express the human HLA-DR4 allele. Infect. Immun. 74:6722-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pammit, M. A., V. N. Budhavarapu, E. K. Raulie, K. E. Klose, J. M. Teale, and B. P. Arulanandam. 2004. Intranasal interleukin-12 treatment promotes antimicrobial clearance and survival in pulmonary Francisella tularensis subsp. novicida infection. Antimicrob. Agents Chemother. 48:4513-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pammit, M. A., E. K. Raulie, C. M. Lauriano, K. E. Klose, and B. P. Arulanandam. 2006. Intranasal vaccination with a defined attenuated Francisella novicida strain induces gamma interferon-dependent antibody-mediated protection against tularemia. Infect. Immun. 74:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkinson, G. N., J. V. Skelly, and S. Neidle. 2000. Crystal structure of FMN-dependent nitroreductase from Escherichia coli B: a prodrug-activating enzyme. J. Med. Chem. 43:3624-3631. [DOI] [PubMed] [Google Scholar]
- 43.Powell, H. J., Y. Cong, J. J. Yu, M. N. Guentzel, M. T. Berton, K. E. Klose, A. K. Murthy, and B. P. Arulanandam. 2008. CD4+ T cells are required during priming but not the effector phase of antibody-mediated IFN-gamma-dependent protective immunity against pulmonary Francisella novicida infection. Immunol. Cell Biol. 86:515-522. [DOI] [PubMed] [Google Scholar]
- 44.Ray, H. J., Y. Cong, A. K. Murthy, D. M. Selby, K. E. Klose, J. R. Barker, M. N. Guentzel, and B. P. Arulanandam. 2009. Oral live vaccine strain-induced protective immunity against pulmonary Francisella tularensis challenge is mediated by CD4+ T cells and antibodies, including immunoglobulin A. Clin. Vaccine Immunol. 16:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohmer, L., C. Fong, S. Abmayr, M. Wasnick, T. J. L. Freeman, M. Radey, T. Guina, K. Svensson, H. S. Hayden, M. Jacobs, L. A. Gallagher, C. Manoil, R. K. Ernst, B. Drees, D. Buckley, E. Haugen, D. Bovee, Y. Zhou, J. Chang, R. Levy, R. Lim, W. Gillett, D. Guenthener, A. Kang, S. A. Shaffer, G. Taylor, J. Z. Chen, B. Gallis, D. A. D'Argenio, M. Forsman, M. V. Olson, D. R. Goodlett, R. Kaul, S. I. Miller, and M. J. Brittnacher. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez-Ferrer, A., R. Bru, and F. Garcia-Carmona. 1994. Phase separation of biomolecules in polyoxyethylene glycol nonionic detergents. Crit. Rev. Biochem. Mol. Biol. 29:275-313. [DOI] [PubMed] [Google Scholar]
- 47.Santic, M., M. Molmeret, and K. Y. Abu. 2005. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 7:957-967. [DOI] [PubMed] [Google Scholar]
- 48.Saslaw, S., H. T. Eigelsbach, J. A. Prior, H. E. Wilson, and S. Carhart. 1961. Tularemia vaccine study. II. Respiratory challenge. Arch. Intern. Med. 107:134-146. [DOI] [PubMed] [Google Scholar]
- 49.Sebastian, S., J. T. Pinkham, J. G. Lynch, R. A. Ross, B. Reinap, L. T. Blalock, J. W. Conlan, and D. L. Kasper. 2009. Cellular and humoral immunity are synergistic in protection against types A and B Francisella tularensis. Vaccine 27:597-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinha, S., K. Kosalai, S. Arora, A. Namane, P. Sharma, A. N. Gaikwad, P. Brodin, and S. T. Cole. 2005. Immunogenic membrane-associated proteins of Mycobacterium tuberculosis revealed by proteomics. Microbiology 151:2411-2419. [DOI] [PubMed] [Google Scholar]
- 51.Sjostedt, A. 2005. Francisella, p. 200-210. In D. J. Brenner, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology. The proteobacteria, part B, vol. 2, 2nd ed. Springer, New York, NY. [Google Scholar]
- 52.Sjostedt, A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105:1-29. [DOI] [PubMed] [Google Scholar]
- 53.Stuart, B. M., and R. L. Pullen. 1945. Tularemic pneumonia: review of American literature and report of 15 additional cases. Am. J. Med. Sci. 210:223-236. [Google Scholar]
- 54.Surcel, H. M. 1990. Diversity of Francisella tularensis antigens recognized by human T lymphocytes. Infect. Immun. 58:2664-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surcel, H. M., J. Ilonen, K. Poikonen, and E. Herva. 1989. Francisella tularensis-specific T-cell clones are human leukocyte antigen class II restricted, secrete interleukin-2 and gamma interferon, and induce immunoglobulin production. Infect. Immun. 57:2906-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarnvik, A., M. L. Lofgren, S. Lofgren, G. Sandstrom, and H. Wolf-Watz. 1985. Long-lasting cell-mediated immunity induced by a live Francisella tularensis vaccine. J. Clin. Microbiol. 22:527-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 58.Wang, L., and R. L. Coppel. 2002. Triton X-114 phase partitioning for antigen characterization. Methods Mol. Med. 72:581-585. [DOI] [PubMed] [Google Scholar]
- 59.Wu, T. H., J. A. Hutt, K. A. Garrison, L. S. Berliba, Y. Zhou, and C. R. Lyons. 2005. Intranasal vaccination induces protective immunity against intranasal infection with virulent Francisella tularensis biovar A. Infect. Immun. 73:2644-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida, Y., Y. Nakano, Y. Yamashita, and T. Koga. 1998. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycetemcomitans. Infect. Immun. 66:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]