Abstract
Notch receptors are transmembrane receptors that regulate cell fate decisions. There are four Notch receptors in mammals. Upon binding to members of the Delta and Jagged family of transmembrane proteins, Notch is cleaved and the Notch intracellular domain (NICD) is released. NICD then translocates to the nucleus, where it associates with the CBF-1, Suppressor of Hairless, and Lag-2 (CSL) and Mastermind-Like (MAML) proteins. This complex activates the transcription of Notch target genes, such as Hairy Enhancer of Split (Hes) and Hes-related with YRPF motif (Hey). Notch signaling is critical for the regulation of mesenchymal stem cell differentiation. Misexpression of Notch in skeletal tissue indicates a role as an inhibitor of skeletal development and postnatal bone formation. Overexpression of Notch inhibits endochondral bone formation and osteoblastic differentiation, causing severe osteopenia. Conditional inactivation of Notch in the skeleton causes an increase in cancellous bone volume and enhanced osteoblastic differentiation. Notch ligands are expressed in the hematopoietic stem cell niche and are critical for the regulation of hematopoietic stem cell self-renewal. Dysregulation of Notch signaling is the underlying cause of diseases affecting the skeletal tissue, including Alagille syndrome, spondylocostal dysostosis, and possibly, osteosarcoma.
Bone remodeling is a temporally and spatially regulated process carried out in discrete multicellular units where osteoblasts form bone and osteoclasts resorb bone in a continuous effort to renew skeletal tissue (14). Osteoblasts are derived from mesenchymal cells, whereas osteoclasts are derived from multipotent hematopoietic cells (13). The number of osteoblasts and osteoclasts is governed by extracellular and intracellular signals that act in a coordinated fashion to maintain skeletal homeostasis. Precursor mesenchymal cells differentiate into cells of various lineages, including osteoblasts, chondrocytes, and adipocytes (9). The differentiation of mesenchymal cells into cells of the osteoblastic lineage is regulated by bone morphogenetic proteins (BMPs) and Wnt (36, 70). The activity of BMPs and Wnt is controlled by extracellular and intracellular proteins, often coexpressed with BMPs and Wnt. The extracellular proteins preclude ligand-receptor interactions by binding to BMPs or Wnt or to their receptors. Extracellular and intracellular proteins are critical to temper the activity of BMPs and Wnt and ensure coordinated skeletal development and function.
Notch is a family of evolutionarily conserved receptors that determine cell fate. As such, they play a role in the differentiation of mesenchymal cells toward osteoblasts. Notch was identified in Drosophila melanogaster, where its inactivation causes notches in the wing blade. In mammals, there are four receptors, termed Notch1 through -4, which are activated following direct contact with their ligands. In vertebrates, there are five Delta/Serrate/Lag-2 (DSL) ligands that are orthologues of Drosophila Delta and Serrate and Caenorhabditis elegans Lag-2. They are known as Jagged 1 (Jag1) and Jag2 and Delta-like 1 (Dll1), Dll3, and Dll4 (76). Notch and DSL ligands are single-pass transmembrane proteins that mediate cell-to-cell signaling. Following ligand receptor interactions, Notch is cleaved and the notch intracellular domain (NICD) is released (107). In Notch canonical signaling, NICD translocates to the nucleus, where it associates with the DNA binding protein Epstein-Barr virus latency C promoter binding factor 1 (CBF1; also known as RBP-Jk in mice), Suppressor of Hairless (identified in Drosophila), and Lag1 (identified in Caenorhabditis) (CSL) to activate transcription (64). In the noncanonical pathway, NICD does not translocate to the nucleus and interacts with Deltex to regulate transcription in a CSL-independent manner (43). Notch interacts with Wnt signaling, and recently Notch was found to be a determinant of osteoblastic cell fate, chondrogenesis, and osteoclastogenesis. As a result, Notch has emerged as an important signal that governs skeletal development, skeletal cell fate, and bone remodeling.
NOTCH RECEPTORS, LIGANDS, AND SIGNALING
Notch receptors.
Notch receptors are highly conserved single-pass transmembrane proteins consisting of an extracellular, a transmembrane, and an intracellular domain (Fig. 1). In the Golgi network, mammalian Notch is cleaved by furin-like pro-protein convertases, and the Notch precursor matures into a heterodimer where the C terminus of the extracellular region binds the N-terminal end of the transmembrane domain. The extracellular domain contains multiple epidermal growth factor (EGF)-like tandem repeats that mediate interactions between Notch and its ligands. EGF repeats bind to calcium ions, and these determine the conformation and affinity of Notch for its ligands (16). The C terminus of the extracellular domain is characterized by a negative regulatory region which prevents receptor activation in the absence of ligands. Point mutations in this region cause unregulated activation of Notch and lead to T-cell acute lymphoblastic leukemia in humans (79, 130). The N terminus of the transmembrane domain of Notch is extracellular and is constituted of a stop translocation signal and the heterodimerization domain. The transmembrane domain is followed by the intracellular domain, which consists of an RBP-Jk association module (RAM) domain linked to seven ankyrin repeats by a nuclear localization sequence. The ankyrin repeats are followed by an additional nuclear localization sequence and a transactivation domain that differs among the four Notch paralogues. The C terminus of the intracellular region contains the proline (P)-, glutamic acid (E)-, serine (S)-, and threonine (T)-rich motifs (PEST) domain that acts as a signal peptide for ubiquitinylation and degradation of NICD (Fig. 1).
FIG. 1.
Domain organization of the four mammalian Notch receptors. Abbreviations: ANK (ankyrin domains), EGF (epidermal growth factor-like), HD (heterodimerization domain), NLS (nuclear localization sequence), PEST (proline-, glutamic acid-, serine-, and threonine-rich domain), RAM (Rbp-Jk association module), TD (transmembrane domain). The number of EFG repeats is 36 in Notch1 and Notch2, 34 in Notch3, and 29 in Notch4. The dotted lines represent interactions between the 2 halves of the Notch heterodimer; the yellow segment represents an additional NLS.
Notch receptors have both redundant and unique functions (131). Notch1 and Notch2 share the highest degree of similarity and are necessary for survival, since inactivation of Notch1 leads to early embryonic death and a hypomorphic allele of Notch2 causes perinatal lethality due to kidney defects (82, 118). Notch3 has a slightly different structural organization than Notch1 and Notch2, and it is characterized by a less active transactivation domain. Notch3 expression is limited to vascular smooth muscle cells, the central nervous system, and selected populations of thymocytes and osteoclasts. Because of its restricted distribution, Notch3 deletion in mice is not lethal, but its constitutive activation phenocopies the cerebral autosomal-dominant arteriopathy with subcortical infarcts (CADASIL) syndrome in humans (7, 86). Activating mutations of Notch3 have been identified in human ovarian and lung carcinomas (59, 100). Notch4, also known as Int3, plays a role in embryonic vascular morphogenesis, but it is dispensable for development since its functions overlap with those of Notch1 (69). Notch4 overexpression targeted to endothelial cells causes brain arteriovenous malformations in mice (88).
Canonical Notch ligands.
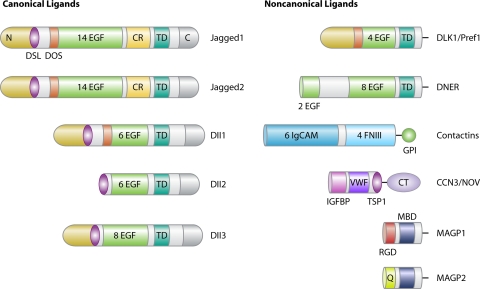
Canonical Notch ligands are transmembrane proteins characterized by multiple tandem EGF repeats in the extracellular domain. Canonical ligands are Jag1 and Jag2, which contain a cysteine-rich (CR) domain, and Dll1, -2, and -3, which lack the CR domain. The Notch binding domain comprises an N-terminal domain followed by a Delta/Serrate/Lag2 (DSL) motif and two characteristic tandem EGF repeats termed Delta and OSM11-like (DOS) protein domains (Fig. 2) (58). The intracellular regions of the canonical ligands are less conserved, and the C-terminal domain can interact with the cytoskeleton (101) (Fig. 2). With the exception of Dll3, the inactivation of Notch ligands causes developmental defects and embryonic lethality, indicating that DSL ligands have no overlapping functions (35, 44, 49, 136). Mice with mutations in Dll3 are viable and present a “pudgy” phenotype, characterized by vertebral and rib deformities secondary to defects in somite patterning (71).
FIG. 2.
Domain organization of the Notch ligands. In the left panel (canonical ligands), abbreviations are as follows: C (C-terminal region), CR (cysteine-rich domain), DOS (Delta and OSM-11-like proteins), DSL (Delta/Serrate/Lag2 motif), EGF (epidermal growth factor-like repeat), N (N-terminal signal), and TD (transmembrane domain). In the right panel (noncanonical ligands), abbreviations are as follows: CT (cysteine knot domain), FNIII (fibronectin type III domain), GPI (glycosylphosphatidylinositol anchor), IgCAM (immunoglobulin-containing cell adhesion molecule domain), IGFBP (insulin-like growth factor binding protein domain), MBD (matrix binding domain), Q (glutamine-rich region), RGD (integrin binding motif), TSP1 (thrombospondin1-like domain), and VWF (Von Willebrand factor type-C-like domain).
Noncanonical notch ligands.
Additional Notch ligands, often termed noncanonical ligands, are structurally heterogeneous transmembrane or soluble proteins that can regulate canonical and noncanonical Notch signaling (19) (Fig. 2). Delta homologue-like 1 (Dlk1), also known as Pref1, is similar to Dll ligands but lacks the DSL domain and inhibits Notch signaling by binding to Notch receptors (6, 127). A second Delta-like protein is Delta/Notch-like EGF-related receptor (DNER), which has the tandem EGF repeats typical of the DSL proteins but lacks the DSL domain. DNER binds and activates Notch in neighboring cells and activates the noncanonical Deltex-dependent pathway (25). F3 and NB3, also known as contactin 1 and 6, respectively, consist of an extracellular domain formed by six repeats of the immunoglobulin-containing cell adhesion molecule domain and four repeats of the fibronectin type III domain. Both contactins activate the noncanonical Deltex-dependent pathway (18, 45). Nephroblastoma overexpressed (CCN3/NOV) and the microfibril-associated glycoprotein (MAGP) family of proteins, MAGP1 and MAGP2, are secreted ligands that can either activate or suppress Notch canonical signaling (1, 85, 104).
Ligand binding and canonical notch signaling.
Notch receptor maturation and activation require several proteolytic events, and the sites for cleavage are sequentially numbered S1 to S4 (61). The S1 site is recognized by furin-like pro-protein convertases in the Golgi network, and it is necessary for the maturation of functional Notch heterodimeric receptors (10). In mammalian cells, internalization of the ligand bound to the Notch extracellular domain, a process known as trans-endocytosis, is necessary to activate Notch signaling (61, 66, 94). These studies have established a model of Notch activation in which a pulling force applied by ligand endocytosis on the Notch extracellular domain is necessary to promote dissociation of the receptor heterodimer. Trans-endocytosis exposes the heterodimerization domain and allows recognition of the S2 site and cleavage of Notch by the tumor necrosis factor α conversion enzyme, a member of the a disintegrin and metalloprotease domain (ADAM) family of metalloproteases (24). ADAM activity generates an unstable intermediate peptide that is recognized by the γ-secretase complex and cleaved at the S3 and S4 intramembranous sites, producing the NICD. The γ-secretase complex is formed by the protease Presenilin and by the regulatory components Nicastrin, Presenilin Enhancer 2, and Anterior pharynx defective 1 (Aph1) (106). Mammals have two different Presenilin isoforms and at least two Aph1 isoforms, and different combinations of the proteins have different functions in vivo (122). Presenilins are critical components of Notch signaling, and deletion of Presenilin1 is lethal due to major abnormalities in the skeleton and central nervous system, whereas Presenilin2 null mice are viable and do not exhibit developmental defects (40, 110). The various proteolytic events result in the release of the NICD, which translocates to the nucleus to regulate transcription (107, 113).
The NICD transcriptional complex.
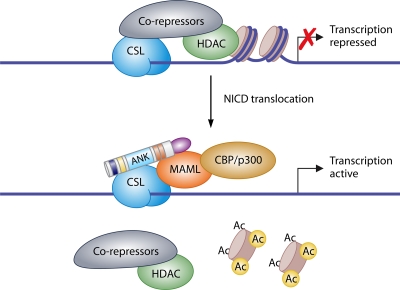
In the absence of NICD, CSL is bound to DNA and to corepressor proteins that recruit histone deacetylase complexes (HDAC) to suppress transcription. NICD displaces the corepressors and forms a ternary complex with CSL and Mastermind-Like (MAML), converting CSL proteins from transcriptional repressors to activators (63). As a consequence, canonical Notch signaling induces the expression of Hairy Enhancer of Split 1 (Hes1), Hes5, Hes6, and Hes7 and HES-related with YRPF motif 1 (Hey1), Hey2, and HeyL (3, 47, 98) (Fig. 3).
FIG. 3.
Activation of transcription by the Notch intracellular domain (NICD). Under basal, unstimulated conditions, corepressors of transcription are bound to Epstein-Barr virus latency C promoter binding factor 1, Suppressor of Hairless, and Lag1 (CSL) and recruit histone deacetylase (HDAC) to suppress transcription. Following translocation of NICD (see Fig. 1 for domain organization) to the nucleus, a ternary complex composed of NICD, CSL, and Mastermind-Like (MAML) is formed. This complex displaces the corepressors and HDAC, resulting in recruitment of CBP/p300, acetylation (Ac) of histones, and gene expression.
CSL is characterized by three domains that are conserved from nematodes to mammals: the N-terminal domain, the β-trefoil domain, and the C-terminal domain (65). The N-terminal and the β-trefoil domains are necessary for DNA binding, and the β-trefoil domain also mediates binding to the RAM domain of the NICD. The C-terminal domain binds to the ANK domain of NICD and to the N-terminal domain of MAML, so that the C-terminal domain of CSL is critical for the formation of the ternary complex (92). In mice and humans, there is a single Csl gene, and its inactivation precludes canonical Notch signaling and causes lethality during early embryogenesis due to vascular abnormalities (68). There are three MAML proteins in mammals which share a helical structure composed of an N-terminal region necessary for binding to CSL and NICD and a C-terminal region that interacts with CBP/p300 and is necessary for transcriptional activity (31, 93, 133). The duration of the Notch transcriptional event is limited, and it is regulated by MAML, which promotes phosphorylation of the PEST domain and the subsequent degradation of the NICD (32).
Regulation of Notch signaling.
The expression of Notch receptors and ligands is regulated in time and space during development and tissue maintenance. This ensures that only certain receptors are activated by specific ligands, providing a basic mechanism of signal regulation (131). Posttranslational modifications, such as glycosylation and regulation of the levels of receptors and ligands at the cell surface through membrane trafficking, are additional mechanisms employed to regulate signaling (20, 126).
The specificity and intensity of binding between Notch and its ligands is partially regulated through glycosylation of the EGF repeats in the Notch extracellular domain. Notch glycosylation by the O-fucosyltransferase Pofut1 is necessary for proper receptor-ligand interactions and trafficking of Notch to the membrane (102). The inactivation of Pofut1 in mice is lethal and phenocopies the deletion of Csl and Presenilins (108, 112). Pofut1 also functions as a chaperone protein necessary for proper Notch folding, and in the absence of Pofut1, Notch signaling is blocked (115). O-Fucose residues are recognized by members of the Fringe family, which are a group of β-1,3-N-acetylglucosaminotransferases that elongate the glycosaminoglycan chain by the addition of N-acetylglucosamines. There are three Fringe proteins in mammals, Lunatic fringe (Lfng), Manic fringe (Mfng), and Radical fringe (Rfng) (126). Glycosylation mediated by Fringe promotes the interaction between Notch and Dll1 ligand and inhibits interactions between Jag1 and Notch (116).
Endocytosis of Notch receptors and ligands maintains optimal levels of signaling proteins, and ligands become competent for signaling after endocytosis and recycling to the plasma membrane. Notch protein levels at the cell surface are regulated by ubiquitin ligases that target Notch for endocytosis (11, 73, 95). In the absence of ligand binding, Notch1 is targeted for degradation in the lysosome by Cbl, which is a Really Interesting New Gene (RING) finger E3 ubiquitin ligase (48). Itch is a murine E3 ubiquitin ligase, a homologue of human AIF4, which acts after the early steps of endocytosis by elongating the ubiquitin chain and promoting Notch1 degradation in the lysosome (15). After ubiquitinylation, Notch can be directed either to the lysosome for degradation or to the recycling endosome to be presented again to the cell surface. The way this process is regulated in mammals is unclear, although a primary role is played by Deltex, a RING E3 ubiquitin ligase involved in Notch noncanonical signaling (87).
Endocytosis and recycling to the plasma membrane are necessary for Dll1 and Dll3 to acquire sufficient affinity for the Notch receptor and to initiate signaling; however, the mechanism involved is unknown (19, 41). In mammals, ubiquitinylation of the intracellular domain of the ligands is critical for the endocytic process, and two DSL ligand ubiquitin ligases have been identified, Neuralized1 and Mind Bomb 1 (Mib1), which ubiquitinylate Jag1 and Dll1, respectively (50, 60, 62, 91). Neuralized1 null mice are viable, whereas the inactivation of Mib1 phenocopies the loss of Notch signaling and causes defects in somitogenesis and impaired vascular and neuronal development that leads to embryonic lethality (60, 103).
TARGET GENES OF CANONICAL NOTCH SIGNALING
Hes and Hey are two families of evolutionarily conserved basic helix-loop-helix (bHLH) transcription factors, which are homologues of Drosophila Hairy and Enhancer of Split and Drosophila Hey, respectively. The Hes family comprises seven members, termed Hes1 through Hes7. With the exception of Hes2 and Hes3, these proteins are established targets of Notch canonical signaling (2, 46, 54). Hes1, -3, and -5 maintain precursor cells in an undifferentiated state during development and adult life in several tissues (39, 51, 89). Hes7 plays a critical role in somite segmentation and regulates the expression of Lfng during mouse development (8). Hes6, a suppressor of Hes1 activity, is expressed during neural development (4). The Hey family is comprised of three members, Hey1 and -2 and HeyL, which are required for normal vascular development (28). The deletion of Hey2 or the combined deletion of Hey1 and HeyL impairs vascular development in mice, whereas the dual inactivation of Hey1 and Hey2 phenocopies the loss of Notch1 (29, 30, 57, 135).
The Hes and Hey proteins have a high degree of structural similarity. Homo- and heterodimerization of Hes and Hey and their dimerization with other bHLH proteins occurs at the bHLH domain. The bHLH domain is followed by an Orange domain which enhances the strength of Hes/Hey interactions (74). The specificity of DNA binding is influenced by the dimerization partner. Heterodimerization of Hes and Hey or heterodimerization with distantly related bHLH transcription factors adds a layer of complexity to the biological impact of Notch signaling (28). Hes proteins can be distinguished by the presence of a conserved proline in the basic domain, whereas Hey proteins have a glycine in the corresponding position. Another difference lies in the C-terminal tetrapeptide motif, where the WRPW and YXXW sequences are present in the Hes and Hey families, respectively (47).
Generally, Hes and Hey proteins act as transcriptional repressors. A common mechanism of action is mediated by the bHLH domain, which confers the ability to recruit HDAC to specific gene promoters. The WRPW motif allows Hes proteins to recruit Transducin-Like Enhancer of Split factors in mammals and, as a consequence, induce the formation of a transcriptional repressor complex (46). Different mechanisms of transcriptional regulation involve interactions with other bHLH factors and the core transcriptional machinery (28). Although Hes and Hey are the better known effectors of Notch signaling, CSL binding sites have been identified in a number of additional promoters, indicating that other genes are potential targets of Notch signaling (28).
NOTCH IN SKELETAL DEVELOPMENT AND BONE REMODELING
Notch and endochondral bone formation.
During development, the appendicular skeleton and parts of the axial skeleton are derived from a template of hyaline cartilage, which arises from the condensation of precursor mesenchymal cells that differentiate into chondrocytes. During endochondral ossification, the chondrocytes in the hyaline cartilage undergo hypertrophy, deposit a mineralized matrix, and become apoptotic. At this stage, blood vessels enter the cartilage template and allow skeletal cell precursors to complete the process of endochondral bone formation.
Notch1 is detected at early stages of chondrocyte proliferation, and activation of Notch signaling in chondrocytic ATDC5 cells suppresses chondrogenic differentiation (17, 128). Conversely, inactivation of Notch signaling with an inhibitor of γ-secretase activity in mouse limb micromass cultures enhances chondrogenic differentiation (33). Hes1 and Hey1 inhibit the activity of the chondrocyte-specific collagen type II α(I) promoter by binding to DNA sequences in close proximity to a SOX9 enhancer recognition site (38). Suppression of chondrogenesis by Notch has been confirmed in vivo. In the chicken embryo, overexpression of Dll1 arrests chondrocyte differentiation and maintains the growth plate chondrocytes in a prehypertrophic state (17). Studies of mice have confirmed the suppressive role of Notch in chondrogenesis (42, 84). In one study, the Prx1 enhancer, which is critical for Prx1 expression in the developing forelimb, was used to delete Presenilin1 or Notch1 and Notch2 in the limb bud, starting from embryonic day 10.5 (78). The conditional deletion of Presenilin1, coupled with a global inactivation of Presenilin2 to prevent Notch cleavage and signaling, caused an accumulation of hypertrophic chondrocytes in the growth plate, resulting in severe skeletal malformations. Mice with a dual Notch1 and Notch2 deletion had a similar phenotype, demonstrating that the phenotype observed following the Presenilin1 and Presenilin2 inactivation was due to the loss of Notch signaling. The conditional deletion of Notch2 caused a developmental phenotype similar to the phenotype observed with the dual deletion of Notch1 and Notch2, suggesting that Notch2 is the predominant regulator of endochondral bone formation (42). In accordance with these results, overexpression of NICD under the control of the Prx1 enhancer suppressed endochondral bone formation in the developing limb (22). Suppression of chondrogenesis was abolished by the conditional deletion of Csl in the context of Notch overexpression, indicating that canonical Notch signaling mediates the effect observed (22). However, the expression of the Prx1 enhancer is not specific to cells of the chondrocytic lineage, and the phenotype observed could be due to indirect effects secondary to the dysregulation of Notch signaling in the limb bud.
Additional studies, using models of overexpression of NICD or conditional deletion of Csl under the control of the chondrocyte-specific collagen type II α(I) promoter, confirmed the inhibitory role of Notch in endochondral bone formation. Notch impaired chondrocyte proliferation and differentiation and caused skeletal abnormalities, whereas the inactivation of Csl caused an enlargement of the hypertrophic chondrocyte zone (84). The involvement of the canonical Notch pathway in endochondral bone formation was confirmed by the results of a subsequent study where Csl was inactivated in cartilage tissue (56, 90). It is of interest that the deletion of Csl in vitro delays and does not accelerate BMP-induced chondrocytic maturation (56) (Fig. 4). There is no information on the role of the noncanonical Notch signaling pathway in chondrocyte differentiation or formation.
FIG. 4.
The diagram represents the effects of Notch in the skeleton. CSL (Epstein-Barr virus latency C promoter binding factor 1, Suppressor of Hairless, and Lag1) indicates involvement of the canonical Notch signaling pathway. Notch suppresses chondrocyte proliferation and differentiation during the formation of the hyaline cartilage and inhibits hypertrophic differentiation. Notch inhibits osteoblast differentiation by interacting with β-catenin (β-Cat), Runt-related transcription factor-2 (Runx-2), and Osterix (Osx). Notch suppresses osteoclastogenesis directly in mononuclear precursors and indirectly by inducing osteoprotegerin (OPG) expression and, as a consequence, suppresses bone remodeling. Notch signaling sustains hematopoietic stem cell self-renewal.
Notch and osteoblast differentiation and function.
Osteoblasts are bone-forming cells that in postnatal development and adult life differentiate from mesenchymal precursors residing in the bone microenvironment (13). The role played by the Notch pathway in the commitment of mesenchymal precursors to the osteoblastic lineage and the subsequent osteoblastic differentiation has been examined in several in vitro and in vivo models. In vitro studies have yielded somewhat controversial results, and Notch was found to both suppress and induce osteoblastic differentiation (21, 96, 97, 109, 121, 125). Activation of Notch achieved by coculturing C2C12 cells, an immortalized murine premyoblast cell line, with cells stably expressing Jag1 maintained mesenchymal cells in an undifferentiated state (97). Overexpression of NICD in ST-2 stromal and MC3T3 osteoblastic cell lines, achieved by transducing retroviral expression vectors, suppressed osteoblastic differentiation secondary to an inhibition of Wnt/β-catenin signaling (21, 109). In contrast, transient induction of Notch signaling by adenoviral vector delivery of NICD or by using immobilized Dll1 or Jag1 was found to enhance selected effects of BMP on osteoblastic cells (96, 121). The discrepancies in these results could be due to differences between the immortalized cell line cultures or the conditions used to induce osteoblast differentiation. It is interesting to note that in the context of BMP stimulation, Notch appears to promote the commitment of mesenchymal cells to the osteoblastic fate, which mirrors the stimulatory effects of Notch on chondrocyte differentiation under conditions of BMP stimulation (56, 96, 121).
In vivo studies of mice have helped to clarify the role of Notch in the skeleton by manipulation of Notch signaling at defined stages of development (27, 42, 140). Transgenic mice expressing NICD under the control of the 2.3-kb type I collagen promoter exhibit increased bone volume and growth retardation. However, this is due to the deposition of woven bone by immature or dysfunctional osteoblasts (27). These effects were mediated by canonical Notch signaling, since the conditional deletion of Csl in the context of NICD induction reversed the phenotype (119). In contrast to the phenotype of transgenics expressing NICD under the control of the 2.3-kb type I collagen promoter, the expression of NICD under the control of the 3.6-kb type I collagen promoter causes a decrease in bone volume that is secondary to a decrease in osteoblast number (140). The differences in the two phenotypes can be explained by the different times of activation for the 2.3-kb and 3.6-kb fragments of the type I collagen promoter, resulting in the arrest of osteoblastic cell differentiation at different stages of maturation (52). It is possible that NICD expression under the control of the 2.3-kb type I collagen promoter, which is restricted to more mature osteoblasts, represses terminal osteoblastic differentiation, allowing for the proliferation of immature, dysfunctional cells (27). Conversely, NICD expression under the control of the 3.6-kb type I collagen promoter would repress osteoblastic differentiation at an earlier stage, leading to a decreased number of mature cells and an osteopenic phenotype (140). Furthermore, when Notch1 and Notch2 or Csl were conditionally deleted by expressing the Cre recombinase under the control of the 2.3-kb type I collagen promoter, no skeletal phenotype was observed, indicating that Notch canonical signaling affects not the function of mature osteoblasts but the differentiation of their precursors (42, 123, 140).
Notch1 overexpression under the control of the Prx1 enhancer induced mesenchymal precursor cell proliferation and suppressed their differentiation. These effects were rescued by the conditional inactivation of Csl in the limb bud, indicating a critical role for canonical Notch signaling in mesenchymal cell differentiation (22). Accordingly, bone marrow stromal cells from Notch1/2 null mice are depleted from osteoblast progenitors, confirming that Notch maintains mesenchymal precursor cells in an undifferentiated state (42). Notch possibly acts by transactivating the Sp7/Osterix promoter and by upregulating Cyclin D and Cyclin E to enhance osteoblast proliferation, as well as by binding to Runt-related transcription factor 2 (Runx-2) to repress osteoblast maturation (27). This occurs by direct interactions between either Notch and Runx-2 or between Hes1, Hey1, and Runx-2 (42). In addition, Notch inhibits the Wnt/β-catenin canonical signaling pathway, explaining the inhibition of osteoblastogenesis (140). The results from these in vivo studies offer an alternative explanation for the conflicting results obtained in vitro. It is conceivable that the stimulatory effects of Notch on osteoblastic differentiation are due to the transient nature of signal activation which is inherent in the use of adenoviral vectors or immobilized Jag1 ligand. This could allow the proliferation of immature osteoblastic cells that, upon attenuation of the Notch stimulus, would proceed with the osteoblastic differentiation program.
Hes and Hey proteins appear to suppress osteoblastic differentiation, but Hes1 interacts with Runx-2 to regulate osteocalcin and osteopontin promoter activity, suggesting additional functions (83, 111, 138, 143). The role of Hes and Hey in osteoblasts has been studied in different in vivo models. Transgenic mice overexpressing Hes1 under the control of the 3.6-kb type I collagen promoter exhibit osteopenia due to impaired osteoblastic function. Conversely, conditional deletion of Hes1 in mature osteoblasts resulted in increases in trabecular bone volume, osteoblast number, and mineral apposition rate (139). These results are in line with those observed following the misexpression of Notch and could indicate that Hes1 plays a role mediating the effects of Notch in the skeleton. The skeletal phenotype of transgenic mice, where Hey1 is ubiquitously overexpressed, was analyzed. Data from this model are difficult to interpret, since the effects observed could be due to secondary, nonskeletal effects. Hey1 transgenic mice display mild impairment of osteoblastic function, which is consistent with the effects of Notch signaling in the skeleton (105). In accordance with the skeletal phenotype of Hey1 transgenics, mice heterozygous for a Hey1 null allele in a HeyL null background exhibited increased bone mass (123) (Fig. 4). Whereas the Notch canonical signaling pathway is active in skeletal cells and is responsible for the inhibitory effects of Notch on osteoblastogenesis, there is no evidence that the noncanonical signaling pathway plays a role determining the fate or function of osteoblasts.
Notch and the hematopoietic stem cell niche.
Hematopoiesis is a process during which hematopoietic cells of different lineages are formed from a common precursor stem cell. The mammalian bone marrow sustains hematopoiesis during late embryonic and postnatal life, starting from the formation of the bone marrow cavity. The microenvironment supporting the self-renewal and the regulation of the differentiation of hematopoietic stem cells is defined as the hematopoietic stem cell niche, and it is located in the endosteal surface of the cortical bone (77, 129, 132, 134, 141). Regulation of Notch signaling during hematopoiesis is critical, and the expression of NICD in murine hematopoietic precursor cells or the exposure of the precursors to Notch ligands can lead to their enhanced self-renewal (53, 117).
Recent studies suggest that cells present in the hematopoietic niche induce Notch signaling in hematopoietic cells by expressing Notch ligands. Transgenic mice expressing a constitutively active parathyroid hormone (PTH) receptor under the control of the 2.3-kb type I collagen promoter display enhanced Jag1 expression and Notch signaling in osteoblasts and bone marrow stromal cells. An ex vivo coculture system, using bone marrow stromal cells from the transgenic mice or treatment of bone marrow stromal cells with PTH, caused the expansion of the hematopoietic stem cell population. These effects were suppressed by treatment with a γ-secretase inhibitor to prevent Presenilin activity and Notch signaling, demonstrating that Notch canonical signaling was involved in the process (12). In a subsequent study, the conditional inactivation of Mib1 in the bone marrow microenvironment, to suppress endocytosis and the activation of DSL ligands, impaired hematopoiesis, causing a myeloproliferative disorder and confirming a critical role for Notch and its ligands in the hematopoietic stem cell niche (55). However, contradictory results were obtained with an alternate approach. The conditional deletion of Jag1 and Notch1 in the bone marrow microenvironment did not affect the hematopoietic stem cell self-renewal, indicating that Jag1 and Notch1 are dispensable for this process (80). It is possible that other Notch ligands or paralogues are responsible for the effects of Notch signaling on the hematopoietic stem cell niche, and the exact mechanism remains to be defined (Fig. 4).
Notch and osteoclastogenesis.
Osteoclasts are multinucleated cells that form through the aggregation of bone marrow mononuclear cell precursors. Osteoclast and osteoblast activities are coordinated through the Receptor Activator of NF-κB-Ligand (RANKL)-Osteoprotegerin axis (72, 120). RANKL and macrophage colony-stimulating factor (M-CSF) are required for osteoclastogenesis, and RANKL activity is opposed by osteoprotegerin, a soluble decoy receptor. RANKL and osteoprotegerin are expressed by bone marrow stromal cells and osteoblasts, and their ratio is critical for the regulation of osteoclastic activity.
Notch suppresses the differentiation of bone marrow osteoclast precursors (5, 27, 137). The deletion of Notch1, Notch2, and Notch3 in murine myeloid cells enhances osteoclast precursor proliferation and differentiation in vitro. Accordingly, exposure of osteoclast precursors to immobilized Dll1, retroviral infection with NICD, or coculture of mononuclear precursors with stromal cells expressing Jag1 suppresses M-CSF- and RANKL-induced osteoclastogenesis (5, 137). Inactivation of Notch1 in osteoblasts causes enhanced osteoclastic differentiation and resorptive activity by suppressing the expression of osteoprotegerin, confirming that Notch inhibits osteoclastogenesis (5). The deletion of Presenilin1 and Presenilin2 in osteoblasts also results in suppressed osteoprotegerin levels, increasing osteoclastogenesis and causing osteopenia (Fig. 4) (27). In contrast, the induction of Notch2 in osteoclast precursors can enhance RANKL-induced osteoclastogenesis. Notch2 acts in conjunction with nuclear factor κB, possibly by regulating the nuclear factor of activated T cells c1 (NFAT-c1) promoter during the terminal phases of osteoclast differentiation (34). This suggests that, under specific conditions, Notch2 can induce osteoclastogenesis.
SKELETAL DISORDERS ASSOCIATED WITH NOTCH SIGNALING
Developmental disorders.
Consistent with the role of Notch signaling in patterning and somitogenesis, mutations in Notch ligands or in other Notch signaling components lead to skeletal developmental defects (124). Mutations of the Notch ligand Jag1 and, rarely, of Notch2 cause Alagille syndrome. The syndrome follows an autosomal dominant inheritance and is characterized by defective bile duct formation leading to cholestatic liver disease and by congenital heart disease characterized most frequently by pulmonary artery and pulmonary valve stenosis and tetralogy of Fallott. Patients with Alagille syndrome also present with defects of the anterior chamber of the eye and developmental skeletal defects. There is widespread involvement of the skeleton, characterized by vertebral segmentation defects presenting as butterfly vertebrae or hemivertebrae and absence of sacrum. The facies are dysmorphic, the forehead is high and broad, and the eyes are deep set. Alagille syndrome can also present with vascular abnormalities and intracranial bleeding, craniosynostosis, and digit abnormalities. Children with Alagille syndrome have decreased bone area and decreased bone mineralization. This can be attributed to the cholestatic liver disease, which can cause liver failure and malnutrition (99).
The Jag1 mutations associated with Alagille syndrome lead to premature termination codons, splice mutations, missense mutations, and gene deletions. Alagille syndrome has high penetrance but variable expressivity, indicating the existence of genetic modifiers of the disease phenotype. Often the mutations cause disease in one or a few organ systems. Jag1 mutations in mice are lethal, and heterozygous mice do not express most of the phenotypes associated with the human syndrome (37, 67). Moreover, the Jag1 heterozygous mice containing a Notch2 hypomorphic allele exhibit growth retardation and bile duct, heart, eye, and kidney abnormalities resembling those occurring in Alagille syndrome (37, 67, 81).
Spondylocostal dysostosis is caused by mutations of Dll3 that lead to the expression of a truncated protein or to missense mutations. Both autosomal dominant and recessive forms have been reported (37, 124). Spondylocostal dysostosis is characterized by vertebral segmentation defects and rib anomalies, presenting with hemivertebrae and rib fusions and deletions, leading to trunk dwarfism. A spontaneous mutation in Dll3 in the mouse, the pudgy mutation, causes a similar phenotype (71). Mesoderm posterior 2 (Mesp2) is an HLH transcription factor regulated by Notch signaling. Its inactivation in the mouse is lethal and leads to abnormal segmentation. In humans, mutations of Mesp2 cause spondylocostal dysostosis type 2, which is manifested by marked segmentation abnormalities of the thoracic spine (124). Lfng deletion in the mouse causes costovertebral abnormalities, and gene mutations of Lfng in humans are associated with spondylocostal dysostosis type 3, which is characterized by hemivertebrae and rib anomalies (23, 114).
Osteosarcoma.
Notch signaling plays a role in a variety of hematologic and solid tumor malignancies, and Notch activation can be oncogenic (75). For example, aberrant expression of constitutively active Notch1 is associated with T-cell acute lymphoblastic leukemia and lymphoma, and dysregulated Notch signaling is involved in multiple myeloma. The mechanisms that have been postulated are induction of cell proliferation and inhibition of apoptosis. Recently, enhanced Notch signaling was found to be associated with osteosarcoma. In human osteosarcoma, increased expression of Jag1 and Notch1 and the target genes Hes1 and Hey1 were reported (26). Activation of Notch signaling is also associated with the invasive potential of osteosarcoma cells (142). Inhibition of Notch signaling reduces tumor growth in vitro and tumor burden in vivo. These results indicate a possible role of Notch signaling in the pathogenesis of osteosarcoma and in its metastatic potential.
CONCLUSIONS
In conclusion, Notch is a novel signaling pathway that plays a critical role in cell fate. In the skeleton, Notch inhibits chondrogenesis, osteoblastogenesis, and osteoclastogenesis. As a consequence, it has an inhibitory function in bone formation and bone remodeling. The expression of Notch ligands in the hematopoietic stem cell niche is necessary for self-renewal of the hematopoietic stem cells. Alterations in Notch signaling lead to developmental skeletal disorders and may play a role in the pathogenesis of osteosarcoma.
Acknowledgments
This work was supported by grant DK045227 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Biography
 Stefano Zanotti has been a postdoctoral fellow in Dr. Canalis's laboratory, located at Saint Francis Hospital and Medical Center in Hartford, CT, since the summer of 2006. He graduated in biological sciences at the University of Genova, Italy, in 2002, and in 2006 obtained a doctoral degree in cellular biology at the University of Bari in Italy. Dr. Zanotti has been involved in the study of skeletal biology since the beginning of his career, while working on the experimental thesis for his master's degree. His graduate work was centered on the study of metalloprotease expression. After initiating his postdoctoral training, his interest shifted to the regulation of skeletal development and bone remodeling, with a focus on the role of the Notch signaling network. Dr. Zanotti is involved in this research due to his desire to understand the effects of regulatory networks in biological systems.
Stefano Zanotti has been a postdoctoral fellow in Dr. Canalis's laboratory, located at Saint Francis Hospital and Medical Center in Hartford, CT, since the summer of 2006. He graduated in biological sciences at the University of Genova, Italy, in 2002, and in 2006 obtained a doctoral degree in cellular biology at the University of Bari in Italy. Dr. Zanotti has been involved in the study of skeletal biology since the beginning of his career, while working on the experimental thesis for his master's degree. His graduate work was centered on the study of metalloprotease expression. After initiating his postdoctoral training, his interest shifted to the regulation of skeletal development and bone remodeling, with a focus on the role of the Notch signaling network. Dr. Zanotti is involved in this research due to his desire to understand the effects of regulatory networks in biological systems.
 Ernesto Canalis, a Professor in the Departments of Medicine and Orthopedic Surgery at the University of Connecticut, has dedicated his career to studies pertinent to skeletal cell biology. His laboratory, located at Saint Francis Hospital and Medical Center in Hartford, has been funded by the NIH since 1981. The Canalis laboratory discovered the existence of skeletal growth factors, and his interests center around growth factors and their antagonists, anabolic agents, and determinants of osteoblastic cell fate. In pursuit of these interests, his laboratory investigates the effects of Notch and its target genes in skeletal cells. Dr. Canalis has served as President of the American Society for Bone and Mineral Research (ASBMR) and is a member of the American Society for Clinical Investigation and the Association of American Physicians. He is a recipient of the Endocrine Society Aurbach Award and the ASBMR Avioli award for his contributions to research.
Ernesto Canalis, a Professor in the Departments of Medicine and Orthopedic Surgery at the University of Connecticut, has dedicated his career to studies pertinent to skeletal cell biology. His laboratory, located at Saint Francis Hospital and Medical Center in Hartford, has been funded by the NIH since 1981. The Canalis laboratory discovered the existence of skeletal growth factors, and his interests center around growth factors and their antagonists, anabolic agents, and determinants of osteoblastic cell fate. In pursuit of these interests, his laboratory investigates the effects of Notch and its target genes in skeletal cells. Dr. Canalis has served as President of the American Society for Bone and Mineral Research (ASBMR) and is a member of the American Society for Clinical Investigation and the Association of American Physicians. He is a recipient of the Endocrine Society Aurbach Award and the ASBMR Avioli award for his contributions to research.
Footnotes
Published ahead of print on 7 December 2009.
REFERENCES
- 1.Albig, A. R., D. J. Becenti, T. G. Roy, and W. P. Schiemann. 2008. Microfibril-associate glycoprotein-2 (MAGP-2) promotes angiogenic cell sprouting by blocking notch signaling in endothelial cells. Microvasc. Res. 76:7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Androutsellis-Theotokis, A., R. R. Leker, F. Soldner, D. J. Hoeppner, R. Ravin, S. W. Poser, M. A. Rueger, S. K. Bae, R. Kittappa, and R. D. McKay. 2006. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442:823-826. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 4.Bae, S., Y. Bessho, M. Hojo, and R. Kageyama. 2000. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 127:2933-2943. [DOI] [PubMed] [Google Scholar]
- 5.Bai, S., R. Kopan, W. Zou, M. J. Hilton, C. T. Ong, F. Long, F. P. Ross, and S. L. Teitelbaum. 2008. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J. Biol. Chem. 283:6509-6518. [DOI] [PubMed] [Google Scholar]
- 6.Baladron, V., M. J. Ruiz-Hidalgo, M. L. Nueda, M. J. Diaz-Guerra, J. J. Garcia-Ramirez, E. Bonvini, E. Gubina, and J. Laborda. 2005. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp. Cell Res. 303:343-359. [DOI] [PubMed] [Google Scholar]
- 7.Bellavia, D., S. Checquolo, A. F. Campese, M. P. Felli, A. Gulino, and I. Screpanti. 2008. Notch3: from subtle structural differences to functional diversity. Oncogene 27:5092-5098. [DOI] [PubMed] [Google Scholar]
- 8.Bessho, Y., R. Sakata, S. Komatsu, K. Shiota, S. Yamada, and R. Kageyama. 2001. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15:2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianco, P., and R. P. Gehron. 2000. Marrow stromal stem cells. J. Clin. Invest. 105:1663-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaumueller, C. M., H. Qi, P. Zagouras, and S. Artavanis-Tsakonas. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90:281-291. [DOI] [PubMed] [Google Scholar]
- 11.Brou, C. 2009. Intracellular trafficking of Notch receptors and ligands. Exp. Cell Res. 315:1549-1555. [DOI] [PubMed] [Google Scholar]
- 12.Calvi, L. M., G. B. Adams, K. W. Weibrecht, J. M. Weber, D. P. Olson, M. C. Knight, R. P. Martin, E. Schipani, P. Divieti, F. R. Bringhurst, L. A. Milner, H. M. Kronenberg, and D. T. Scadden. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425:841-846. [DOI] [PubMed] [Google Scholar]
- 13.Canalis, E. 2005. The fate of circulating osteoblasts. N. Engl. J. Med. 352:2014-2016. [DOI] [PubMed] [Google Scholar]
- 14.Canalis, E., A. Giustina, and J. P. Bilezikian. 2007. Mechanisms of anabolic therapies for osteoporosis. N. Engl. J. Med. 357:905-916. [DOI] [PubMed] [Google Scholar]
- 15.Chastagner, P., A. Israel, and C. Brou. 2008. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One 3:e2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordle, J., C. Redfieldz, M. Stacey, P. A. van der Merwe, A. C. Willis, B. R. Champion, S. Hambleton, and P. A. Handford. 2008. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J. Biol. Chem. 283:11785-11793. [DOI] [PubMed] [Google Scholar]
- 17.Crowe, R., J. Zikherman, and L. Niswander. 1999. Delta-1 negatively regulates the transition from prehypertrophic to hypertrophic chondrocytes during cartilage formation. Development 126:987-998. [DOI] [PubMed] [Google Scholar]
- 18.Cui, X. Y., Q. D. Hu, M. Tekaya, Y. Shimoda, B. T. Ang, D. Y. Nie, L. Sun, W. P. Hu, M. Karsak, T. Duka, Y. Takeda, L. Y. Ou, G. S. Dawe, F. G. Yu, S. Ahmed, L. H. Jin, M. Schachner, K. Watanabe, Y. Arsenijevic, and Z. C. Xiao. 2004. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J. Biol. Chem. 279:25858-25865. [DOI] [PubMed] [Google Scholar]
- 19.D'Souza, B., A. Miyamoto, and G. Weinmaster. 2008. The many facets of Notch ligands. Oncogene 27:5148-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dale, J. K., M. Maroto, M. L. Dequeant, P. Malapert, M. McGrew, and O. Pourquie. 2003. Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature 421:275-278. [DOI] [PubMed] [Google Scholar]
- 21.Deregowski, V., E. Gazzerro, L. Priest, S. Rydziel, and E. Canalis. 2006. Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J. Biol. Chem. 281:6203-6210. [DOI] [PubMed] [Google Scholar]
- 22.Dong, Y., A. Jesse, A. Kohn, L. Gunnell, M. Zuscik, R. O'Keefe, T. Honjo, and M. Hilton. 2009. RBPJk-dependent Notch signaling maintains and expands mesenchymal stem cells during skeletal development. J. Bone Miner. Res. 24(Suppl. 1). http://www.asbmr.org/Meetings/AnnualMeeting/Abstracts.aspx.
- 23.Dunwoodie, S. L. 2009. Mutation of the fucose-specific beta1,3 N-acetylglucosaminyltransferase LFNG results in abnormal formation of the spine. Biochim. Biophys. Acta 1792:100-111. [DOI] [PubMed] [Google Scholar]
- 24.Ehebauer, M., P. Hayward, and A. Martinez-Arias. 2006. Notch signaling pathway. Sci. STKE 2006:cm7. [DOI] [PubMed] [Google Scholar]
- 25.Eiraku, M., A. Tohgo, K. Ono, M. Kaneko, K. Fujishima, T. Hirano, and M. Kengaku. 2005. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat. Neurosci. 8:873-880. [DOI] [PubMed] [Google Scholar]
- 26.Engin, F., T. Bertin, O. Ma, M. M. Jiang, L. Wang, R. E. Sutton, L. A. Donehower, and B. Lee. 2009. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum. Mol. Genet. 18:1464-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engin, F., Z. Yao, T. Yang, G. Zhou, T. Bertin, M. M. Jiang, Y. Chen, L. Wang, H. Zheng, R. E. Sutton, B. F. Boyce, and B. Lee. 2008. Dimorphic effects of Notch signaling in bone homeostasis. Nat. Med. 14:299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer, A., and M. Gessler. 2007. Delta-Notch—and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 35:4583-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer, A., N. Schumacher, M. Maier, M. Sendtner, and M. Gessler. 2004. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer, A., C. Steidl, T. U. Wagner, E. Lang, P. M. Jakob, P. Friedl, K. P. Knobeloch, and M. Gessler. 2007. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ. Res. 100:856-863. [DOI] [PubMed] [Google Scholar]
- 31.Fryer, C. J., E. Lamar, I. Turbachova, C. Kintner, and K. A. Jones. 2002. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16:1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fryer, C. J., J. B. White, and K. A. Jones. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16:509-520. [DOI] [PubMed] [Google Scholar]
- 33.Fujimaki, R., Y. Toyama, N. Hozumi, and K. Tezuka. 2006. Involvement of Notch signaling in initiation of prechondrogenic condensation and nodule formation in limb bud micromass cultures. J. Bone Miner. Metab. 24:191-198. [DOI] [PubMed] [Google Scholar]
- 34.Fukushima, H., A. Nakao, F. Okamoto, M. Shin, H. Kajiya, S. Sakano, A. Bigas, E. Jimi, and K. Okabe. 2008. The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol. Cell. Biol. 28:6402-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gale, N. W., M. G. Dominguez, I. Noguera, L. Pan, V. Hughes, D. M. Valenzuela, A. J. Murphy, N. C. Adams, H. C. Lin, J. Holash, G. Thurston, and G. D. Yancopoulos. 2004. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. U. S. A. 101:15949-15954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gazzerro, E., and E. Canalis. 2006. Bone morphogenetic proteins and their antagonists. Rev. Endocr. Metab. Disord. 7:51-65. [DOI] [PubMed] [Google Scholar]
- 37.Gridley, T. 2003. Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 12:R9-R13. [DOI] [PubMed] [Google Scholar]
- 38.Grogan, S. P., T. Olee, K. Hiraoka, and M. K. Lotz. 2008. Repression of chondrogenesis through binding of notch signaling proteins HES-1 and HEY-1 to N-box domains in the COL2A1 enhancer site. Arthritis Rheum. 58:2754-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatakeyama, J., Y. Bessho, K. Katoh, S. Ookawara, M. Fujioka, F. Guillemot, and R. Kageyama. 2004. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131:5539-5550. [DOI] [PubMed] [Google Scholar]
- 40.Herreman, A., D. Hartmann, W. Annaert, P. Saftig, K. Craessaerts, L. Serneels, L. Umans, V. Schrijvers, F. Checler, H. Vanderstichele, V. Baekelandt, R. Dressel, P. Cupers, D. Huylebroeck, A. Zwijsen, L. F. Van, and B. De Strooper. 1999. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc. Natl. Acad. Sci. U. S. A. 96:11872-11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heuss, S. F., D. Ndiaye-Lobry, E. M. Six, A. Israel, and F. Logeat. 2008. The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc. Natl. Acad. Sci. U. S. A. 105:11212-11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hilton, M. J., X. Tu, X. Wu, S. Bai, H. Zhao, T. Kobayashi, H. M. Kronenberg, S. L. Teitelbaum, F. P. Ross, R. Kopan, and F. Long. 2008. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 14:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hori, K., M. Fostier, M. Ito, T. J. Fuwa, M. J. Go, H. Okano, M. Baron, and K. Matsuno. 2004. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 131:5527-5537. [DOI] [PubMed] [Google Scholar]
- 44.Hrabĕ de Angelis, M., J. McIntyre II, and A. Gossler. 1997. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature 386:717-721. [DOI] [PubMed] [Google Scholar]
- 45.Hu, Q. D., B. T. Ang, M. Karsak, W. P. Hu, X. Y. Cui, T. Duka, Y. Takeda, W. Chia, N. Sankar, Y. K. Ng, E. A. Ling, T. Maciag, D. Small, R. Trifonova, R. Kopan, H. Okano, M. Nakafuku, S. Chiba, H. Hirai, J. C. Aster, M. Schachner, C. J. Pallen, K. Watanabe, and Z. C. Xiao. 2003. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell 115:163-175. [DOI] [PubMed] [Google Scholar]
- 46.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237-255. [DOI] [PubMed] [Google Scholar]
- 47.Iso, T., V. Sartorelli, C. Poizat, S. Iezzi, H. Y. Wu, G. Chung, L. Kedes, and Y. Hamamori. 2001. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell. Biol. 21:6080-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jehn, B. M., I. Dittert, S. Beyer, K. von der Mark, and W. Bielke. 2002. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J. Biol. Chem. 277:8033-8040. [DOI] [PubMed] [Google Scholar]
- 49.Jiang, R., Y. Lan, H. D. Chapman, C. Shawber, C. R. Norton, D. V. Serreze, G. Weinmaster, and T. Gridley. 1998. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 12:1046-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin, Y., E. K. Blue, S. Dixon, Z. Shao, and P. J. Gallagher. 2002. A death-associated protein kinase (DAPK)-interacting protein, DIP-1, is an E3 ubiquitin ligase that promotes tumor necrosis factor-induced apoptosis and regulates the cellular levels of DAPK. J. Biol. Chem. 277:46980-46986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kageyama, R., T. Ohtsuka, and T. Kobayashi. 2007. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134:1243-1251. [DOI] [PubMed] [Google Scholar]
- 52.Kalajzic, I., Z. Kalajzic, M. Kaliterna, G. Gronowicz, S. H. Clark, A. C. Lichtler, and D. Rowe. 2002. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J. Bone Miner. Res. 17:15-25. [DOI] [PubMed] [Google Scholar]
- 53.Karanu, F. N., B. Murdoch, L. Gallacher, D. M. Wu, M. Koremoto, S. Sakano, and M. Bhatia. 2000. The notch ligand jagged-1 represents a novel growth factor of human hematopoietic stem cells. J. Exp. Med. 192:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh, M., and M. Katoh. 2004. Identification and characterization of human HES2, HES3, and HES5 genes in silico. Int. J. Oncol. 25:529-534. [PubMed] [Google Scholar]
- 55.Kim, Y. W., B. K. Koo, H. W. Jeong, M. J. Yoon, R. Song, J. Shin, D. C. Jeong, S. H. Kim, and Y. Y. Kong. 2008. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood 112:4628-4638. [DOI] [PubMed] [Google Scholar]
- 56.Kohn, A., Y. Dong, A. Jesse, T. Honjo, R. Kopan, T. Gridley, R. J. O'Keefe, and M. J. Hilton. 2009. RBPJk-dependent Notch signaling regulates chondrocyte maturation in a cartilage specific manner. J. Bone Miner. Res. 24(Suppl. 1). http://www.asbmr.org/Meetings/AnnualMeeting/Abstracts.aspx.
- 57.Kokubo, H., S. Miyagawa-Tomita, M. Nakazawa, Y. Saga, and R. L. Johnson. 2005. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev. Biol. 278:301-309. [DOI] [PubMed] [Google Scholar]
- 58.Komatsu, H., M. Y. Chao, J. Larkins-Ford, M. E. Corkins, G. A. Somers, T. Tucey, H. M. Dionne, J. Q. White, K. Wani, M. Boxem, and A. C. Hart. 2008. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS. Biol. 6:e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konishi, J., K. S. Kawaguchi, H. Vo, N. Haruki, A. Gonzalez, D. P. Carbone, and T. P. Dang. 2007. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 67:8051-8057. [DOI] [PubMed] [Google Scholar]
- 60.Koo, B. K., H. S. Lim, R. Song, M. J. Yoon, K. J. Yoon, J. S. Moon, Y. W. Kim, M. C. Kwon, K. W. Yoo, M. P. Kong, J. Lee, A. B. Chitnis, C. H. Kim, and Y. Y. Kong. 2005. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development 132:3459-3470. [DOI] [PubMed] [Google Scholar]
- 61.Kopan, R., and M. X. Ilagan. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koutelou, E., S. Sato, C. Tomomori-Sato, L. Florens, S. K. Swanson, M. P. Washburn, M. Kokkinaki, R. C. Conaway, J. W. Conaway, and N. K. Moschonas. 2008. Neuralized-like 1 (Neurl1) targeted to the plasma membrane by N-myristoylation regulates the Notch ligand Jagged1. J. Biol. Chem. 283:3846-3853. [DOI] [PubMed] [Google Scholar]
- 63.Kovall, R. A. 2007. Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr. Opin. Struct. Biol. 17:117-127. [DOI] [PubMed] [Google Scholar]
- 64.Kovall, R. A. 2008. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene 27:5099-5109. [DOI] [PubMed] [Google Scholar]
- 65.Kovall, R. A., and W. A. Hendrickson. 2004. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 23:3441-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kramer, H. 2000. RIPping notch apart: a new role for endocytosis in signal transduction? Sci. STKE 2000:E1. [DOI] [PubMed] [Google Scholar]
- 67.Krantz, I. D., D. A. Piccoli, and N. B. Spinner. 1997. Alagille syndrome. J. Med. Genet. 34:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krebs, L. T., J. R. Shutter, K. Tanigaki, T. Honjo, K. L. Stark, and T. Gridley. 2004. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 18:2469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krebs, L. T., Y. Xue, C. R. Norton, J. R. Shutter, M. Maguire, J. P. Sundberg, D. Gallahan, V. Closson, J. Kitajewski, R. Callahan, G. H. Smith, K. L. Stark, and T. Gridley. 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14:1343-1352. [PMC free article] [PubMed] [Google Scholar]
- 70.Krishnan, V., H. U. Bryant, and O. A. MacDougald. 2006. Regulation of bone mass by Wnt signaling. J. Clin. Invest. 116:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kusumi, K., E. S. Sun, A. W. Kerrebrock, R. T. Bronson, D. C. Chi, M. S. Bulotsky, J. B. Spencer, B. W. Birren, W. N. Frankel, and E. S. Lander. 1998. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat. Genet. 19:274-278. [DOI] [PubMed] [Google Scholar]
- 72.Lacey, D. L., E. Timms, H. L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y. X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165-176. [DOI] [PubMed] [Google Scholar]
- 73.Le B, R., A. Bardin, and F. Schweisguth. 2005. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 132:1751-1762. [DOI] [PubMed] [Google Scholar]
- 74.Leimeister, C., K. Dale, A. Fischer, B. Klamt, M. Hrabĕ de Angelis, F. Radtke, M. J. McGrew, O. Pourquie, and M. Gessler. 2000. Oscillating expression of c-Hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev. Biol. 227:91-103. [DOI] [PubMed] [Google Scholar]
- 75.Leong, K. G., and A. Karsan. 2006. Recent insights into the role of Notch signaling in tumorigenesis. Blood 107:2223-2233. [DOI] [PubMed] [Google Scholar]
- 76.Lindsell, C. E., J. Boulter, G. diSibio, A. Gossler, and G. Weinmaster. 1996. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol. Cell. Neurosci. 8:14-27. [DOI] [PubMed] [Google Scholar]
- 77.Lo Celso, C., H. E. Fleming, J. W. Wu, C. X. Zhao, S. Miake-Lye, J. Fujisaki, D. Cote, D. W. Rowe, C. P. Lin, and D. T. Scadden. 2009. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Logan, M., J. F. Martin, A. Nagy, C. Lobe, E. N. Olson, and C. J. Tabin. 2002. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33:77-80. [DOI] [PubMed] [Google Scholar]
- 79.Malecki, M. J., C. Sanchez-Irizarry, J. L. Mitchell, G. Histen, M. L. Xu, J. C. Aster, and S. C. Blacklow. 2006. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 26:4642-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mancini, S. J., N. Mantei, A. Dumortier, U. Suter, H. R. MacDonald, and F. Radtke. 2005. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood 105:2340-2342. [DOI] [PubMed] [Google Scholar]
- 81.McCright, B., J. Lozier, and T. Gridley. 2002. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 129:1075-1082. [DOI] [PubMed] [Google Scholar]
- 82.McCright, B., J. Lozier, and T. Gridley. 2006. Generation of new Notch2 mutant alleles. Genesis 44:29-33. [DOI] [PubMed] [Google Scholar]
- 83.McLarren, K. W., R. Lo, D. Grbavec, K. Thirunavukkarasu, G. Karsenty, and S. Stifani. 2000. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J. Biol. Chem. 275:530-538. [DOI] [PubMed] [Google Scholar]
- 84.Mead, T. J., and K. E. Yutzey. 2009. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc. Natl. Acad. Sci. U. S. A. 106:14420-14425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyamoto, A., R. Lau, P. W. Hein, J. M. Shipley, and G. Weinmaster. 2006. Microfibrillar proteins MAGP-1 and MAGP-2 induce Notch1 extracellular domain dissociation and receptor activation. J. Biol. Chem. 281:10089-10097. [DOI] [PubMed] [Google Scholar]
- 86.Monet, M., V. Domenga, B. Lemaire, C. Souilhol, F. Langa, C. Babinet, T. Gridley, E. Tournier-Lasserve, M. Cohen-Tannoudji, and A. Joutel. 2007. The archetypal R90C CADASIL-NOTCH3 mutation retains NOTCH3 function in vivo. Hum. Mol. Genet. 16:982-992. [DOI] [PubMed] [Google Scholar]
- 87.Mukherjee, A., A. Veraksa, A. Bauer, C. Rosse, J. Camonis, and S. Artavanis-Tsakonas. 2005. Regulation of Notch signalling by non-visual beta-arrestin. Nat. Cell Biol. 7:1191-1201. [DOI] [PubMed] [Google Scholar]
- 88.Murphy, P. A., M. T. Lam, X. Wu, T. N. Kim, S. M. Vartanian, A. W. Bollen, T. R. Carlson, and R. A. Wang. 2008. Endothelial Notch4 signaling induces hallmarks of brain arteriovenous malformations in mice. Proc. Natl. Acad. Sci. U. S. A. 105:10901-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murtaugh, L. C., B. Z. Stanger, K. M. Kwan, and D. A. Melton. 2003. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. U. S. A. 100:14920-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakamura, E., M. T. Nguyen, and S. Mackem. 2006. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 235:2603-2612. [DOI] [PubMed] [Google Scholar]
- 91.Nakamura, H., M. Yoshida, H. Tsuiki, K. Ito, M. Ueno, M. Nakao, K. Oka, M. Tada, M. Kochi, J. Kuratsu, Y. Ushio, and H. Saya. 1998. Identification of a human homolog of the Drosophila neuralized gene within the 10q25.1 malignant astrocytoma deletion region. Oncogene 16:1009-1019. [DOI] [PubMed] [Google Scholar]
- 92.Nam, Y., P. Sliz, L. Song, J. C. Aster, and S. C. Blacklow. 2006. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124:973-983. [DOI] [PubMed] [Google Scholar]
- 93.Nam, Y., A. P. Weng, J. C. Aster, and S. C. Blacklow. 2003. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 278:21232-21239. [DOI] [PubMed] [Google Scholar]
- 94.Nichols, J. T., A. Miyamoto, S. L. Olsen, B. D'Souza, C. Yao, and G. Weinmaster. 2007. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 176:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nichols, J. T., A. Miyamoto, and G. Weinmaster. 2007. Notch signaling—constantly on the move. Traffic 8:959-969. [DOI] [PubMed] [Google Scholar]
- 96.Nobta, M., T. Tsukazaki, Y. Shibata, C. Xin, T. Moriishi, S. Sakano, H. Shindo, and A. Yamaguchi. 2005. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. J. Biol. Chem. 280:15842-15848. [DOI] [PubMed] [Google Scholar]
- 97.Nofziger, D., A. Miyamoto, K. M. Lyons, and G. Weinmaster. 1999. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126:1689-1702. [DOI] [PubMed] [Google Scholar]
- 98.Ohtsuka, T., M. Ishibashi, G. Gradwohl, S. Nakanishi, F. Guillemot, and R. Kageyama. 1999. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18:2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Olsen, I. E., R. F. Ittenbach, A. J. Rovner, M. B. Leonard, A. E. Mulberg, V. A. Stallings, D. A. Piccoli, and B. S. Zemel. 2005. Deficits in size-adjusted bone mass in children with Alagille syndrome. J. Pediatr. Gastroenterol. Nutr. 40:76-82. [DOI] [PubMed] [Google Scholar]
- 100.Park, J. T., M. Li, K. Nakayama, T. L. Mao, B. Davidson, Z. Zhang, R. J. Kurman, C. G. Eberhart, I. Shih, and T. L. Wang. 2006. Notch3 gene amplification in ovarian cancer. Cancer Res. 66:6312-6318. [DOI] [PubMed] [Google Scholar]
- 101.Pintar, A., A. De Biasio, M. Popovic, N. Ivanova, and S. Pongor. 2007. The intracellular region of Notch ligands: does the tail make the difference? Biol. Direct. 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rampal, R., J. F. Arboleda-Velasquez, A. Nita-Lazar, K. S. Kosik, and R. S. Haltiwanger. 2005. Highly conserved O-fucose sites have distinct effects on Notch1 function. J. Biol. Chem. 280:32133-32140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ruan, Y., L. Tecott, M. M. Jiang, L. Y. Jan, and Y. N. Jan. 2001. Ethanol hypersensitivity and olfactory discrimination defect in mice lacking a homolog of Drosophila neuralized. Proc. Natl. Acad. Sci. U. S. A. 98:9907-9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rydziel, S., L. Stadmeyer, S. Zanotti, D. Durant, A. Smerdel-Ramoya, and E. Canalis. 2007. Nephroblastoma overexpressed (Nov) inhibits osteoblastogenesis and causes osteopenia. J. Biol. Chem. 282:19762-19772. [DOI] [PubMed] [Google Scholar]
- 105.Salie, R., M. Kneissel, M. Vukevic, N. Zamurovic, I. Kramer, G. Evans, N. Gerwin, M. Mueller, B. Kinzel, and M. Susa. 24 October 2009. Ubiquitous overexpression of Hey1 transcription factor leads to osteopenia and chondrocyte hypertrophy in bone. [Epub ahead of print.] Bone. doi: 10.1016/j.bone.2009.10.022. [DOI] [PubMed]
- 106.Sato, T., T. S. Diehl, S. Narayanan, S. Funamoto, Y. Ihara, B. De Strooper, H. Steiner, C. Haass, and M. S. Wolfe. 2007. Active gamma-secretase complexes contain only one of each component. J. Biol. Chem. 282:33985-33993. [DOI] [PubMed] [Google Scholar]
- 107.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393:382-386. [DOI] [PubMed] [Google Scholar]
- 108.Schuster-Gossler, K., B. Harris, K. R. Johnson, J. Serth, and A. Gossler. 2009. Notch signalling in the paraxial mesoderm is most sensitive to reduced Pofut1 levels during early mouse development. BMC Dev. Biol. 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sciaudone, M., E. Gazzerro, L. Priest, A. M. Delany, and E. Canalis. 2003. Notch 1 impairs osteoblastic cell differentiation. Endocrinology 144:5631-5639. [DOI] [PubMed] [Google Scholar]
- 110.Shen, J., R. T. Bronson, D. F. Chen, W. Xia, D. J. Selkoe, and S. Tonegawa. 1997. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell 89:629-639. [DOI] [PubMed] [Google Scholar]
- 111.Shen, Q., and S. Christakos. 2005. The vitamin D receptor, Runx2, and the Notch signaling pathway cooperate in the transcriptional regulation of osteopontin. J. Biol. Chem. 280:40589-40598. [DOI] [PubMed] [Google Scholar]
- 112.Shi, S., and P. Stanley. 2003. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 100:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Song, W., P. Nadeau, M. Yuan, X. Yang, J. Shen, and B. A. Yankner. 1999. Proteolytic release and nuclear translocation of Notch-1 are induced by presenilin-1 and impaired by pathogenic presenilin-1 mutations. Proc. Natl. Acad. Sci. U. S. A. 96:6959-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sparrow, D. B., G. Chapman, M. A. Wouters, N. V. Whittock, S. Ellard, D. Fatkin, P. D. Turnpenny, K. Kusumi, D. Sillence, and S. L. Dunwoodie. 2006. Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am. J. Hum. Genet. 78:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stahl, M., K. Uemura, C. Ge, S. Shi, Y. Tashima, and P. Stanley. 2008. Roles of Pofut1 and O-fucose in mammalian Notch signaling. J. Biol. Chem. 283:13638-13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stanley, P. 2007. Regulation of Notch signaling by glycosylation. Curr. Opin. Struct. Biol. 17:530-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stier, S., T. Cheng, D. Dombkowski, N. Carlesso, and D. T. Scadden. 2002. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood 99:2369-2378. [DOI] [PubMed] [Google Scholar]
- 118.Swiatek, P. J., C. E. Lindsell, F. F. del Amo, G. Weinmaster, and T. Gridley. 1994. Notch1 is essential for postimplantation development in mice. Genes Dev. 8:707-719. [DOI] [PubMed] [Google Scholar]
- 119.Tao, J., S. Chen, G. Sule, I. Yang, B. Dawson, and B. Lee. 2009. Notch function in osteoblasts is dependent on canonical Rbpj signaling. J. Bone Miner. Res. 24(Suppl. 1). http://www.asbmr.org/Meetings/AnnualMeeting/Abstracts.aspx.
- 120.Teitelbaum, S. L. 2007. Osteoclasts: what do they do and how do they do it? Am. J. Pathol. 170:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tezuka, K., M. Yasuda, N. Watanabe, N. Morimura, K. Kuroda, S. Miyatani, and N. Hozumi. 2002. Stimulation of osteoblastic cell differentiation by Notch. J. Bone Miner. Res. 17:231-239. [DOI] [PubMed] [Google Scholar]
- 122.Tolia, A., and B. De Strooper. 2009. Structure and function of gamma-secretase. Semin. Cell Dev. Biol. 20:211-218. [DOI] [PubMed] [Google Scholar]
- 123.Tu, X., J. Lim, K. Ganss, K. Surendran, R. Kopan, M. Gessler, and F. Long. 2009. Notch inhibits early stages of osteoblastogenesis through RBP-Jk and Hey proteins. J. Bone Miner. Res. 24(Suppl. 1). http://www.asbmr.org/Meetings/AnnualMeeting/Abstracts.aspx.
- 124.Turnpenny, P. D., B. Alman, A. S. Cornier, P. F. Giampietro, A. Offiah, O. Tassy, O. Pourquie, K. Kusumi, and S. Dunwoodie. 2007. Abnormal vertebral segmentation and the notch signaling pathway in man. Dev. Dyn. 236:1456-1474. [DOI] [PubMed] [Google Scholar]
- 125.Ugarte, F., M. Ryser, S. Thieme, F. A. Fierro, K. Navratiel, M. Bornhauser, and S. Brenner. 2009. Notch signaling enhances osteogenic differentiation while inhibiting adipogenesis in primary human bone marrow stromal cells. Exp. Hematol. 37:867-875. [DOI] [PubMed] [Google Scholar]
- 126.Visan, I., J. B. Tan, J. S. Yuan, J. A. Harper, U. Koch, and C. J. Guidos. 2006. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat. Immunol. 7:634-643. [DOI] [PubMed] [Google Scholar]
- 127.Wang, Y., K. A. Kim, J. H. Kim, and H. S. Sul. 2006. Pref-1, a preadipocyte secreted factor that inhibits adipogenesis. J. Nutr. 136:2953-2956. [DOI] [PubMed] [Google Scholar]
- 128.Watanabe, N., Y. Tezuka, K. Matsuno, S. Miyatani, N. Morimura, M. Yasuda, R. Fujimaki, K. Kuroda, Y. Hiraki, N. Hozumi, and K. Tezuka. 2003. Suppression of differentiation and proliferation of early chondrogenic cells by Notch. J. Bone Miner. Metab. 21:344-352. [DOI] [PubMed] [Google Scholar]
- 129.Weber, J. M., and L. M. Calvi. 11 August 2009. ahead of print. Notch signaling and the bone marrow hematopoietic stem cell niche. Bone. [Epub ahead of print.] doi: 10.1016/j.bone.2009.08.007. [DOI] [PMC free article] [PubMed]
- 130.Weng, A. P., A. A. Ferrando, W. Lee, J. P. Morris, L. B. Silverman, C. Sanchez-Irizarry, S. C. Blacklow, A. T. Look, and J. C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306:269-271. [DOI] [PubMed] [Google Scholar]
- 131.Wu, J., and E. H. Bresnick. 2007. Bare rudiments of notch signaling: how receptor levels are regulated. Trends Biochem. Sci. 32:477-485. [DOI] [PubMed] [Google Scholar]
- 132.Wu, J. Y., D. T. Scadden, and H. M. Kronenberg. 2009. Role of the osteoblast lineage in the bone marrow hematopoietic niches. J. Bone Miner. Res. 24:759-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu, L., J. C. Aster, S. C. Blacklow, R. Lake, S. Artavanis-Tsakonas, and J. D. Griffin. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26:484-489. [DOI] [PubMed] [Google Scholar]
- 134.Xie, Y., T. Yin, W. Wiegraebe, X. C. He, D. Miller, D. Stark, K. Perko, R. Alexander, J. Schwartz, J. C. Grindley, J. Park, J. S. Haug, J. P. Wunderlich, H. Li, S. Zhang, T. Johnson, R. A. Feldman, and L. Li. 2009. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature 457:97-101. [DOI] [PubMed] [Google Scholar]
- 135.Xin, M., E. M. Small, E. van Rooij, X. Qi, J. A. Richardson, D. Srivastava, O. Nakagawa, and E. N. Olson. 2007. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc. Natl. Acad. Sci. U. S. A. 104:7975-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xue, Y., X. Gao, C. E. Lindsell, C. R. Norton, B. Chang, C. Hicks, M. Gendron-Maguire, E. B. Rand, G. Weinmaster, and T. Gridley. 1999. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 8:723-730. [DOI] [PubMed] [Google Scholar]
- 137.Yamada, T., H. Yamazaki, T. Yamane, M. Yoshino, H. Okuyama, M. Tsuneto, T. Kurino, S. Hayashi, and S. Sakano. 2003. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood 101:2227-2234. [DOI] [PubMed] [Google Scholar]
- 138.Zamurovic, N., D. Cappellen, D. Rohner, and M. Susa. 2004. Coordinated activation of notch, Wnt, and transforming growth factor-beta signaling pathways in bone morphogenic protein 2-induced osteogenesis. Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J. Biol. Chem. 279:37704-37715. [DOI] [PubMed] [Google Scholar]
- 139.Zanotti, S., A. Smerdel-Ramoya, D. Durant, and E. Canalis. 2009. Hairy enhancer of Split (Hes)-1 inhibits osteoblastic function and causes osteopenia in vivo. J. Bone Miner. Res. 24(Suppl. 1). http://www.asbmr.org/Meetings/AnnualMeeting/Abstracts.aspx.
- 140.Zanotti, S., A. Smerdel-Ramoya, L. Stadmeyer, D. Durant, F. Radtke, and E. Canalis. 2008. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology 149:3890-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang, J., C. Niu, L. Ye, H. Huang, X. He, W. G. Tong, J. Ross, J. Haug, T. Johnson, J. Q. Feng, S. Harris, L. M. Wiedemann, Y. Mishina, and L. Li. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425:836-841. [DOI] [PubMed] [Google Scholar]
- 142.Zhang, P., Y. Yang, P. A. Zweidler-McKay, and D. P. Hughes. 2008. Critical role of notch signaling in osteosarcoma invasion and metastasis. Clin. Cancer Res. 14:2962-2969. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 143.Zhang, Y., J. B. Lian, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2009. The notch-responsive transcription factor Hes-1 attenuates osteocalcin promoter activity in osteoblastic cells. J. Cell. Biochem. 108:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]