Abstract
The biogenesis of cytochrome c oxidase initiates with synthesis and maturation of the mitochondrion-encoded Cox1 subunit prior to the addition of other subunits. Cox1 contains redox cofactors, including the low-spin heme a center and the heterobimetallic heme a3:CuB center. We sought to identify the step in the maturation of Cox1 in which the redox cofactor centers are assembled. Newly synthesized Cox1 is incorporated within one early assembly intermediate containing Mss51 in Saccharomyces cerevisiae. Subsequent Cox1 maturation involves the progression to downstream assembly intermediates involving Coa1 and Shy1. We show that the two heme a cofactor sites in Cox1 form downstream of Mss51- and Coa1-containing Cox1 intermediates. These Cox1 intermediates form normally in cells defective in heme a biosynthesis or in cox1 mutant strains with heme a axial His mutations. In contrast, the Shy1-containing Cox1 assembly intermediate is perturbed in the absence of heme a. Heme a3 center formation in Cox1 appears to be chaperoned by Shy1. CuB site formation occurs near or at the Shy1-containing Cox1 assembly intermediate also. The CuB metallochaperone Cox11 transiently interacts with Shy1 by coimmunoprecipitation. The Shy1-containing Cox1 complex is markedly attenuated in cells lacking Cox11 but is partially restored with a nonfunctional Cox11 mutant. Thus, formation of the heterobimetallic CuB:heme a3 site likely occurs in the Shy1-containing Cox1 complex.
Cytochrome c oxidase (CcO) is the terminal oxidase in the oxidative phosphorylation chain within mitochondria. Mammalian CcO is a 13-subunit complex in which three mitochondrion-encoded subunits (Cox1 to Cox3) form the catalytic core (14). The catalytic core is surrounded by nucleus-encoded subunits, which confer stability to the holoenzyme and likely provide sites for the regulation of its activity (25). The fully assembled yeast holoenzyme is further organized into supercomplexes with the bc1 cytochrome c reductase (22). The catalytic core subunits contain heme and copper redox cofactors (41). Cox2 binds two copper ions, forming the binuclear CuA center that is reduced by cytochrome c. Electrons from the CuA center are transferred to a low-spin heme a center in Cox1 and subsequently to a heterobimetallic heme a-copper site, designated heme a3:CuB, where molecular oxygen is bound and reduced to water (3, 45).
The heme a cofactor found in CcO differs from protoheme in that a hydroxyethylfarnesyl group replaces a vinyl moiety and a pyrrole methyl group is oxidized to a formyl substituent. Heme a synthesis is catalyzed by two successive enzymes, Cox10 and Cox15, that reside within the inner membrane (IM) (7, 21). Cox10 is a farnesyl transferase that converts protoheme to heme o. Cox15 subsequently catalyzes the oxidation of the C-8 heme methyl group in a reaction that involves matrix Yah1 ferredoxin and Arh1 ferredoxin reductase (8, 11). Yeast cells lacking Cox15 contain no heme a, but show low levels of heme o, suggesting that the activities of the two enzymes are not linked (9). Likewise, Cox15 mutations in patients exhibiting fatal infantile hypertrophic cardiomyopathy result in reduced heme a but elevated heme o levels (2).
In yeast, CcO biogenesis commences with Cox1 synthesis on mitochondrial ribosomes tethered to the IM by IM-associated Pet309 and Mss51 that bind to the 5′ untranslated region (UTR) of the Cox1 transcript (29, 40, 46). Mss51 has a second function in translational elongation of Cox1, and this function occurs within high-mass Mss51 complexes (∼450 and ∼400 kDa) consisting of Mss51, Cox14, and newly synthesized Cox1 (6, 33, 34). Cox1 appears to progress from the Mss51-containing complex to downstream transient assembly complexes involving Shy1 (31, 34). Yeast cells contain another Cox1 maturation factor, Coa1, which also forms an ∼440-kDa Cox1 assembly intermediate (34). The observed interactions of Coa1 with Mss51 and Shy1 suggest that it participates in the early Cox1 maturation pathway. Information on whether Coa1 is an integral component of the Mss51- or Shy1-containing Cox1 complexes is lacking.
The heme a3 cofactor center appears to be inserted in Cox1 associated with the Shy1 complex (26, 35). The evidence for heme a3 site formation within the Shy1 complex is 2-fold. First, CcO assembly stalled at CuB site formation in Cox1 or at the downstream maturation of Cox2 results in accumulation of a transient Cox1 pro-oxidant intermediate that correlates with the presence of a reactive five-coordinate heme a3 cofactor (26). The pro-oxidant heme a3:Cox1 intermediate is absent in cells lacking Shy1, Coa1, or Cox1 (35). Second, isolation of CcO in Rhodobacter or Paracoccus cells lacking Surf1 (a Shy1 ortholog) reveals an enzyme complex deficient in heme a3 but not heme a (12, 37). Shy1 is not likely a heme a3-insertase, since yeast shy1Δ cells and mutant SURF1 human cells retain 10 to 15% residual CcO activity (17, 36, 47). Rather, Shy1 may be a Cox1 chaperone stabilizing the heme a3 site during Cox1 maturation. In the absence of Shy1, it is likely that the heme a3:Cox1 assembly intermediate is destabilized and only a fraction of the intermediate progresses to the final stages of CcO maturation.
CuB site formation in Cox1 requires the assembly factor Cox11. CcO isolated from Rhodobacter sphaeroides cox11Δ cells lacked CuB but contained both hemes and the CuA site (23). However, heme a3 showed an altered environment by electron paramagnetic resonance (EPR) spectroscopy, most likely due to the absence of the CuB site. Assembly of CcO in Rhodobacter differs from that in yeast in that the three-subunit core enzyme can form without heme or Cu cofactors (24). In contrast, yeast cells lacking Cox11 fail to assemble CcO, and the stalled assembly complexes are largely removed by proteolysis, although residual heme a3:Cox1 intermediates persist in cox11Δ yeast cells, resulting in hydrogen peroxide sensitivity (26).
The heme a center in Cox1 may be formed earlier than the CuB-heme a3 center. Studies with fibroblasts from patients with mutant Cox10 or Cox15 reveal limited accumulation of the free Cox1 subunit (1, 2). In contrast, CcO-deficient patients with mutations in SURF1 revealed a Cox1 assembly intermediate with two nuclear CcO subunits, CoxIV and Va (equivalent to yeast subunits Cox5a and Cox6) (39, 43, 47). One interpretation of these results is that heme a insertion may be necessary for formation or stabilization of the S2 intermediate. In addition, studies of the assembly of the bo3 oxidase of Escherichia coli revealed that insertion of heme b (analogous to the heme a site in cytochrome oxidase) was necessary for subunit assembly (38).
Two goals motivated the present work. First, we sought to elucidate the interrelationship of the various Cox1 maturation complexes involving Mss51, Coa1, and Shy1. Second, we wanted to discern the steps in which the heme a and CuB-heme a3 cofactor sites are formed during Cox1 maturation in yeast. We show here that separate Mss51-Cox1, Coa1-Cox1, and Shy1-Cox1 assembly intermediates exist and that the heme a and CuB centers are formed downstream of the Mss51-containing and Coa1-containing Cox1 intermediates.
MATERIALS AND METHODS
Yeast strains and vectors.
Yeast strains used in this study are described in Table 1. The chromosomal loci of the respective genes in DY5113 Saccharomyces cerevisiae were tagged with a 13Myc epitope generated by PCR-based gene modification using the template pFA6a-13Myc-TRP1, pFA6a-13Myc-HIS3MX6, or pFA6a-13Myc-KanMX4 (28). The deletion strains were created by disruption using homologous recombination of KanMX4 or Candida albicans URA3. Proper integration was confirmed by PCR of the locus. We also used plasmids pRS423-MSS51 (35), pRS416-SHY1-13Myc, pRS413-SCO1-HA, pHCDW3, pHCDW3-111, pHCDW3-208, and pHCDW3-210 (15). Plasmid pRS415-COX11-3HA was obtained by cloning a 1,059-bp XhoI/XbaI COX11-3HA fragment from pRS416-COX11-3HA (27) into the single-copy expression vector pRS415 under control of the ADH1 promoter (32).
TABLE 1.
Yeast strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| W303 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | |
| W303-1B | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] | 30 |
| CKWT | MATaleu1 kar1-1 | 30 |
| CW(H62A) | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] [cox1 (H62A)] | This study |
| CW(H376A) | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] [cox1 (H376A)] | This study |
| CW(H62A) SHY1-13Myc | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] [cox1 (H62A)] SHY1-13Myc::HIS3MX6 | This study |
| CW(H376A) SHY1-13Myc | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] [cox1 (H376A)] SHY1-13Myc::HIS3MX6 | This study |
| CW(H62A) MSS51-13Myc | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] [cox1 (H62A)] MSS51-13Myc::HIS3MX6 | This study |
| CW(H376A) MSS51-13Myc | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] [cox1 (H376A)] MSS51-13Myc::HIS3MX6 | This study |
| CW(H62A) COA1-13Myc | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] [cox1 (H62A)] COA1-13Myc::HIS3 | This study |
| CW(H376A) COA1-13Myc | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 [Δi] [cox1 (H376A)] COA1-13Myc::HIS3 | This study |
| MSS51-13Myc COA1-3HA | MATaade2-1 his3-1,15 leu2-3,112 trp1 ura3-1 COA1-3HA::TRP1 MSS51-13Myc::HIS3MX6 | 34 |
| MSS51-13Myc COA1-3HA cox11Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1 ura3-1 COA1-3HA::TRP1 MSS51-13Myc::HIS3MX6 cox11Δ::CaURA3 | This study |
| MSS51-13Myc COA1-3HA sdh2Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1 ura3-1 COA1-3HA::TRP1 MSS51-13Myc::HIS3MX6 sdh2Δ::CaURA3 | This study |
| MSS51-13Myc COA1-3HA cox15Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1 ura3-1 COA1-3HA::TRP1 MSS51-13Myc::HIS3MX6 cox15Δ::CaURA3 | This study |
| MSS51-13Myc COA1-3HA cox10Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1 ura3-1 COA1-3HA::TRP1 MSS51-13Myc::HIS3MX6 cox10Δ::CaURA3 | This study |
| COA1-13Myc | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-13Myc::HIS3MX6 | 34 |
| COA1-13Myc cox11Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-13Myc::HIS3MX6 cox11Δ::CaURA3 | This study |
| COA1-13Myc sdh2Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-13Myc::HIS3MX6 sdh2Δ::CaURA3 | This study |
| COA1-13Myc shy1Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-13Myc::HIS3MX6 shy1Δ::CaURA3 | This study |
| COA1-13Myc cox15Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-13Myc::HIS3MX6 cox15Δ::CaURA3 | This study |
| COA1-13Myc cox10Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COA1-13Myc::HIS3MX6 cox10Δ::CaURA3 | This study |
| SHY1-13Myc | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 | 35 |
| SHY1-13Myc COA1-3HA | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 COA1-13Myc::HIS3MX6 | 35 |
| SHY1-13Myc cox11Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 cox11Δ::HIS3 | This study |
| SHY1-13Myc sco1Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 sco1Δ::CaURA3 | This study |
| SHY1-13Myc sdh2Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 sdh2Δ::CaURA3 | This study |
| SHY1-13Myc cox15Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 cox15Δ::CaURA3 | This study |
| SHY1-13Myc cox10Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 cox10Δ::CaURA3 | This study |
| SHY1-13Myc cox14Δ | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 SHY1-13Myc::TRP1 cox14Δ::CaURA3 | |
| COX14-3HA | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COX14-HA::HIS3MX6 | 34 |
| COX14-13Myc MSS51-3HA | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COX14-13Myc::HIS3MX6 MSS51-3HA::TRP1 | 34 |
| COX14-13Myc COA1-3HA | MATaade2-1 his3-1,15 leu2-3,112 trp1Δ ura3-1 COX14-13Myc::TRP1 COA1-3HA::HIS3MX6 | 34 |
| cox1Δ::ARG8 m | MATalys2 leu2-3,112 arg8::hisG ura3-52 [cox1Δ::ARG8 m] | 33 |
| cox1Δ::ARG8 m SHY1-13Myc | MATalys2 leu2-3,112 arg8::hisG ura3-52 [cox1Δ::ARG8 m] SHY1-13Myc::KanMX4 | This study |
Cells were cultured in either rich medium or synthetic complete (SC) medium lacking the appropriate nutrients for plasmid selection. The carbon source used was either 1% or 2% glucose, 2% glycerol-2% lactate, or 2% raffinose. Yeast strains were transformed using lithium acetate.
Generation of the Cox1 heme a binding mutant strains.
The plasmid pYGT21 carrying the wild-type (WT) intronless sequence of COX1 was kindly provided by J. Lazowska (CNRS). Mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommendations. After verification of the sequence, the plasmids carrying the mutated genes were used for biolistic transformation. Mitochondrial transformation of a rho0 recipient strain by microprojectile bombardment and identification of the mitochondrial transformants were performed as described previously (30) The mutated genes were then introduced into a rho+ mitochondrial genome by recombination by mating the mitochondrial transformants (or synthetic rho−) with CKWT, carrying the wild-type intronless mitochondrial genome and the nuclear mutation kar1-1, which is required for cytoduction (18). Recombinant rho+ colonies with mutated cox1 were identified by crossing with tester strains. The mutated mitochondrial genomes of these strains were then transferred into W303-1B/rho0. The resulting strains were used for analysis.
Mitochondrion purification and assays.
Intact mitochondria were isolated from yeast as described previously (19, 20). The total mitochondrial protein concentration was determined by the Bradford assay (10).
In vivo mitochondrial protein translation assay.
The cells were pregrown overnight in supplemented medium containing 2% raffinose, reinoculated in complete medium with 2% raffinose, and grown to an optical density at 600 nm (OD600) of 1. The labeling and preparation of the samples for 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were done as described previously (34). The gel was dried, and radiolabeled proteins were visualized by autoradiography.
BN-PAGE.
Blue native gel electrophoresis (BN-PAGE) was performed essentially as described previously (44) with slight modifications. Mitochondria (30 to 50 μg) were spun down at 12,000 × g for 10 min at 4°C. Pellets were solubilized in lysis buffer (50 mM NaCl, 5 mM 6-aminocaproic acid, 50 mM imidazole, pH 7.0) in the presence of 1% digitonin and protease inhibitors [1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride and Roche Diagnostics protease inhibitor cocktail]. After incubation for 20 min on ice and centrifugation (20,000 × g for 15 min at 2°C) supernatants were mixed with sample buffer (5% Coomassie brilliant blue G250, 0.5 M 6-aminocaproic acid, pH 7.0) and loaded on a 5 to 13% gradient polyacrylamide gel. Separated complexes were transferred to a polyvinylidene fluoride membrane and detected by immunoblotting with appropriate antibodies. For second-dimension SDS-PAGE, lanes were cut out, incubated in 1% SDS-1% β-mercaptoethanol for 1 h at room temperature, run on a 12% SDS gel, and analyzed by immunoblotting.
Antibody shift native electrophoresis.
Antibody shift BN-PAGE was carried out as described previously (42). Briefly, clarified mitochondrial lysates were prepared as described above, mixed with 30 to 40 μg of either nonspecific preimmune IgG or anti-Myc antibodies, and incubated at 4°C for 1 h with gentle agitation. Following the incubation, lysates were reclarified by centrifugation at 20,000 × g for 15 min at 2°C, mixed with the sample buffer, and subjected to BN-PAGE.
Sucrose gradient centrifugation.
Mitochondria (1,500 to 2,000 μg) were lysed in 1% digitonin, 20 mM HEPES (pH 7.4), 150 mM KCl, 1.2 mM MgCl2, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) for 15 min at 4°C. The cleared lysates (centrifuged for 15 min at 20,000 × g and 4°C) were subjected to the antibody shift as described above, loaded onto continuous 7 to 30% sucrose gradients (5 ml; 20 mM HEPES [pH 7.4], 150 mM KCl, 1.2 mM MgCl2, 0.5 mM PMSF, and 0.1% digitonin), and centrifuged at 148,000 × g for 8 h at 2°C. Collected fractions were analyzed by SDS-PAGE and immunoblotting.
Immunoassays.
Immunoprecipitations (IP) were performed as described previously (34). For immunoblotting, mitochondrial protein was loaded onto a 12% polyacrylamide gel, separated by SDS-PAGE, and transferred onto a nitrocellulose membrane. Membranes were probed with the indicated primary antibody and visualized with ECL reagents (Pierce) following incubation with horseradish peroxidase-conjugated secondary antibodies or with the Odyssey infrared imaging system (Li-Cor Biosciences) with fluorescent secondary antibodies. Anti-Myc and antihemagglutinin (anti-HA) antibodies were obtained from Roche Diagnostics and Molecular Probes; antiporin was from Molecular Probes; and antisera to Cox1, Cox2, and Cox3 subunits were from Mitosciences. We also used anti-Cox11 antiserum (15). Antiserum to Sod2 was a kind gift from Val Culotta. Alex Tzagoloff provided antiserum to F1 ATP synthase. Bernard Trumpower kindly provided antiserum to Rip1.
RESULTS
Early Cox1 maturation complexes.
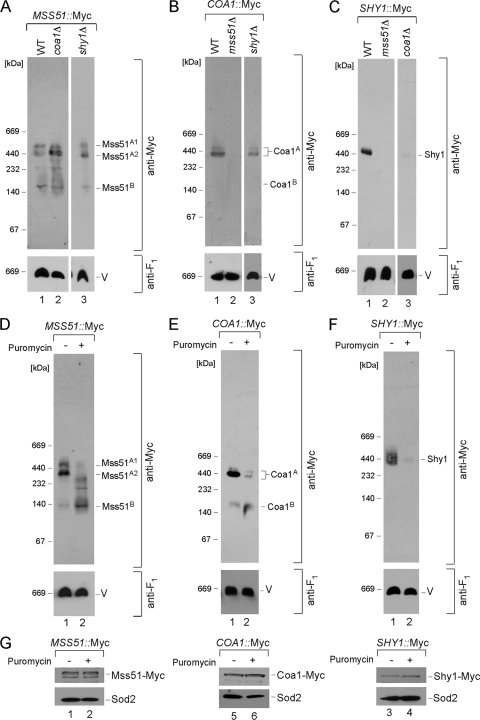
The biogenesis of CcO commences with Cox1 synthesis on IM-associated mitoribosomes. The newly synthesized Cox1 is rapidly captured by a protein complex containing Mss51 and Cox14 (6, 33, 34). Blue native polyacrylamide gel electrophoresis (BN-PAGE) of digitonin-solubilized mitochondria isolated from cells containing a chromosomally epitope-tagged Mss51 reveals two high-mass protein complexes (Fig. 1A, lane 1, complexes A1 and A2) (34). Related high-mass complexes are seen in cells with chromosomally tagged Coa1 or Shy1 (Fig. 1B and C, lanes 1) (34). The Mss51-containing and Coa1-containing Cox1 complexes are independent of Shy1, as the complexes are present in greater abundance in shy1Δ cells (Fig. 1A and B, lanes 3). Likewise, the A2 Mss51-containing Cox1 complex is enhanced in abundance in coa1Δ cells (Fig. 1A). Neither the Coa1 nor the Shy1 complex is present in cells lacking Mss51 (Fig. 1B and C, lanes 2). These results are consistent with the Mss51-, Coa1-, and Shy1-containing Cox1 complexes being distinct entities.
FIG. 1.
Cox1 maturation complexes. (A) Mitochondria (30 to 50 μg) containing a 13Myc epitope-tagged version of MSS51 were isolated from wild-type (WT), coa1Δ, and shy1Δ cells and lysed with 1% digitonin. Clarified lysates were loaded onto a continuous 5 to 13% gradient gel and subjected to BN-PAGE. The distribution of the respective complexes was analyzed by immunoblotting with anti-Myc antibodies. Anti-F1 serum was used to visualize the monomeric form of complex V (V), which served as a loading control. (B) COA1-13Myc-tagged mitochondria isolated from WT, mss51Δ, and shy1Δ cells, analyzed by native electrophoresis. (C) BN-PAGE analysis of WT, mss51Δ, and coa1Δ mitochondria containing Shy1-13Myc. (D to F) Mitochondria (30 to 50 μg) were isolated from WT strains expressing MSS51 (D), COA1 (E), and SHY1 (F) genes endogenously tagged with a 13Myc epitope tag. Isolated organelles were incubated for 30 to 45 min on ice in the presence (+) or absence (−) of puromycin (250 μM). Followed incubation, reisolated mitochondria were solubilized and subjected to BN-PAGE. (G) Steady-state levels of the indicated proteins were assessed by SDS-PAGE in both puromycin-treated (+) and untreated (−) mitochondria. Matrix protein Sod2 visualized by the respective antiserum was used as a loading control.
Each of the three complexes contains newly synthesized Cox1 (34). The Mss51, Coa1, and Shy1 complexes are absent in the mitochondria lacking Cox1 or its translation activator Pet309 or in rho0 cells (reference 34 and data not shown). The three Mss51, Coa1, and Shy1 complexes are dynamic in nature. Purified mitochondria treated for 30 min with puromycin to prematurely terminate mitochondrial translation attenuated each complex (Fig. 1D, E, and F, lanes 2). Neither treatment attenuated the steady-state levels of the tagged proteins (Fig. 1G). The complexes were also diminished by inhibition of mitochondrial protein synthesis by a 3-h treatment of the cultures with chloramphenicol (data not shown). These results suggest that the stability of the high-mass Mss51, Coa1, and Shy1 complexes is sensitive to changes in Cox1 synthesis and that they likely represent transient assembly intermediates.
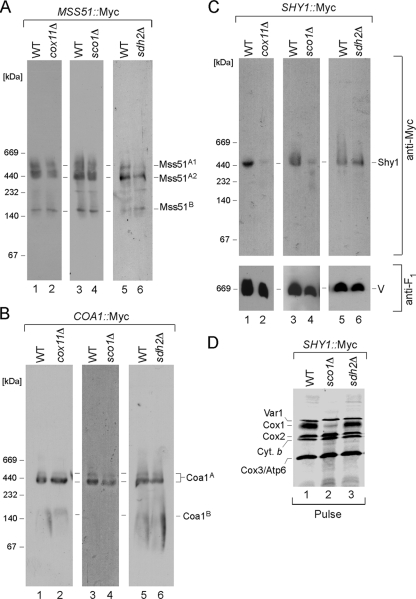
The Mss51, Coa1, and Shy1 complexes were assessed in cells arrested in CcO maturation at later stages. Cells lacking Cox11 fail to assemble the CuB center in Cox1. The Mss51-containing and Coa1-containing Cox1 complexes are WT in cox11Δ cells, although the Shy1-containing Cox1 complex is markedly attenuated (Fig. 2A, B, and C, lanes 2). Although the ∼450-kDa Shy1 complex is attenuated, steady-state levels of Shy1 are normal. A similar situation exists in cells stalled in Cox2 maturation by the deletion of SCO1, resulting in a lack of CuA site formation. The Shy1-containing Cox1 complex is diminished, whereas the Mss51- and Coa1-containing Cox1 complexes are normal (Fig. 2A, B, and C, lanes 4). To assess whether the attenuation of the Shy1-containing Cox1 complex is simply due to the respiratory deficiency of the cells, we evaluated the three complexes in respiration-deficient sdh2Δ cells that lack succinate dehydrogenase (Fig. 2A, B, and C, lanes 6). All three complexes are present at WT levels. One difference between respiration-deficient cells with defects in succinate dehydrogenase and those with CcO deficiency is that many CcO assembly mutants result in an attenuation in Cox1 translation (6). As can be seen in Fig. 2D, sco1Δ cells show a significant diminution in newly synthesized Cox1 in the pulse phase of a mitochondrial translation assay, whereas sdh2Δ cells show normal Cox1 levels. The diminution in Shy1-containing Cox1 complex levels in cox11Δ and sco1Δ cells may arise from diminished newly synthesized Cox1 levels caused by decreased translation (6) as well as enhanced degradation. Assessment of Cox1 translation by monitoring expression of a Cox1-Arg8 fusion in shy1Δ cells revealed WT translation (33), so the reduced Cox1 levels in cox11Δ and sco1Δ cells may arise in part from facilitated Cox1 degradation.
FIG. 2.
The Shy1 complex is affected in CcO assembly mutants. Mitochondria were prepared from the wild-type (WT), cox11Δ, sco1Δ, and sdh2Δ strains with the genomically tagged versions of the respective proteins and analyzed by BN-PAGE as described for Fig. 1. (A to C) Distributions of Mss51-Myc (A), Coa1-Myc (B), and Shy1-Myc (C) complexes in cox11Δ, sco1Δ, and sdh2Δ mitochondria. (D) In vivo labeling of mitochondrial translation products. WT, sco1Δ, and sdh2Δ cells were pulsed for 15 min with [35S]methionine at 30°C. The reaction was stopped by addition of cold methionine, and samples were analyzed by 12% SDS-PAGE followed by autoradiography.
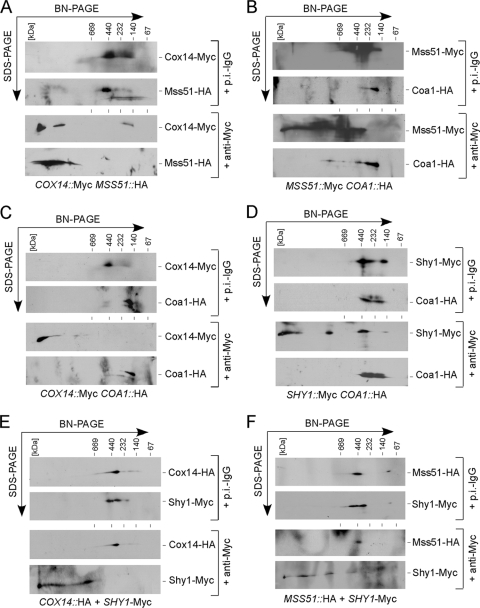
To further clarify the interrelationship of the three Cox1 assembly complexes, we conducted a series of antibody shift studies using BN-PAGE. Specifically, mitochondria isolated from cells containing dually tagged CcO assembly factors were solubilized with digitonin and fractionated by two-dimensional BN-PAGE. Two-dimensional analysis was required since the HA epitope tag was poorly visible upon immunoblotting of one-dimensional gels. In each case, anti-Myc monoclonal antibodies were used to shift one chromosomally tagged factor and the position of the HA-tagged protein was assessed in the second SDS-PAGE dimension (Fig. 3). Mss51, Coa1, and Shy1 are all known to form lower-mass components (the B complexes seen in Fig. 1 and 2), so antibody shifts will also result in retardation of these components. As a positive control, we evaluated the interaction of Cox14 and Mss51, which is well established (6). Anti-Myc antibodies yield a dramatic mobility shift of Cox14-Myc, resulting in a corresponding shift in Mss51-HA, whereas no shift in either is seen with nonspecific antibodies (Fig. 3A). To address the putative interaction of Mss51 and Coa1, Mss51-Myc and Coa1-HA mitochondrial lysates were fractionated (Fig. 3B). The addition of anti-Myc antibodies yields a clear shift of Mss51, but only a small fraction of Coa1-HA is shifted. This observation is consistent with the above-mentioned WT Mss51-containing Cox1 complexes in coa1Δ cells (Fig. 1A). Whereas Cox14 is clearly a component of the Mss51-containing Cox1 complex, Cox14 does not appear to be a stable component of the Coa1 complex (Fig. 3C) or the Shy1-containing Cox1 complex (Fig. 3E). Likewise, despite a weak interaction of Coa1 and Shy1 seen by co-IP (34), antibody shifting of the Shy1-containing Cox1 complex did not result in a coshift of Coa1 (Fig. 3D). Finally, Mss51 and Shy1 were reported to interact (31, 34), but antibody shifting of the Shy1 complex failed to shift Mss51 (Fig. 3F). This last observation is consistent with a second co-IP study that failed to detect a Mss51/Shy1 interaction (5).
FIG. 3.
Antibody gel shift BN studies. Mitochondria (0.3 mg) from yeast strains coexpressing COX14-Myc and MSS51-HA (A), MSS51-Myc and COA1-HA (B), COX14-Myc and COA1-HA (C), SHY1-Myc and COA1-HA (D), COX14-HA and SHY1-Myc (E), and MSS51-HA and SHY1-Myc (F) were solubilized with 1% digitonin, and clarified lysates were incubated with either nonspecific preimmune (p.i.-IgG) or anti-Myc antibodies for 60 min on ice. Followed the incubation, reclarified lysates were separated by native electrophoresis in the first dimension and SDS-PAGE in the second dimension and analyzed by Western blotting.
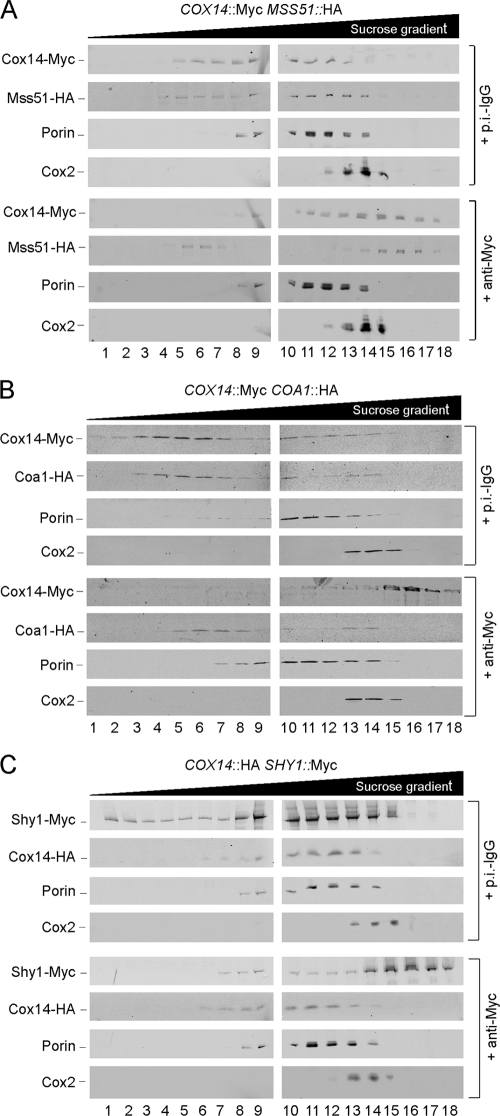
The antibody shift experiment was also evaluated by sucrose density gradient fractionation (Fig. 4). The coshifting of Mss51 and Cox14 was corroborated as a positive control (Fig. 4A). The independence of the Cox14, Coa1, and Shy1 complexes was corroborated, as antibody shifting of Cox14 had minimal effect on Coa1 (Fig. 4B) or Shy1 (Fig. 4C). The shifting of Cox14 also did not appear to perturb assembled CcO within a bc1:CcO supercomplex as marked by Cox2. Thus, the reported presence of Cox14 in the supercomplexes (31) needs further verification.
FIG. 4.
Antibody gel shift sucrose gradients. Mitochondria (1.5 to 2 mg) from yeast strains coexpressing COX14-Myc and MSS51-HA (A), COX14-Myc and COA1-HA (B), and COX14-HA and SHY1-Myc (C) were lysed with 1% digitonin, and clarified lysates were incubated with the respective antibodies as described for Fig. 3. Followed the incubation, reclarified lysates were loaded onto continuous 7 to 30% sucrose gradients and subjected to high-velocity centrifugation. Fractions were collected and analyzed by SDS-PAGE and immunoblotting.
The antibody shift experiments support the postulate that three separate newly synthesized Cox1 maturation complexes exist. The first complex consists of Mss51 and Cox14. The Coa1-containing Cox1 complex is distinct from the Mss51- or the Shy1-containing complexes. We next sought to assess whether the Cox1 present in either the Mss51, Coa1, or Shy1 assembly intermediate complexes contained the redox cofactors.
Effect of Cox10 on early Cox1 maturation complexes.
Cox1 maturation involves the addition of two heme a cofactors and the formation of a mononuclear CuB center. Cox1 consists of 12 transmembrane helices organized in six successive helical pairs forming a closed bundle. The heme a site farnesyl tail is packed between helices 1,11, and 12. The axial His ligands for heme a are His-62 in helix 2 and His-378 in helix 10. Thus, heme a binding may be important to stabilize the Cox1 helical bundle. We took two approaches to assess whether the Mss51, Coa1, and Shy1 complexes were influenced by heme a cofactor insertion into Cox1. First, we tested whether the Mss51, Coa1, and Shy1 complexes formed in cells lacking Cox10. The second approach focused on related complexes in mutant Cox1 strains with substitutions at heme a axial His ligands.
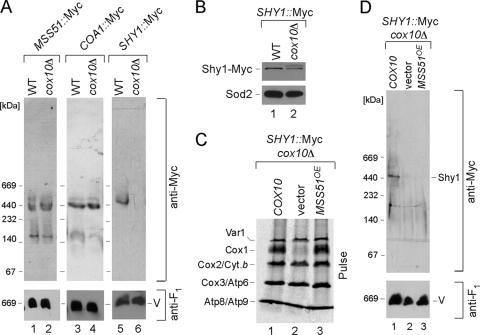
In the first approach, the COX10 locus was deleted in cells with tagged Mss51-Myc, Coa1-Myc, or Shy1-Myc. Mitochondria were isolated from the tagged cox10Δ strains and subjected to BN-PAGE. As can be seen in Fig. 5A, the two high-mass A1/A2 Mss51 complexes were intact in cells lacking Cox10. Coa1-Myc cells lacking Cox10 exhibited a WT Coa1 BN complex (Fig. 5A). In contrast, loss of Cox10 had a profound effect on the Shy1 complex. Cells containing Shy1-Myc but lacking Cox10 fail to show the high-mass Shy1 BN complex. The absence of Cox10 does not attenuate the level of Shy1 in yeast as assessed by SDS-PAGE (Fig. 5B). The lack of perturbation of Mss51 or Coa1 complexes in cells lacking Cox10 suggests that either the Coa1- and Mss51-containing Cox1 complexes lack the two heme a centers or, alternatively, heme a binding is not critical for the stability of these complexes. In contrast, the Shy1-Myc BN complex is sensitive to the absence of Cox10.
FIG. 5.
Cox1 assembly intermediates in cells compromised in heme a synthesis. (A) Lysates from wild-type (WT) or cox10Δ mitochondria with genomically tagged Mss51-Myc, Coa1-Myc, or Shy1-Myc were subjected to BN-PAGE, and the distribution of the respective complexes was analyzed by immunoblotting as described for Fig. 1. (B) The steady-state levels of Shy1-Myc were assessed as described for Fig. 1. (C) In vivo labeling of mitochondrial translation products in SHY1::Myc cox10Δ cells expressing either empty vector or COX10 or overexpressing MSS51. Cells were treated and analyzed as for Fig. 2D. (D) Distribution of Shy1-Myc complexes in SHY1::Myc cox10Δ cells carrying either COX10 or empty vector or overexpressing MSS51, analyzed by native electrophoresis.
Since Cox1 translation is impaired in cox10Δ cells (6), the attenuation in the Shy1 BN complex may arise merely from reduced Cox1 levels. Overexpression of MSS51 reverses the Cox1 translation block in CcO assembly mutants (4, 6). Elevated Mss51 levels in cox10Δ cells restored Cox1 translation (Fig. 5C), but restoration of newly synthesized Cox1 levels did not restore the high-mass Shy1 complex (Fig. 5D). Thus, the destabilization of the Shy1 complex may relate to the absence of the heme a in Cox1.
Effect of perturbation of heme a binding sites in Cox1 on assembly intermediates.
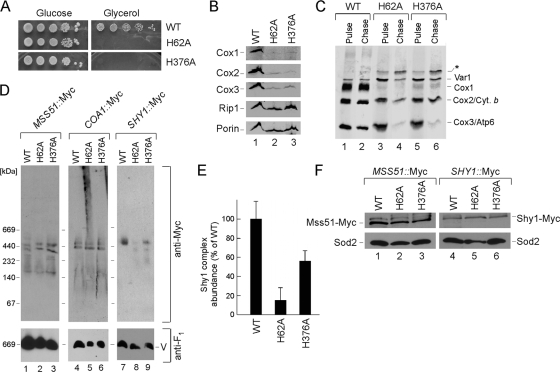
A second strategy to assess the role of heme a in the early Cox1 subassembly complexes was to assess formation of Mss51, Coa1, and Shy1 assembly complexes in cells containing mutant alleles of Cox1 in which the axial heme a ligands were mutated. One of the two His ligands of the heme a site (His-62) and the single axial His-376 ligand of the heme a3 site were each changed to Ala. The two resulting mutant Cox1 strains were respiration deficient and steady-state levels of CcO subunits were attenuated, consistent with the lack of CcO activity (Fig. 6A and B). The newly synthesized Cox1 was markedly attenuated in the mitochondrial translation assay, although limited levels were present (Fig. 6C). Dimeric complex III and monomeric complex V were intact in the mutant cells, also suggesting that the mitochondrial genome was intact (data not shown). Chromosomal Myc epitope tags were engineered into MSS51, COA1, and SHY1 loci, generating C-terminally Myc-tagged molecules in the cox1 mutant strains. Detergent-solubilized mitochondrial lysates from each strain were subjected to BN-PAGE (Fig. 6D). Whereas both strains showed the normal A1/A2 Mss51 and Coa1 complexes, the Shy1 complexes were barely detectable in the H62A cox1 mutant and were attenuated in the H376A mutant. However, steady-state levels of Shy1 were normal (Fig. 6F). In repeated analyses of the abundance of the Shy1 BN complex, the level was markedly attenuated in cells with the H62A Cox1 mutant (Fig. 6E). We conclude that the failure to bind heme a in either cofactor site in Cox1 has no apparent effect on the high-mass Mss51 and Coa1 complexes but results in a destabilization of the Shy1 complexes.
FIG. 6.
Cox1 assembly intermediates in cells compromised in heme a coordination. (A) Respiratory growth of cox1 heme-binding mutants. Cells were pregrown in complete liquid medium, serially diluted, and spotted onto plates containing 2% glucose or glycerol/lactate as a carbon source. (B) Steady-state levels of Cox1 to -3 subunits as well as Rip1 were assessed in mitochondria (80 μg) from the WT or cox1 H62A and cox1 H376A mutants. The outer mitochondrial membrane porin served as a loading control. (C) In vivo labeling of mitochondrial translation products. WT, cox1 H62A, and cox1 H376A cells were pulsed for 15 min with [35S]methionine at 30°C. The reaction was stopped by addition of cold methionine. Followed a 90-min chase at 30°C, samples were subjected to SDS-PAGE and analyzed by autoradiography. The asterisk designates an unidentified translation product. (D) WT, cox1 H62A, and cox1 H376A mitochondria with endogenously tagged Mss51-Myc, Coa1-Myc, and Shy1-Myc were analyzed by BN-PAGE. (E) Bar graph showing abundance (normalized to the monomeric ATPase level using ImageJ software) of Shy1 BN complexes in the WT and the cox1 H62A and cox1 H376A mutants (error bars indicate standard deviations; n = 3). (F) Steady-state levels of Mss51-Myc and Shy1-Myc were assessed as described for Fig. 1.
Formation of the CuB site in Cox1 maturation.
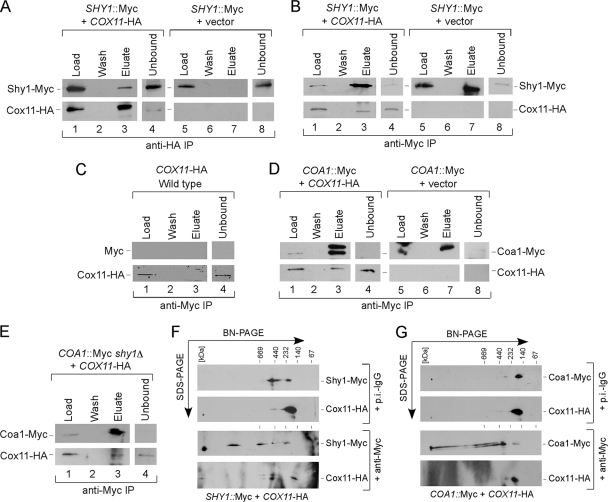
If Shy1 is a Cox1 chaperone stabilizing the heme a3 center, the prediction is that the Shy1-containing Cox1 complex may be the intermediate in which the CuB site is formed, leading to the heterobimetallic CuB-heme a3 center. The only CuB-specific assembly factor known to date is the Cu(I) binding Cox11 protein tethered to the IM by an N-terminal transmembrane domain. To assess whether Cox11 interacts with Shy1, we performed a series of co-IP studies with epitope-tagged Shy1-Myc and Cox11-HA. Both tagged proteins are fully functional in mediating CcO assembly. Immunoprecipitation of Cox11 led to the co-IP of limited quantities of Shy1 (Fig. 7A). Limited interaction also occurred in the reciprocal IP (Fig. 7B), and no nonspecific bead binding was observed (Fig. 7C). Whereas Cox11 showed a co-IP with Shy1, no such interaction was observed between Shy1 and the CuA metallochaperone Sco1 or between Cox11 and Sco1 (data not shown). Cox11 was also found to partially interact with Coa1 by IP (Fig. 7D), and this interaction persisted in the absence of Shy1 (Fig. 7E). To assess whether the Cox11 interactions with Shy1 and Coa1 led to stable complexes, antibody gel shift studies were performed. Anti-Myc antibodies successfully shifted Shy1-Myc on BN-PAGE, and limited amounts of Cox11-HA were shifted (Fig. 7F). Similar antibody shift experiments with Coa1-Myc and Cox11-HA mitochondrial detergent lysates revealed no appreciable modulation of Cox11 (Fig. 7G). These experiments suggest that Cox11 may transiently associate with the Shy1-containing and Coa1-containing Cox1 complexes, but Cox11 is not an integral component of either complex.
FIG. 7.
Analysis of the Cox11 interactome. (A) Mitochondria (0.45 mg) from Shy1-Myc wild-type cells transformed with an empty vector or Cox11-HA-expressing plasmid were solubilized in buffer containing 1% digitonin, and clarified extracts were immunoprecipitated with goat polyclonal anti-HA beads. The load, representing 1% of the total fraction and the entire fractions of the last wash and bead eluate, as well as the fraction of the unbound material (1%) were analyzed by immunoblotting with the respective antibodies. (B) Mitochondrial lysates from Shy1-Myc wild-type cells transformed with an empty vector or Cox11-HA-expressing plasmid were solubilized, immunoprecipitated with goat polyclonal anti-Myc beads, and analyzed as described for panel A. (C) Mitochondria from untagged wild-type cells transformed with Cox11-HA-expressing plasmid were lysed and immunoprecipitated with anti-Myc beads as described for panel B. (D) Mitochondria isolated from Coa1-Myc wild-type cells expressing episomal Cox11-HA were lysed, immunoprecipitated with anti-HA beads, and analyzed as described for panel B. (E) Lysates from Coa1-Myc shy1Δ mitochondria containing Cox11-HA were immunoprecipitated and analyzed as described above. (F) Mitochondria (0.3 mg) from Shy1-Myc cox11Δ cells transformed with Cox11-HA-expressing plasmid were solubilized in buffer containing 1% digitonin, and clarified extracts were subjected to an antibody shift followed by two-dimensional BN-PAGE/SDS-PAGE analysis as described for Fig. 3. (G) Mitochondrial lysates from Coa1-Myc wild-type cells expressing episomal Cox11-HA were subjected to antibody shift native electrophoresis, followed by SDS-PAGE separation in the second dimension as described for Fig. 3.
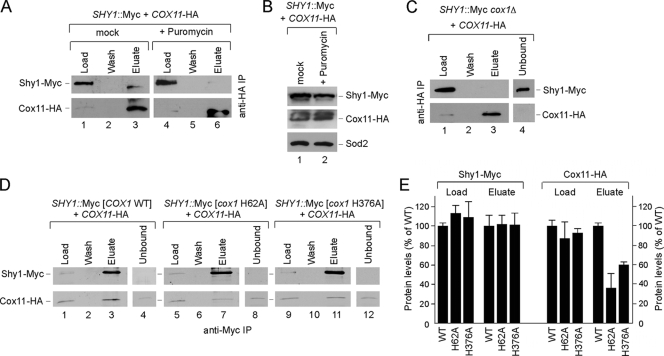
To assess whether the observed Cox11/Shy1 interaction was dependent on Cox1, we tested whether the interaction persisted in mitochondria treated with puromycin to prematurely terminate mitochondrial translation. The 30-min puromycin treatment abrogated the Cox11/Shy1 interaction (Fig. 8A), although the treatment did not alter the steady-state levels of either protein (Fig. 8B). The Cox11/Shy1 interaction was not evident in mitochondria from cox1Δ cells (Fig. 8C) or in cells lacking Cox14 that fail to form either the Coa1- and Shy1-containing Cox1 complexes (data not shown). The Cox11/Shy1 interaction was also attenuated in cells harboring the mutant Cox1 alleles, especially the H62A Cox1 mutant (Fig. 8D and E).
FIG. 8.
Cox11 interaction with Shy1 depends on the translation and early maturation of Cox1. (A) Mitochondria prepared from untreated Shy1-Myc wild-type cells expressing Cox11-HA cells were incubated for 30 to 45 min on ice in the presence of puromycin (250 μM) or were mock treated. Followed the incubation, mitochondria were solubilized in buffer containing 1% digitonin and clarified extracts were immunoprecipitated with anti-HA beads and analyzed as for Fig. 6. (B) Equal amounts of each type of mitochondria used in IP experiment were subjected to SDS-PAGE and analyzed by Western blotting with the indicated antibodies. (C) Lysates from Shy1-Myc cox1Δ mitochondria containing Cox11-HA were immunoprecipitated with anti-HA beads and analyzed as described above. (D) WT, cox1 H62A, and cox1 H376A mitochondria with endogenously tagged Shy1-Myc and Cox11-HA were solubilized and immunoprecipitated with anti-Myc beads. (E) Bar graph showing normalized levels of immunoprecipitated Shy1-Myc and Cox11-HA in the WT and the cox1 H62A and cox1 H376A mutants (error bars indicate standard deviations; n = 3).
Involvement of Cox11 in stabilization of the Shy1-Cox1 complex.
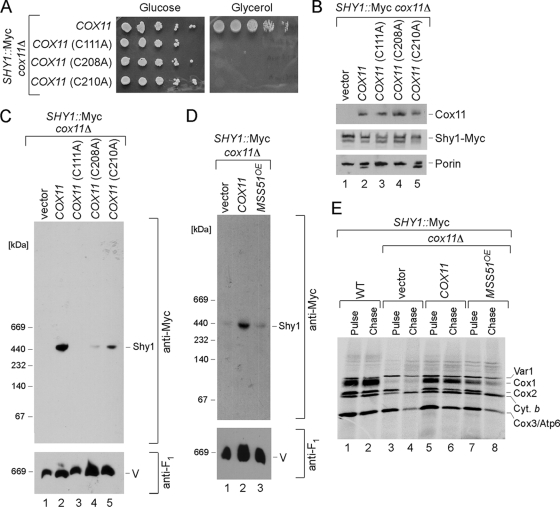
The ∼450-kDa Shy1 complex is unstable in cells lacking Cox11. To assess whether the Shy1 complex was dependent on a functional Cox11, we tested three mutant alleles of Cox11 with Cys substitutions. We reported previously that C111A, C208A, and C210A substitution in yeast Cox11 resulted in a CcO deficiency. Cys208 and Cys210 are two Cu(I) binding residues (15). The respiratory deficiency of cells containing these mutant Cox11 alleles is shown in Fig. 9A. The mutant proteins accumulate to normal levels (Fig. 9B). BN-PAGE of detergent lysates of mitochondria isolated from mutant Cox11 cells showed that the marked attenuation of the ∼450-kDa Shy1 complex in cox11Δ cells was reversed by the introduction of WT COX11 (Fig. 9C). Residual Shy1 complexes were evident in cells containing the nonfunctional C208A or C210A Cox11 mutant. In contrast, no enhanced Shy1 complex is seen with a C111A Cox11 mutant. It is not clear whether Cys111 is involved in Cu(I) binding or Cox11 dimer formation (4, 15). Thus, formation of the Shy1-containing Cox1 complex is not strictly dependent on a functional Cox11. As mentioned, the ∼450-kDa Shy1 complex may be influenced by the level of Cox1 translation. We tested whether overexpression of MSS51 in cox11Δ cells would restore a normal Shy1 complex. Although high Mss51 levels increase Cox1 translation seen in the pulse phase of a translation assay (Fig. 9E), the Shy1 complex was not restored in cox11Δ cells (Fig. 9D, lane 3). Therefore, formation of the Shy1-containing complex is dependent on numerous factors that stabilize newly synthesized Cox1, including Cox11.
FIG. 9.
Role of Cox11 in formation of the high-mass Shy1-containing Cox1 intermediate. (A) Respiratory growth of SHY1::Myc cox11Δ cells carrying wild-type COX11 or mutant versions of it. Cells were handled and tested as for Fig. 5C. (B) Steady-state levels of Cox11, Shy1-Myc, and porin in the transformants, analyzed by SDS-PAGE as described above. (C) BN-PAGE analysis of Shy1 complexes in cox11Δ cells transformed with either the COX11 gene or its mutant forms was performed as described above. (D) Distribution of Shy1-Myc complexes in SHY1::Myc cox11Δ cells carrying either COX11 or empty vector or overexpressing MSS51, analyzed by native electrophoresis. (E) In vivo labeling of mitochondrial translation products in SHY1::Myc cox11Δ cells expressing either empty vector or COX11 or overexpressing MSS51. Cells were treated and analyzed as described for Fig. 6C.
DISCUSSION
Cox1 maturation initiates CcO biogenesis. In mitochondrial genomes sequenced to date, Cox1, Cox2, and Cox3 are commonly mitochondrially encoded. However, Cox1 is the only universally encoded mitochondrial subunit of CcO, with one example being chlamydomonads, which contain only Cox1 of CcO subunits (13). The presence of COX1 in the mitochondrial genome enables a rapid biosynthetic response to a CcO deficiency. Newly synthesized Cox1 is captured in early high-mass assembly intermediates containing Mss51 and Cox14 (6, 33, 34). Cox1 maturation appears to involve a progression from the Mss51 complex to two downstream assembly intermediates involving Coa1 and Shy1 (31, 34). All three complexes are dependent on newly synthesized Cox1 and are abrogated in mitochondria treated with puromycin. The three steady-state Cox1 assembly complexes are distinct components in the Cox1 maturation process. This conclusion is based on two lines of evidence. First, the two high-mass Mss51- and ∼440-kDa Coa1-containing Cox1 complexes persist and actually accumulate in cells lacking Shy1. The Mss51 and Coa1 complexes are therefore upstream of the Shy1 complex. The two high-mass Mss51 complexes are also stable in cells lacking Coa1, suggesting that they are upstream of the Coa1 complex. Second, antibody shift experiments confirm the three separate Cox1-containing complexes. The high-mass Mss51 complexes are independent of Shy1 but contain Cox14. A minor fraction of Mss51 associates with Coa1. The only BN complex that contains Cox14 is the high-mass Mss51 complex(es). The ∼440-kDa Coa1-containing Cox1 complex does not contain significant levels of Shy1 or Cox14. The Coa1-containing complex fails to form in cells lacking Cox14 (34). This is rational, as Cox14 is upstream within the Mss51 complex.
The conclusion that separate Cox1 complexes exist with Mss51, Coa1, or Shy1 as components is complicated by the observed copurification or co-IP results showing interactions between Mss51 and Shy1 (31, 34) and by the observation that Coa1 interacts with Mss51, Cox14, and Shy1 (31, 34). These weak interactions may suggest that the separate Cox1 assembly intermediates transiently interact or that the BN procedure destabilizes these interactions. Destabilization by the BN procedure is unlikely, as we fail to see Cox14 interacting with either Coa1 or Shy1 by antibody shift studies using sucrose density gradients. Cox1 maturation from the Mss51-containing complexes may involve Coa1 binding, thereby inducing progression to the next assembly intermediate. Likewise, progression from the Coa1-containing Cox1 complex may involve the Shy1-induced progression to the high-mass Shy1-dominated complex. This situation may yield the observed transient interactions of Coa1 and Mss51 as well as Coa1 and Shy1. These transient interactions are depicted in the scheme shown in Fig. 10.
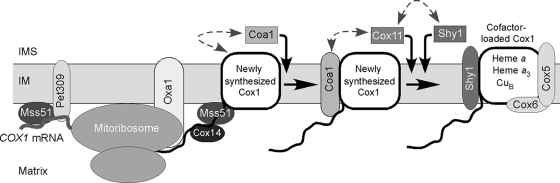
FIG. 10.
Model for involvement of the CcO assembly factors in the early maturation steps. The first apparent newly synthesized Cox1 assembly complex involves Mss51 and Cox14. We propose that Coa1 inserts into this complex to drive Cox1 maturation to the next step. The insertion of Cox11 and Shy1 drives Cox1 to the Shy1 complex in which the redox cofactor sites are populated.
We show here that the two heme a cofactors are likely added to Cox1 within or near the high-mass Shy1 complex and not the early Mss51 or Coa1 assembly intermediates. The evidence for this conclusion is 2-fold. First, the high-mass Mss51 and Coa1 complexes are unaffected in cells lacking Cox10 that generates heme o from heme b. In contrast, the high-mass Shy1 BN complex is attenuated when COX10 is deleted. Stimulation of Cox1 translation through overexpression of MSS51 does not restore the Shy1 complex. Second, Cox1 mutants with Ala substitutions of axial His ligands to heme a and a3 show WT Mss51 and Coa1 BN complexes, whereas the Shy1 complex was markedly perturbed in cells containing a mutant Cox1 lacking the His62 axial heme a ligand. Since the heme a cofactor appears to have a stabilization role in the Cox1 helical bundle, we conclude that the heme a site in Cox1 is populated not in the Mss51 or Coa1 complexes but in the downstream Shy1-containing complex or in the transition to the Shy1 complex. Our assumption that heme a has a structural role in Cox1 stabilization may not be true if the Cox1 conformer within the Mss51 complex is significantly different from that in the mature Cox1. However, the most likely interpretation of the present data is that both heme a cofactor sites are populated in Cox1 during the Shy1-associated phase of Cox1 maturation. Formation of the two heme a centers may occur stepwise, with the heme a site being populated in a transition to the Shy1-containing complex whereas the heme a3 center may be populated within the Shy1 complex. This would be consistent with studies of R. sphaeroides cells lacking the Shy1 homolog Surf1, which showed a selective depletion of heme a3 (12, 37). Alternatively, both heme a centers may be populated within the Shy1 complex, but in the absence of Shy1 only the heme a3 center with a single axial ligand is labile.
We demonstrated previously that formation of the heme a3 center is dependent on Shy1 (35). The heme a3 center does not appear to have the same major structural effect on Cox1. This may account for the presence of the residual Shy1 BN complex in the Cox1 mutant lacking the heme a3 single axial His. Mutations that stall CcO assembly at later steps, such as maturation of the CuA site, result in hydrogen peroxide sensitivity of the mutant cells (26). The hydrogen peroxide sensitivity arises from a transient heme a3:Cox1 pro-oxidant intermediate. Shy1 appears to be important for the stability of this assembly intermediate. Shy1 is not absolutely required for this step, since cells lacking Shy1 have residual assembled and active CcO.
CuB site formation also appears to involve the Shy1-containing Cox1 assembly intermediate. The CuB metallochaperone Cox11 interacts with Shy1 by co-IP but does not form a stable complex. The ∼450-kDa Shy1-containing Cox1 complex is markedly attenuated in cells lacking Cox11 but is partially restored with a nonfunctional Cox11 mutant compromised in Cu(I) binding. An antibody-induced mass shift in the Shy1 complex shows a partial gel shift in Cox11, suggesting that the two proteins transiently interact. Thus, formation of the heterobimetallic CuB-heme a3 site likely occurs in the Shy1-containing Cox1 complex. The observed Cox11/Shy1 interaction is dependent on the presence of Cox1 and is attenuated in Cox1 heme a/a3 site mutants. The formation of the CuB-heme a3 site within the Shy1-containing Cox1 intermediate is supported by the observation that accumulation of the nonfunctional CcO complexes in Rhodobacter and Paracoccus surf1Δ cells is compromised in both CuB and heme a3 (12, 37).
The stability of the Shy1-containing Cox1 complex is dependent on numerous factors. First, the complex is attenuated in cells with mutant Cox1 alleles and is absent in cells lacking Cox1. Second, the complex is destabilized in cells devoid of either Cox11 or Sco1 and is not restored by merely enhancing Cox1 translation. Thus, Cox11 has a stabilizing role in the maintenance of the important Shy1 complex. The destabilization of the Shy1 complex in sco1Δ cells awaits further investigation but may be related to enhanced proteolysis.
Formation of the CuB site in Cox1 within the Shy1-containing Cox1 complex is linked to our earlier observation that cells lacking Shy1 or Coa1 are attenuated in mitochondrial copper levels (16). We postulate that Cox1 maturation through the Coa1 and Shy1 complexes is linked to the translocation of copper from the matrix copper pool to the intermembrane space (IMS) for Cox11-mediated formation of the CuB site. The Coa1-containing Cox1 complex may be a signaling complex for Cu(I) translocation. The precise function of the high-mass Mss51 complexes remains unclear.
In summary, we postulate that Cox1 maturation initiates with an early Cox1 assembly intermediate containing Mss51 and Cox14 (Fig. 10). This may represent a reservoir of Cox1 poised for CcO biogenesis. This complex does not appear to have the cofactor sites populated. This may be a favorable state of Cox1, as it would preclude any deleterious oxidative damage from the presence of solvent-accessible heme a3. When a need for additional CcO exists, a signaling event may induce the progression of Cox1 from the reservoir Mss51 complex to downstream complexes containing Coa1 and Shy1. Within the Shy1 complex, the heme a3:CuB site center is formed, although it remains unclear whether the heme a site is populated within the Shy1 complex or just prior to that step. These steps are likely concurrent with the addition of the Cox5a and Cox6 subunits. Upon completion of these steps, Cox1 is likely competent to receive Cox2 and the additional CcO subunits. Future studies will address the maturation steps downstream of the Shy1 complex.
Acknowledgments
This work was supported by grant ES03817 from the National Institutes of Environmental Health Sciences, NIH, to D.R.W.
Footnotes
Published ahead of print on 7 December 2009.
REFERENCES
- 1.Antonicka, H., S. C. Leary, G. H. Guercin, J. N. Agar, R. Horvath, N. G. Kennaway, C. O. Harding, M. Jaksch, and E. A. Shoubridge. 2003. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 12:2693-2702. [DOI] [PubMed] [Google Scholar]
- 2.Antonicka, H., A. Mattman, C. G. Carlson, D. M. Glerum, K. C. Hoffbuhr, S. C. Leary, N. G. Kennaway, and E. A. Shoubridge. 2003. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 72:101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babcock, G. T., and M. Wikstrom. 1992. Oxygen activation and the conservation of energy in cell respiration. Nature 356:301-309. [DOI] [PubMed] [Google Scholar]
- 4.Banci, L., I. Bertini, F. Cantini, S. Ciofi-Baffoni, L. Gonnelli, and S. Mangani. 2004. Solution structure of Cox11: a novel type of beta-immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. J. Biol. Chem. 279:34833-34839. [DOI] [PubMed] [Google Scholar]
- 5.Barrientos, A., D. Korr, and A. Tzagoloff. 2002. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 21:43-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrientos, A., A. Zambrano, and A. Tzagoloff. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23:3472-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barros, M. H., C. G. Carlson, D. M. Glerum, and A. Tzagoloff. 2001. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme o. FEBS Lett. 492:133-138. [DOI] [PubMed] [Google Scholar]
- 8.Barros, M. H., F. G. Nobrega, and A. Tzagoloff. 2002. Mitochondrial ferredoxin is required for heme A synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 277:9997-10002. [DOI] [PubMed] [Google Scholar]
- 9.Barros, M. H., and A. Tzagoloff. 2002. Regulation of the heme a biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 516:119-123. [DOI] [PubMed] [Google Scholar]
- 10.Bradford, N. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Brown, K. R., B. M. Allan, P. Do, and E. L. Hegg. 2002. Identification of novel hemes generated by heme A synthase: evidence for two successive monooxygenase reactions. Biochemistry 41:10906-10913. [DOI] [PubMed] [Google Scholar]
- 12.Bundschuh, F. A., K. Hoffmeier, and B. Ludwig. 2008. Two variants of the assembly factor Surf1 target specific terminal oxidases in Paracoccus denitrificans. Biochim. Biophys. Acta 1777:1336-1343. [DOI] [PubMed] [Google Scholar]
- 13.Burger, G., M. W. Gray, and B. F. Lang. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19:709-716. [DOI] [PubMed] [Google Scholar]
- 14.Capaldi, R. A. 1990. Structure and function of cytochrome c oxidase. Annu. Rev. Biochem. 59:569-596. [DOI] [PubMed] [Google Scholar]
- 15.Carr, H. S., G. N. George, and D. R. Winge. 2002. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I) binding protein. J. Biol. Chem. 277:31237-31242. [DOI] [PubMed] [Google Scholar]
- 16.Cobine, P. A., F. Pierrel, M. L. Bestwick, and D. R. Winge. 2006. Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J. Biol. Chem. 281:36552-36559. [DOI] [PubMed] [Google Scholar]
- 17.Coenen, M. J., J. A. Smeitink, J. M. Pots, E. van Kaauwen, F. J. Trijbels, F. A. Hol, and L. P. van den Heuvel. 2006. Sequence analysis of the structural nuclear encoded subunits and assembly genes of cytochrome c oxidase in a cohort of 10 isolated complex IV-deficient patients revealed five mutations. J. Child Neurol. 21:508-511. [DOI] [PubMed] [Google Scholar]
- 18.Conde, J., and G. R. Fink. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. U. S. A. 73:3651-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daum, G., P. C. Bohni, and G. Schatz. 1982. Import of proteins into mitochondria, cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257:13028-13033. [PubMed] [Google Scholar]
- 20.Diekert, K., A. I. De Kroon, G. Kispal, and R. Lill. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65:37-51. [DOI] [PubMed] [Google Scholar]
- 21.Glerum, D. M., and A. Tzagoloff. 1994. Isolation of a human cDNA for heme a:farnesyltransferase by functional complementation of a yeast cox10 mutant. Proc. Natl. Acad. Sci. U. S. A. 91:8452-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinemeyer, J., H. P. Braun, E. J. Boekema, and R. Kouril. 2007. A structural model of the cytochrome c reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 282:12240-12248. [DOI] [PubMed] [Google Scholar]
- 23.Hiser, L., M. Di Valentin, A. G. Hamer, and J. P. Hosler. 2000. Cox11p is required for stable formation of the CuB and magnesium centers of cytochrome c oxidase. J. Biol. Chem. 275:619-623. [DOI] [PubMed] [Google Scholar]
- 24.Hiser, L., and J. P. Hosler. 2001. Heme a is not essential for assembly of the subunits of cytochrome c oxidase of Rhodobacter sphaeroides. J. Biol. Chem. 276:45403-45407. [DOI] [PubMed] [Google Scholar]
- 25.Kadenbach, B., M. Huttemann, S. Arnold, I. Lee, and E. Bender. 2000. Mitochondrial energy metabolism is regulated via nuclear-coded subunits of cytochrome c oxidase. Free Radic. Biol. Med. 29:211-221. [DOI] [PubMed] [Google Scholar]
- 26.Khalimonchuk, O., A. Bird, and D. R. Winge. 2007. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282:17442-17449. [DOI] [PubMed] [Google Scholar]
- 27.Khalimonchuk, O., K. Ostermann, and G. Rodel. 2005. Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the Cu(B) site formation of cytochrome c oxidase. Curr. Genet. 47:223-233. [DOI] [PubMed] [Google Scholar]
- 28.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 29.Manthey, G. M., and J. E. McEwen. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14:4031-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meunier, B. 2001. Site-directed mutations in the mitochondrially encoded subunits I and III of yeast cytochrome oxidase. Biochem. J. 354:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mick, D. U., K. Wagner, M. van der Laan, A. E. Frazier, I. Perschil, M. Pawlas, H. E. Meyer, B. Warscheid, and P. Rehling. 2007. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26:4347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use of heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez-Martinez, X., S. A. Broadley, and T. D. Fox. 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22:5951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierrel, F., M. L. Bestwick, P. A. Cobine, O. Khalimonchuk, J. A. Cricco, and D. R. Winge. 2007. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26:4335-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierrel, F., O. Khalimonchuk, P. A. Cobine, M. Bestwick, and D. R. Winge. 2008. Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis facilitating the maturation of Cox1. Mol. Cell. Biol. 28:4927-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poyau, A., K. Buchet, M. F. Bouzidi, M.-T. Zabot, B. Echenne, J. Yao, E. A. Shoubridge, and C. Godinot. 2000. Missense mutations in SURF1 associated with deficient cytochrome c oxidase assembly in Leigh syndrome patients. Hum. Genet. 106:194-205. [DOI] [PubMed] [Google Scholar]
- 37.Smith, D., J. Gray, L. Mitchell, W. E. Antholine, and J. P. Hosler. 2005. Assembly of cytochrome c oxidase in the absence of the assembly protein Surf1p leads to loss of the active site heme. J. Biol. Chem. 280:17652-17656. [DOI] [PubMed] [Google Scholar]
- 38.Stenberg, F., G. von Heijne, and D. O. Daley. 2007. Assembly of the cytochrome bo3 complex. J. Mol. Biol. 371:765-773. [DOI] [PubMed] [Google Scholar]
- 39.Stiburek, L., K. Vesela, H. Hansikova, P. Pecina, M. Tesarova, L. Cerna, J. Houstek, and J. Zeman. 2005. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 392:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavares-Carreon, F., Y. Camacho-Villasana, A. Zamudio-Ochoa, M. Shingu-Vazquez, A. Torres-Larios, and X. Perez-Martinez. 2008. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 283:1472-1479. [DOI] [PubMed] [Google Scholar]
- 41.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Hakashima, R. Yaono, and S. Yoshikawa. 1995. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8A. Science 269:1069-1074. [DOI] [PubMed] [Google Scholar]
- 42.Waizenegger, T., and D. Rapaport. 2007. Analyzing import intermediates of mitochondrial proteins by blue native gel electrophoresis. Methods Mol. Biol. 372:287-295. [DOI] [PubMed] [Google Scholar]
- 43.Williams, S. L., I. Valnot, P. Rustin, and J.-W. Taanman. 2004. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1 or SURF1. J. Biol. Chem. 279:7462-7469. [DOI] [PubMed] [Google Scholar]
- 44.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1:418-428. [DOI] [PubMed] [Google Scholar]
- 45.Yoshikawa, S., K. Muramoto, K. Shinzawa-Itoh, H. Aoyama, T. Tsukihara, T. Ogura, K. Shimokata, Y. Katayama, and H. Shimada. 2006. Reaction mechanism of bovine heart cytochrome c oxidase. Biochim. Biophys. Acta 1757:395-400. [DOI] [PubMed] [Google Scholar]
- 46.Zambrano, A., F. Fontanesi, A. Solans, R. Leite de Oliveira, T. D. Fox, A. Tzagoloff, and A. Barrientos. 2007. Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 18:523-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, Z., J. Yao, T. Johns, K. Fu, I. De Bie, C. Macmillan, A. P. Cuthbert, R. F. Newbold, J. Wang, M. Chevrette, G. K. Brown, R. M. Brown, and E. A. Shoubridge. 1998. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 20:337-343. [DOI] [PubMed] [Google Scholar]