Abstract
Chromatin boundaries facilitate independent gene regulation by insulating genes from the effects of enhancers or organized chromatin. However, the mechanisms of boundary action are not well understood. To investigate whether boundary function depends on a higher order of chromatin organization, we examined the function of several Drosophila melanogaster insulators in cells with reduced chromatin-remodeling activities. We found that knockdown of NURF301 and ISWI, key components of the nucleosome-remodeling factor (NURF), synergistically disrupted the enhancer-blocking function of Fab7 and SF1 and augmented the function of Fab8. Mutations in Nurf301/Ebx and Iswi also affected the function of these boundaries in vivo. We further show that ISWI was localized on the endogenous Fab7 and Fab8 insulators and that NURF knockdown resulted in a marked increase in the nucleosome occupancy at these insulator sites. In contrast to the effect of NURF knockdown, reduction in dMi-2, the ATPase component of the Drosophila nucleosome-remodeling and deacetylation (NuRD) complex, augmented Fab7 and suppressed Fab8. Our results provide the first evidence that higher-order chromatin organization influences the enhancer-blocking activity of chromatin boundaries. In particular, the NURF and NuRD nucleosome-remodeling complexes may regulate Hox expression by modulating the function of boundaries in these complexes. The unique responses by different classes of boundaries to changes in the chromatin environment may be indicative of their distinct mechanisms of action, which may influence their placement in the genome and selection during evolution.
Chromatin boundaries, or insulators, play important roles in gene regulation in a wide variety of organisms. These specialized regulatory DNAs can block transcriptional influences from enhancers or silencers or from active or silent chromatin (15, 17, 20, 30, 49). The best-characterized vertebrate insulators include the mammalian ICR boundary and the beta-globin HS4 boundary; both require the CTCF zinc finger protein for their enhancer-blocking function (8, 10, 11, 38, 51). The Drosophila melanogaster genome contains the most diverse boundaries reported to date. Among these are the Mcp-1, Fab6, Fab7, and Fab8 insulators from the Bithorax complex (BX-C); the SF1 insulator from the Antennapedia complex (ANT-C); the suHw insulator from the Gypsy retrotransposon; and the scs and scs′ insulators from the hsp70 loci. These different insulators depend on distinct protein factors for their enhancer-blocking activity. For example, the Fab7 and SF1 boundaries require the GAF protein, whereas the Fab8 insulator depends on dCTCF, the ortholog of the vertebrate CTCF protein, for its insulator activity (12, 29, 61, 78). The Gypsy and the heat shock locus insulators also require unique DNA-binding proteins for their function (31, 34, 43, 77). Recent studies indicate that insulator-like sequences are widely present in genomes and are likely involved in both gene regulation and genome organization (2, 9, 16, 40).
The mechanisms underlying insulator activities are not well understood. Several models have been proposed based on functional and structural characteristics of insulators and their binding proteins (3, 19, 24, 32, 55, 60). However, the cellular machinery that facilitates and regulates boundary function remains poorly characterized. For example, although boundaries are believed to limit the spread of organized chromatin, it is not clear how a higher order of chromatin organization in turn influences boundary function.
Here we show that nucleosome-remodeling factor (NURF), an ATP-dependent nucleosome-remodeling complex, modulates the function of several insulators from the Drosophila homeotic gene complexes (HOM/HOX) in an insulator-specific fashion. Using a cell-based enhancer-blocking assay, we show that knockdown of NURF301, the largest subunit of NURF, or ISWI, the ATPase component of NURF, disrupts the enhancer-blocking activity of Fab7 and SF1 and enhances that of Fab8. Double knockdown of NURF301 and ISWI synergistically affects the function of these insulators, suggesting that they do so in the context of the NURF complex. Mutations in the Nurf301/Ebx and Iswi genes also affect the SF1 and Fab8 insulators similarly in transgenic embryos. We further show that ISWI is localized on the endogenous Fab7 and Fab8 insulators and that NURF knockdown causes in a marked increase in the nucleosome occupancy at these insulator sites. In contrast to the effect of NURF knockdown, reducing dMi-2, the ATPase component of the Drosophila nucleosome-remodeling and deacetylation (NuRD) complex, augments Fab7 and suppresses Fab8. Our results show that nucleosome organization can affect the enhancer-blocking activity of multiple insulators from the Drosophila Hox clusters and that regulation of Hox genes by the NURF and NuRD nucleosome-remodeling complexes may involve modulation of boundary activity in these complexes. Our data suggest that the Fab7 class chromatin boundaries are compatible with open and active chromatin, whereas the Fab8 class insulators may function in a more repressed chromatin environment. The unique responses by different classes of boundary elements to changes in chromatin environment may be indicative of their distinct mechanisms of action and may determine their placement in the genome and selection during evolution (see Discussion).
MATERIALS AND METHODS
Cell culture insulator assay, RNAi, and qRT-PCR.
Construction of the transgenes used in the cell-based enhancer-blocking assay, S2 cell culture and transfection, and double-stranded RNA (dsRNA) treatment procedures were as described previously (52). All experiments including RNA interference (RNAi) knockdown, fluorescence-activated cell sorting (FACS) analysis, quantitative reverse transcription-PCR (qRT-PCR), and semiquantitative duplex RT-PCR were done in triplicates or more. For dsRNA synthesis, sense and antisense RNAs were transcribed in vitro and annealed (MEGAscript kit; Ambion). Two different dsRNA preparations were made to target NURF301 or ISWI. For the first, DNAs corresponding to selected exons of Nurf301 or Iswi were PCR cloned into pCRII-TOPO (Invitrogen) and used as templates for in vitro transcription from the T7 and SP6 RNA polymerases. The primers used were 5′CGAGTCGGAGTATCACTACG3′ and 5′ACCAGCATGTAAAGTTCTCC3′ (for Nurf301) and 5′CGCTCAATATCCTGCAACTCAG and 5′CATCGTCATCGTGCCAAAGT3′ (for Iswi). We also designed RNAi probes according to the recommendation of the E-RNAi, an online dsRNA design tool (http://www.dkfz.de/signaling2/e-rnai/). The primers used for these probes contained T7 promoter sequences and were 5′TAATACGACTCACTATAGGGTGCATTTGCTGATTCTCGAC3′ and 5′TAATACGACTCACTATAGGGATGTCCCTCAGGCAAGAAAA3′ (for Nurf301), 5′TAATACGACTCACTATAGGGCTGCTTAATTTCCTGCTGCC3′ and 5′TAATACGACTCACTATAGGGGCAGCTTGTCCAGAATAGCC3′ (for Iswi), 5′TAATACGACTCACTATAGGGCAAGGGACATGGTGAGTGTG3′ and 5′TAATACGACTCACTATAGGGGTCCTCATCGGCTACGGATA3′ (for Hdac3), and 5′TAATACGACTCACTATAGGGGCTCTCGAGTCCTTGTCACC3′ and 5′TAATACGACTCACTATAGGGGCAAGTCCAAGCTCAAGGTC3′ (for dMi-2). The PCR products were purified and used directly as templates for in vitro transcription.
Total RNA was isolated using Trizol (Invitrogen) as previously described (52). Real-time qRT-PCR data analysis and acquisition were done on an ABI 7500 real-time PCR system (Applied Biosystems). Relative quantitation (RQ) of the target mRNA level was calculated with the ABI 7500 software V2.0.
Briefly, first-strand cDNA synthesis was done using Superscript III (Invitrogen). The synthesized cDNAs were subsequently used as templates for the Qiagen QuantiTect SYBR green PCR system. Primers for qRT-PCR were designed using the Primer3 program to yield products of approximately100 bp in length (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The following gene-specific primers were used: 5′TTTCTCAGCCATCACAGTCG3′ and 5′GGGTCGAATCGAACTTGAAC3′ (for Gapdh), 5′GCAACTAATGAAGGCTCAAGA3′ and 5′GAACAGGGGTTAGAGGAGGTG3′ (for Nurf301), 5′AACAAGGGCAAGAATTACACTGA3′ and 5′GGAATTGCGGAGAAGCTCTTA3′ (for Iswi), 5′CAGCCTGGTAATGAACTACGG3′ and 5′CTTCGTATAGGCCACGGAAT3′ (for Hdac3), and 5′ATGCCTCCAATGCAGCTC3′ and 5′TGCATGGAATACGAATGCTC3′ (for dMi-2). The specificity of each reaction was confirmed by a single sharp dissociation curve and a single DNA product of the expected size on agarose gels. Duplicate qRT-PCRs were carried out using cDNAs prepared from two independent experiments.
The semiquantitative duplex RT-PCR was performed as previously described (52). The following primers were used in RT-PCRs: 5′ATAAGCGACGCCATCTGTCC3′ and 5′TTACGTTCTTCCTCATCATCC3′ (for Nurf301), 5′TTAAGACCGCGAATCTGGTAGTC3′ and 5′AAGAGAACTCGAACGAGACGACT3′ (for Iswi), 5′GAGGATTTGGTCCACAACTAGG3′ and 5′CTTCGTATAGGCCACGGAAT3′ (for Hdac3), and 5′GATGGTGTCTCCCACACCGT3′ and 5′CGATCGGCAATACCAGGGT3′ (for actin 88F).
Flow cytometry.
Twenty thousand to 50,000 cells were analyzed in each FACS experiment, using a FACSCalibur flow cytometer (Becton Dickinson). The fluorescence excitation frequency was 488 nm, and the photomultiplier detection voltages were 400 V for FL1 and 375 V for FL2. Fluorescence was detected using FL1 530/30 BP (green fluorescent protein [GFP]) and FL2 585/42 BP (red fluorescent protein [RFP]) filters. Data analysis was done using the FloJo software. The GFP/RFP ratio (R) in the dual-reporter experiments was calculated as (GFP cell % × GFP cell mean fluorescence)induced/(RFP cell % × RFP cell mean fluorescence)induced. The change in GFP/RFP ratio (C) in Fig. 1 and 2 was calculated as RRNAi/Rno RNAi × 100%. The standard error of the mean (SEM) was calculated as ±standard deviation (SD)/n−2. The significance of the change of GFP/RFP ratio was determined using Student's t test.
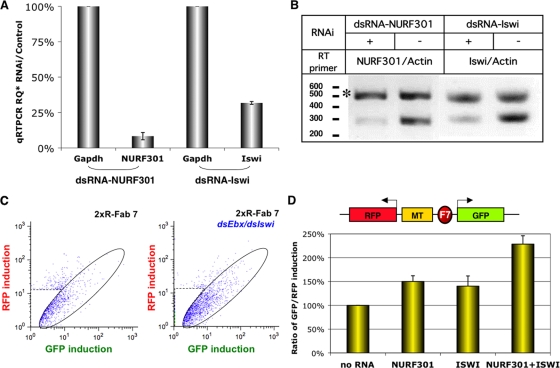
FIG. 1.
NURF301 and ISWI knockdown disrupts the enhancer-blocking activity of the Fab7 insulator. (A) Ratio of mRNA levels in Drosophila S2 cells with and without RNAi treatment as assayed by qRT-PCR. Transcripts of Gapdh (bars 1 and 3) and NURF301 (bar 2) or ISWI (bar 4) were quantitated in cells treated with dsRNA-NURF301 (left two bars) or dsRNA-ISWI (right two bars) and compared to a no-RNAi mock control (see Materials and Methods). (B) Duplex RT-PCR evaluation of mRNA levels of the RNAi targets and actin 88F. Left two lanes, NURF301 mRNA levels in cells with (lane 1) or without (lane 2) dsRNA-NURF301 treatment. Right two lanes, ISWI mRNA levels in cells with (lane 3) or without (lane 4) dsRNA-ISWI treatment. A DNA size marker (100-bp ladder) is shown on the left, and asterisks indicate the product of actin 88F. (C) FACS flow charts of 2×R-Fab7-containing cells before (left) and after (right) double RNAi with dsRNA-NURF301and dsRNA-ISWI. Open ovals highlight cells exhibiting changes in the GFP/RFP ratio, and dashed rectangles gate out untransfected cells. (D) Top, diagram of the 2×R-Fab7 transgene. Rectangles represent regulatory elements as indicated. F7, Fab7. Bottom, GFP/RFP ratio in 2×R-Fab7-containing cells treated with, from left to right, medium, dsRNA-NURF301, dsRNA-ISWI, and dsRNA-NURF301 together with dsRNA-ISWI. Error bars indicate standard errors.
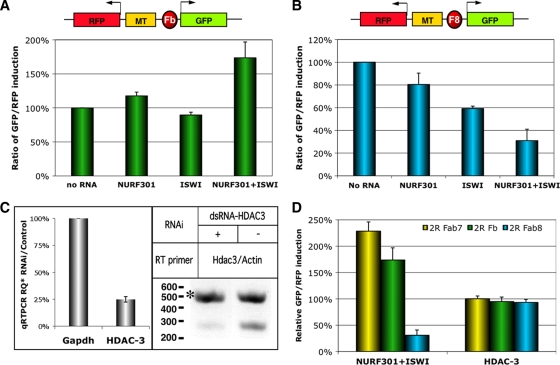
FIG. 2.
NURF components specifically modulate the enhancer-blocking activity of multiple Drosophila insulators. (A) NURF301 and ISWI facilitate insulator function of SF1b. Top, diagram of the 2×R-SF1b transgene. Rectangles represent regulatory elements as indicated. Fb, SF1b. Bottom, GFP/RFP ratio in 2×R-SF1b-containing cells treated with, from left to right, medium, dsRNA-NURF301, dsRNA-ISWI, and dsRNA-NURF301 together with dsRNA-ISWI. (B) NURF301 and ISWI suppress insulator function of Fab8. Top, diagram of the 2×R-Fab8 transgene. F8, Fab8. Bottom, GFP/RFP ratio in 2×R-Fab8-containing cells treated with, from left to right, medium, dsRNA-NURF301, dsRNA-ISWI, and dsRNA-NURF301 together with dsRNA-ISWI. (C) Evaluation of HDAC3 transcript level before and after knockdown. Left, ratio of transcript levels of Gapdh (bar 1) and HDAC3 (bar 2) in cells with and without dsRNA-HDAC3 treatment as assayed by qRT-PCR (see Materials and Methods). Right, duplex RT-PCR assessment of HDAC3 and actin 88F mRNAs in cells with (lane 1) or without (lane 2) dsRNA-HDAC3 treatment. A DNA size marker is shown on the left, and asterisks indicate the product of actin 88F. (D) Comparison between NURF and HDAC3 knockdown effects on the enhancer-blocking function of Drosophila insulators. Left, changes in GFP/RFP ratio after double knockdown of NURF301 and ISWI in S2 cells containing Fab7, SF1b, or Fab8 transgenes. Right, changes in GFP/RFP ratio after HDAC3 knockdown in S2 cells containing the same transgenes. Error bars indicate standard errors.
Enhancer-blocking assay with wild-type and Ebxry122 and Iswi1 transgenic Drosophila embryos.
The construction of the enhancer-blocking transgenes containing the SF1b insulator was described previously (12, 18). The Fab8 element was PCR cloned into pCRII-TOPO (Invitrogen) using the primers 5′ATTCAAATTTATAATGAAAGTT3′ and 5′GCGGCCGCTTCCATAAATTATTATAT3′ and subsequently cloned into the NotI site between the NEE and H1 enhancers in pEbNH (12, 18). The Ebxry122 and Iswi1 mutant strains were kindly provided by the Bloomington Drosophila Stock Center (Bloomington, IN) and by John Tamkun (University of California, Santa Cruz), respectively. Males homozygous for the enhancer-blocking transgenes were mated with virgin females, either wild type (wt) or heterozygous for the Ebxry122 and Iswi1 mutations, respectively. Reporter expression in F1 embryos from all crosses were examined under the same staining condition by whole-mount in situ hybridization using anti-lacZ RNA probes according to a previously described protocol (67). The extent of enhancer blocking was evaluated in a double-blind fashion by at least two authors and categorized into no-block and strong-block groups.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were performed using the EZ-Magna ChIP G kit (Millipore Corporation) according to the manufacturer's protocol with slight modifications. Approximately 108 S2 cells were used for each experiment. Two milliliters of cell lysate was sonicated using a Branson Sonifier 450 sonicator. Samples were kept cool in an ethanol-ice-water bath and sonicated for six 30-s pulses with 1-min pauses between them. Fragmented chromatin was diluted to ∼5 × 107 cells/ml in ChIP dilution buffer (Millipore EZ-Magna ChIP kit), distributed in 300-μl aliquots, and used directly or stored at −80°C. The anti-ISWI (ab10748) and anti-histone H3 (ab1791) antibodies were purchased from Abcam. Following immunoprecipitation, DNA was analyzed by real-time PCR using the following primers: 5′GGAAGAGAGCGGAAAGTG3′ and 5′TACACTCAAGCAGTCGCTCT3′ for Fab7, 5′GCACAATCAAGTTAATGTTGG3′ and 5′AAGCGAAGAGTTCCATTCT3′ for Fab8, and 5′CTACCCAGAAGACCGTCGAT3′ and 5′CGACCTCCTCATCGGTGTAT3′ for a control region in the coding sequence of the Gapdh gene.
RESULTS
NURF301 and ISWI facilitate the enhancer-blocking activity of the Drosophila Fab7 chromatin boundary.
To test whether chromatin organization affects insulator activity, we examined the activity of several Drosophila insulators in cells with reduced chromatin-remodeling activities. We first employed a cell-based enhancer-blocking assay, which uses a transgene containing the metallothionein (MT) enhancer flanked by divergently transcribed GFP and RFP reporters (2×R) (52). In transiently transfected Drosophila S2 cells, insulators such as suHw, SF1b, Fab7, or Fab8, when inserted between MT and GFP, can dramatically reduce the Cu2+-mediated induction of GFP compared to that of RFP (52). The enhancer-blocking activity, indicated by the ratio of GFP/RFP induction, can be readily quantitated by FACS analysis. We have combined this assay with treatment with double-stranded RNA (dsRNA), which induces powerful and specific gene interference and is the predominant agent for RNAi-mediated gene knockdown in Drosophila. We have shown that knockdown of insulator proteins such as SuHw and dCTCF specifically disrupts the Gypsy and Fab8 insulator activities, respectively (21, 52).
The Drosophila NURF nucleosome-remodeling complex disrupts ordered nucleosomes and increases DNA accessibility by “sliding” nucleosomes (69-71, 73). NURF is also required for the activation of numerous genes, including the heat shock and Hox genes (5, 6, 25, 73). NURF is of particular interest to us because it has been shown to partner with GAF, a protein implicated in the insulator function of Fab7 and SF1b (12, 63, 65, 69). Therefore, we first knocked down NURF301, the largest subunit of NURF, and examine the effect on the activity of the Fab7 insulator.
Drosophila S2 cells were transiently transfected with the Fab7-containing enhancer-blocking plasmid (2×R-Fab7) (Fig. 1A). We have previously shown that the GFP/RFP induction ratio in these cells was greatly reduced compared to that with the no-insulator or spacer controls (2×R or 2×R spacer) (52). Transfected cells were treated for 96 h with dsRNA against NURF301 (dsRNA-NURF), followed by Cu2+ induction and FACS analysis. Quantitative RT-PCR (qRT-PCR) showed that the NURF301 mRNA level was reduced by 92% compared to that for the Gapdh control (Fig. 1A). Semiquantitative RT-PCR also confirmed an approximately 80% reduction in NURF301 mRNA compared to the actin control (Fig. 1B, asterisk). The NURF301 knockdown significantly increased the number and fluorescence level of GFP-positive cells but not RFP-positive cells (Fig. 1C). The resulting 50% increase in the GFP/RFP ratio (n ≥ 3) is indicative of a significant loss of the Fab7 enhancer-blocking activity (Fig. 1D). This result suggests that NURF301 facilitates the enhancer-blocking function of the Fab7 insulator.
To probe whether the disruption of the Fab7 insulator following NURF301 knockdown is due to its role in the NURF nucleosome-remodeling complex, we also knocked down ISWI, the catalytic subunit of a number of chromatin-remodeling complexes, including NURF, and examined whether it affected the Fab7 insulator. Treatment with dsRNA-ISWI resulted in a 68% reduction in the ISWI mRNA level, as assayed by qRT-PCR and semiquantitative RT-PCR (Fig. 1A and B). It led to a significant loss of Fab7 enhancer-blocking activity, as shown by the 40% increase in the GFP/RFP ratio (Fig. 1D). This indicates that ISWI indeed influences the Fab7 activity in a similar fashion as NURF301. Furthermore, double knockdown of NURF301 and ISWI elicited a dramatic loss of the enhancer-blocking activity of the Fab7 insulator, as shown by the 140% increase in GFP/RFP ratio (Fig. 1D). The extent of disruption in Fab7 insulator activity by double knockdown of NURF301/ISWI is comparable to the knockdown effect of essential insulator components (52). This indicates that the two NURF factors may facilitate the Fab7 insulator in a synergistic or cooperative fashion. Our results suggest that the NURF nucleosome-remodeling activity may participate in the function of Fab7.
NURF components modulate the enhancer-blocking activity of multiple Drosophila insulators in an insulator-specific fashion.
The role of NURF in Fab7 activity is intriguing, because Fab7 has been shown to depend on the GAGA-binding protein GAF, which is known to interact with the NURF complex (65, 69). GAF is a member of the Trithorax group (Trx-G) proteins, which are essential for Hox gene regulation (26, 59). It has also been implicated in the function of multiple Drosophila insulators, including Fab7, SF1, and Mcp1, all of which are located in the Drosophila Hox gene clusters (12, 58, 65). However, the Fab8 insulator, located next to Fab7 in the regulatory region of the Hox gene Abd B, requires a different protein, dCTCF, the Drosophila ortholog of the vertebrate CTCF insulator protein (61).
To test whether NURF components participate in the function of other Drosophila insulators, we examined SF1 and Fab8 insulators in cells with reduced levels of NURF301 and ISWI. The 0.7-kb SF1b element, which contains the bulk of the enhancer-blocking activity of the SF1 insulator, can strongly block the MT enhancer in S2 cells (52). Knockdown of NURF301 resulted in a strong reduction of the target mRNA (not shown). This elicited a smaller but significant loss of the SF1b insulator activity, as indicated by the 20% increase in the GFP/RFP ratio. A similar knockdown of ISWI resulted in a mild increase of the insulator activity (10% decrease of the GFP/RFP ratio). However, double knockdown of both genes resulted in a strong disruption of SF1b activity (75% increase in GFP/RFP ratio) (Fig. 2A). This result indicates again that the two NURF factors may synergistically regulate SF1b insulator function. The similar responses of SF1b and Fab7 to NURF301 RNAi and double NURF301/ISWI knockdown, together with their dependence on GAF, further suggest that SF1 and Fab7 may share an enhancer-blocking mechanism.
In contrast, the Fab8 insulator responded to the knockdown of NURF components differently from Fab7 and SF1b. Single knockdown of NURF301 or ISWI mRNAs reduced the GFP/RFP ratio by 20% and 40%, respectively, suggesting a significant increase in the Fab8 enhancer-blocking activity. Double knockdown of both NURF301 and ISWI dramatically augmented the Fab8 insulator activity, as shown by the 70% reduction in the GFP/RFP ratio (Fig. 2B).
These results provide the first evidence that the Drosophila NURF components modulate the enhancer-blocking activity of multiple Drosophila insulators, despite their distinct DNA-binding components. Our findings also suggest that NURF activity may modulate the enhancer-blocking activity in an insulator-specific fashion. While the Fab7/SF1 class insulators are facilitated by NURF factors, possibly through increased nucleosome mobility and chromatin flexibility, the Fab8 insulator may require a more rigid organization of nucleosomes arrays and therefore be suppressed by the NURF activity.
In addition to nucleosome-remodeling complexes, histone-modifying enzymes, such as histone acetyl- and methyltransferases also play important roles in regulating chromatin structure and organization (44). To test whether histone modifiers, such as Drosophila histone deacetylase 3 (HDAC3), also influence insulator function, we examined the activities of Fab7, SF1, and Fab8 in S2 cells with reduced HDAC3 activity (41). Treatment with dsRNA-HDAC3 resulted in a 75% reduction of the HDAC3 mRNA level, as quantitated by qRT-PCR and semiquantitative RT-PCR (Fig. 2C). However, this change does not significantly affect the enhancer-blocking function of the Fab7, SF1b, or Fab8 insulator (P = 0.45, 0.16, and 0.08, respectively) (Fig. 2D). This result is in contrast to that for the NURF knockdown, suggesting that all chromatin modifiers do not participate in chromatin boundary function.
NURF mutations disrupt insulator function in transgenic Drosophila embryos.
Next we validated the role of NURF factors in insulator function in vivo. NURF301 and ISWI activities are essential, and both are provided maternally (6, 23). We tested whether reducing the NURF301 and ISWI maternal input by half would influence the enhancer-blocking activity of SF1 and Fab8 in early embryos.
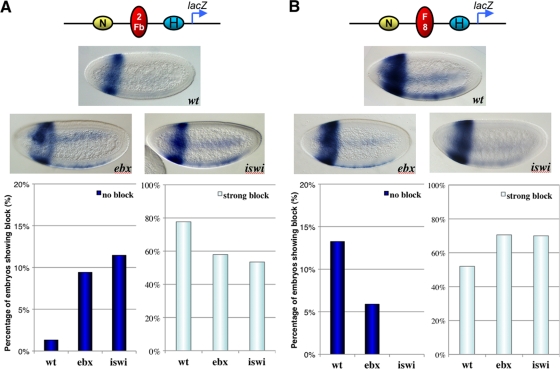
The NbbH transgene used in the embryo enhancer-blocking assay contains the divergently transcribed lacZ reporter and the miniwhite eye color marker (Fig. 3A, map). The neuroectoderm enhancer (NEE) from the rhomboid gene and the stripe 1 enhancer (H1) from the hairy gene are placed between the two reporters (Fig. 3A) (18). Two copies of SF1b inserted between NEE and H1 strongly block the distal NEE enhancer, resulting in a loss of lacZ expression in the ventral lateral neuroectoderm in late-blastoderm-stage embryos (Fig. 3A, wt). Note that the H1-driven expression in the head is not affected (Fig. 3A). Fifty to 100 transgenic embryos were scored in a double-blind fashion for enhancer blocking (see Materials and Methods) (12). Approximately 78% of embryos from wild-type females showed a strong block, whereas only 1% showed no block. Embryos produced by females heterozygous for Ebxry122, a hypomorphic mutation of Nurf301, showed a significant increase in the NEE-directed lacZ expression, with 58% of embryos scored as having a strong block and only 9% as having no block (6). This is indicative of a loss of SF1b insulator function and is consistent with our observation with S2 cells. Embryos produced by female heterozygous for the Iswi1 null mutation also showed a significant loss of SF1b insulator function, with only 53% of embryos showing a strong block and 11% showing no block (Fig. 3A) (23).
FIG. 3.
Nur301 and Iswi mutations disrupt enhancer-blocking activity in transgenic Drosophila embryos. (A) SF1b activity is disrupted in NURF mutant embryos. Top, diagram of the NbbH transgene containing the lacZ reporter (blue arrow) and the NEE (N) and H1 (H) enhancers. Two copies of the SF1b insulator are inserted between N and H (2Fb, red oval). Middle, lacZ expression in NbbH transgenic embryos produced by wild-type (top), Ebxry122 (left), or Iswi1 (right) females. Embryos are oriented dorsal up and anterior to the left. Lack of lacZ expression in the ventral lateral region, seen in a wild-type embryo, is indicative of a strong block of the distal NEE enhancer. NEE-directed lacZ expression in mutant embryos indicates a weaker enhancer block. Bottom, quantitation of the enhancer-blocking activity of SF1b. Fifty to 100 NbbH embryos from wild-type and NURF mutant females were scored in a double-blind fashion for strong blocking and no blocking of the NEE enhancer (see Materials and Methods). (B) Fab8 activity is enhanced in NURF mutant embryos. Top, diagram of the NF8H transgene containing the lacZ reporter (blue arrow) and the NEE (N) and H1 (H) enhancers. A single Fab8 insulator is inserted between N and H (F8, oval). Middle, lacZ expression in NF8H transgenic embryos produced by wild-type (top), Ebxry122 (left), or Iswi1 (right) females. lacZ expression is seen in the ventral lateral region in a wild-type embryo, indicative of a weak enhancer block. NEE-directed lacZ expression in mutant embryos was reduced, indicative of a strong enhancer block. Bottom, quantitation of the enhancer-blocking activity of Fab8 using 50 to 100 transgenic embryos as described for panel A (see Materials and Methods).
Next, we used an NF8H transgene to test the ability of Fab8 to block enhancers in NURF mutant embryos. In wild-type embryos, the Fab8 insulator blocked the distal NEE enhancer less effectively than two copies of the SF1b insulator, with 52% of embryos showing a strong block and 13.3% showing no block (Fig. 3B, wt). The enhancer-blocking efficiency was significantly increased in embryos produced by females heterozygous for the Ebxry122 mutation. The lacZ expression was significantly repressed in the NEE region, with 72% of embryos showing a strong block and 5.6% showing no block (Fig. 3B, Ebx). Embryos produced by females heterozygous for the Iswi1 null mutation also showed a significant enhancement of the Fab8 insulator activity, with 70% of embryos showing a strong block and 0% showing no block (Fig. 3B, Iswi). Our enhancer-blocking results with transgenic embryos indicate that Nurf301 and Iswi differentially modulate the SF1b and the Fab8 insulator functions in vivo. These results are consistent with the NURF knockdown results from S2 cells and support a role of the Drosophila NURF nucleosome-remodeling complex in modulating the enhancer-blocking activity in an insulator-specific fashion.
The Gypsy insulator suHw responds distinctly to NURF knockdown.
We have so far shown that NURF knockdown affects the function of several Drosophila insulators from the Hox gene complexes. Another well-characterized Drosophila insulator, the Gypsy insulator suHw, is distinct from the above-described insulators in its retrotransposon origin and its unique transacting factors. We tested whether the suHw insulator is also affected by the knockdown of the NURF factors. The 340-bp suHw element can effectively block the MT enhancer in S2 cells (reference 52 and data not shown). Knockdown of NURF301 and ISWI resulted in a significant reduction of the insulator activity, as indicated by the 30 to 40% increase in the GFP/RFP ratio (data not shown). However, unlike the case for Fab7, with SF1b and Fab8, for which double knockdown of both NURF301 and ISWI caused a synergistic change in enhancer-blocking activity, reducing both NURF components affected the suHw function to the same extent as single knockdown. These results suggest that the two factors may affect the suHw function separately, rather than as part of the NURF complex. Furthermore, knockdown of HDAC3, which had little effect on Fab7, SF1b, and Fab8, disrupted the suHw activity as did knockdown of NURF, suggesting that the suHw function may be sensitive to general perturbation of the chromatin structure rather than to NURF-mediated nucleosome remodeling (data not shown). It is unclear why the suHw insulator responds differently to changes in chromatin remodeling than Fab7, SF1b, and Fab8. A possible explanation may lie in the different transacting factors associated with these insulators. Both GAF and dCTCF, which mediate the activity of the three Hox insulators, are known to directly interact with the NURF complex or to modulate nucleosome structure (1, 28, 64).
NURF knockdown alters nucleosome occupancy at endogenous insulator sites.
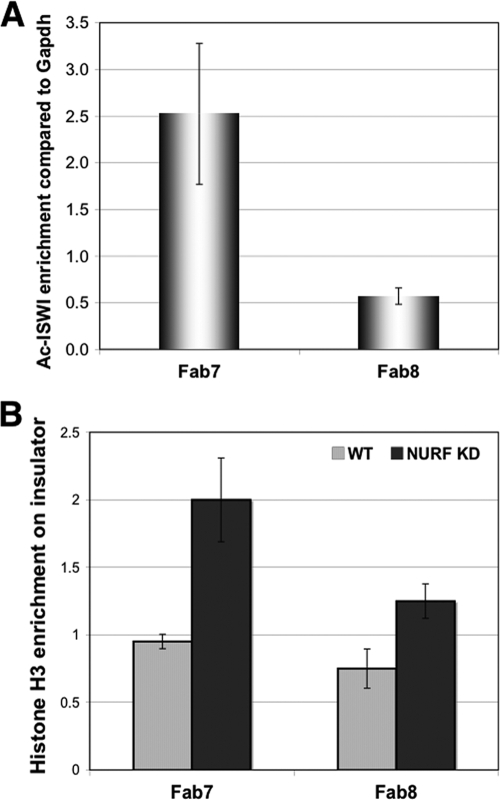
Our results suggest that the NURF nucleosome-remodeling complex may be part of the enhancer-blocking mechanism. To test whether the NURF complex is specifically enriched at the endogenous insulators, we probed the association of NURF component ISWI on the endogenous Fab7 and Fab8 elements. It has been shown that the ISWI protein in Drosophila can be acetylated at K753 by the GCN5 acetyltransferase (27). Although the acetylated form represents only a minor fraction of the total ISWI in the cell, it appears to specifically reside within the NURF complex but not the CHRAC/ACF nucleosome-remodeling complexes (27). We conducted chromatin immunoprecipitation (ChIP) using an antibody that is specific for the acetylated form of ISWI (Ac-ISWI) to test the association of the NURF complex on the insulator elements. We included the Gapdh exon as a baseline control on the assumption that coding regions are less biased for NURF enrichment or depletion than regulatory sequences. We found that Ac-ISWI was differentially associated with the two insulators: whereas the protein was enriched on Fab7 by 2.5-fold over the Gapdh control, it was depleted (0.5-fold of the value for the Gapdh control) at the Fab8 site (Fig. 4A). This observation is consistent with a recent report that the CTCF protein, the vertebrate ortholog of dCTCF, contains a strong nucleosome-positioning activity and can organize arrays of up to 20 nucleosomes around its binding sites (28). It is possible that dCTCF may position nucleosomes by precluding NURF binding on Fab8. To examine whether effects of NURF knockdown correlate with changes in local nucleosome structure, we probed the nucleosome occupancy on the Fab7 and Fab8 insulators before and after NURF knockdown. S2 cells were treated with dsRNAs against both NURF301 and ISWI, followed by ChIP using anti-H3 antibody (Fig. 4B). Fab7 and Fab8 elements exhibit similar nucleosome occupancy in wild-type cells. After knockdown, there is a dramatic increase or stabilization of nucleosomes on the Fab7 element (+110%; P = 0.0002) (Fig. 4B). NURF knockdown also induced a significant increase in nucleosome occupancy on the Fab8 insulator (+67%; P = 0.02). Interestingly, while the increase in nucleosome occupancy coincided with a loss of the Fab7 activity, it resulted in a marked enhancement of Fab8 activity, further strengthening the role of nucleosome organization in the enhancer-blocking mechanism of this insulator.
FIG. 4.
NURF localization at endogenous insulators and knockdown effect on nucleosome occupancy at these sites. (A) Differential localization of ISWI at endogenous Fab7 and Fab8 insulator elements. Fold enrichment of the Fab7 and Fab8 sequences over that for a control coding region (Gapdh exon) after chromatin immunoprecipitation (ChIP) in S2 cells using an anti-ISWI antibody is shown. Data represent the averages and standard deviations from three independent ChIP experiments, each with triplicate qPCR quantitation. (B) NURF knockdown increases nucleosome occupancy at the endogenous Fab7 and Fab8 sites. Fold enrichment of the Fab7 and Fab8 sequences over that for a control coding region (Gapdh exon) was determined by ChIP using an anti-H3 antibody before (green bars; n = 3) and after (brown bars; n = 3) NURF knockdown.
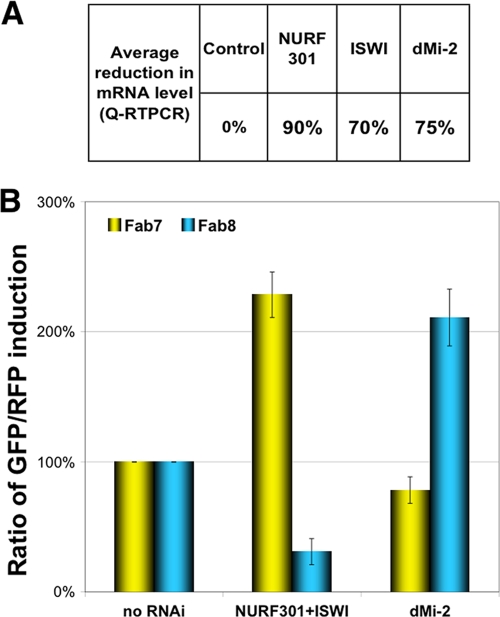
NURF and dMi-2 nucleosome-remodeling activities affect Fab7 and Fab8 insulators in opposite directions.
The NURF complex belongs to one of the three major classes of ATP-dependent chromatin-remodeling activities that utilize ISWI, SWI/SNF, or Mi2, respectively, as their ATPase subunits (for reviews, see references 46, 56, and 57). Many of the functional characteristics of nucleosome-remodeling complexes are determined by their ATPase subunits. The NURF complex and the SWI/SNF complexes have well-documented roles in transcription activation. In contrast, the nucleosome-remodeling and deacetylation (NuRD) complex, which uses dMi-2 as the ATPase subunit, is often associated with transcription repression (6, 7, 23, 37, 42, 50, 74). To investigate whether other ATP-dependent chromatin-remodeling enzymes exhibit an effect on insulator function similar to that of NURF, we decided to knock down dMi-2, the Drosophila ortholog of Mi-2, and examine its effect on the activity of Fab7 and Fab8 insulators (Fig. 5). A 70% reduction of the dMi-2 transcript level coincided with a moderate enhancement of the Fab7 activity (22% reduction in GFP/RFP ratio) and a strong disruption in the Fab8 function, as seen in the 110% increase in the GFP/RFP ratio (Fig. 5A). Our results show that the dMi-2-mediated nucleosome-remodeling activity affects the function of the Fab7 and Fab8 insulators in the opposite direction from NURF. It is unclear why the two nucleosome remodelers exert opposite effects on insulator function, but several differences between ISWI and dMi-2 have been previously documented. For example, dMi-2 can interact with both chromatosomes and nucleosomes, whereas ISWI binding requires the 248-bp nucleosomes that contain free DNA. Further, the ATPase activities of the two factors also show different histone tails or nucleosome dependence (22). The two factors also differ in their nucleosome-remodeling mechanisms: while ISWI-mediated remodeling moves a nucleosome to the center of the DNA template, dMi-2 moves it to the ends of DNA (14). It is also possible that NURF and NuRD complexes antagonize each other for interaction with the cis or trans components of the insulators. The opposing effects of the two complexes are consistent with their functional roles in Hox gene regulation: NURF is known to collaborate with GAF, a Trithorax group (Trx-G) protein, in enhancing Hox gene expression, whereas dMi-2 plays important role in Polycomb group (PcG)-mediated repression of Hox genes (5, 6, 42).
FIG. 5.
Knockdown of NURF and dMi-2 affects Fab7 and Fab8 insulators in opposite directions. (A) Average reduction in mRNA level caused by dsRNA-mediated knockdown of NURF301, ISWI, and dMi-2 as determined by qRT-PCR. (B) Opposite effects of NURF and dMi-2 knockdown on the enhancer-blocking activity of Fab7 and Fab8. Bars show the GFP/RFP ratio in S2 cells containing Fab7 (yellow) or Fab8 (blue) transgenes before knockdown (left) and after double knockdown of NURF301 and ISWI (middle) or knockdown of dMi-2 (right). Error bars indicate standard errors.
DISCUSSION
Insulator function is influenced by higher-order chromatin structure.
Chromatin boundaries organize the genome into functional units by limiting the range of organized chromatin and/or regulating enhancers. Understanding the interplay between boundary function and chromatin structure could provide further insights into their regulatory mechanisms. Previous studies have revealed that chromatin barriers in yeast and vertebrates utilize chromatin-remodeling factors to establish centers of active chromatin that impede the spread of silent chromatin (72). However, it is unclear whether the enhancer-blocking function of boundary elements is also influenced by the structure of the surrounding chromatin. We showed that NURF and dMi-2 nucleosome-remodeling activities modulate the enhancer-blocking function of multiple Drosophila insulators from the homeotic gene complexes, providing the first evidence that a higher order of chromatin organization influences the enhancer-blocking activity of chromatin boundaries. Compared to that of Fab7 and Fab8, the effect of NURF knockdown on the enhancer-blocking activity of the suHw insulator appears to be less specific, possibly mediated by mechanisms that are independent of nucleosome organization.
Fab 7 and Fab8 insulators utilize different enhancer-blocking mechanisms.
We showed that the NURF component ISWI was differentially localized to the endogenous Fab7 and Fab8 insulators. Reduction of NURF function stabilized or increased nucleosome occupancy on both insulators. While this change disrupted the enhancer-blocking function of Fab7, it enhanced the function of Fab8. These results indicate that the two insulators differ in their enhancer-blocking mechanisms. It appears that the activity of Fab7 is compatible with or enhanced by an open and more flexible nucleosome structure. The Fab7/SF1 class insulators have been previously shown to depend on GAF, a protein known to partner with NURF during transcription activation, prompting the hypothesis that transcription activation and enhancer blocking may be related in mechanism (12, 23, 63, 65, 73). Open chromatin and transcription initiation, facilitated by GAF and NURF, may functionally mimic aspects of a promoter and “trap” enhancers or activators (promoter decoy model [4, 13, 33, 39, 43, 62, 66]). Alternatively, NURF may facilitate GAF-dependent insulators at the level of chromatin structure. GAF has been postulated to facilitate enhancer blocking by dimerizing with other GAFs bound at remote DNA sites to form chromatin loops (35, 55, 58). It is possible that increased nucleosome mobility and chromatin flexibility facilitate chromatin loop formation and enhance the activity of these insulators. In contrast to the case for Fab7, NURF knockdown enhances the insulator activity of Fab8, suggesting that Fab8, and possibly other dCTCF/CTCF-dependent insulators, may depend on highly organized nucleosome arrays for their enhancer-blocking activity. Indeed, a recent study indicated that CTCF exhibits a striking ability to position up to 20 nucleosomes surrounding its binding sites in the human genome (25). However, the functional significance and mechanism of such nucleosome remodeling by the CTCF insulators were not clear. Our current data suggest that dCTCF/CTCF may confer enhancer-blocking activity by positioning nucleosomes, possibly by recruiting repressive nucleosome- or chromatin-remodeling enzymes (54, 75).
Opposite roles of NURF and NuRD in modulating insulator activity.
We showed that knockdown of dMi-2, the ATPase component of the Drosophila NuRD complex, enhanced the activity of Fab7 while suppressing that of Fab8. This is opposite to the effect of NURF knockdown. Both NURF and NuRD have been implicated in homeotic gene regulation. NURF301, the largest subunit of the NURF complex, was originally identified as enhancer of Bithorax (Ebx), an activator of the Ubx and other homeotic genes (5, 6). Conversely, dMi-2 was identified by its function in Polycomb-mediated repression of the homeotic genes (42). Our findings suggest that NURF and NuRD nucleosome-remodeling activities may regulate the homeotic genes by modulating the function of chromatin boundaries within the HOM/HOX complexes.
It is unclear how nucleosome remodeling by these two complexes confers opposite effects on insulator function. Transcription repression by NuRD was originally attributed to its histone deacetylase activity (68, 74, 76). However, recent studies have shown that dMi-2-related complexes without the histone deacetylase, such as dMec and dCHD (chromodomain-helicase-DNA-binding) complexes, could also mediate transcriptional repression (45, 47). Mechanistic differences between ISWI and dMi-2 nucleosome-remodeling factors have been documented. For example, ISWI binding to nucleosomes requires the 248-bp nucleosomes that contain free DNA, whereas dMi-2 can interact with both chromatosomes and nucleosomes (14). The ATPase activities of the two factors also show different histone tails or nucleosome dependence (22). Interestingly, the two factors slide nucleosomes in opposite directions: while ISWI moves a nucleosome to the center of the DNA template, dMi-2 moves it to the ends of DNA (14).
Our results show that the NURF nucleosome-remodeling activity facilitates the enhancer-blocking activity of SF1 and Fab7 boundaries, both of which also require GAF, a Trx-G protein, for their insulator function. These two elements may represent a class of Drosophila boundaries, which also include Mcp-1 and Evenskipped insulator, both GAF dependent, that function in an active and open chromatin environment (48, 58, 62). Consistent with this notion, SF1, Mcp-1, and Fab7 boundaries are located between homeotic enhancers that are active in multiple body segments, possibly to insulate them from each other (8, 29, 36, 53). In contrast, Fab8 is the most distal chromatin boundary element in the Drosophila Hox cluster, and its neighboring enhancers are silenced in all but the most posterior cells. Fab8 might function in such a repressive chromatin environment to limit the extent of PcG-mediated silencing encroaching into the Abd-B transcription unit. Interestingly, the CTCF/dCTCF class of insulators appear to be the only enhancer-blocking activities remaining in the vertebrate lineage. It is possible that changes in the genomic and higher-order chromatin organization played a role in the selection of chromatin boundaries during evolution.
Acknowledgments
We thank John Tamkun for the Iswi1 stock and the Bloomington Stock Center for the Ebxry122 stock.
Footnotes
Published ahead of print on 7 December 2009.
REFERENCES
- 1.Adkins, N. L., T. A. Hagerman, and P. Georgel. 2006. GAGA protein: a multi-faceted transcription factor. Biochem. Cell. Biol. 84:559-567. [DOI] [PubMed] [Google Scholar]
- 2.Adryan, B., G. Woerfel, I. Birch-Machin, S. Gao, M. Quick, L. Meadows, S. Russell, and R. White. 2007. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 8:R167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ameres, S. L., L. Drueppel, K. Pfleiderer, A. Schmidt, W. Hillen, and C. Berens. 2005. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. EMBO J. 24:358-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avramova, Z., and A. Tikhonov. 1999. Are scs and scs′ ‘neutral’ chromatin domain boundaries of the locus? Trends Genet. 15:138-139. [DOI] [PubMed] [Google Scholar]
- 5.Babu, P., K. S. Selvakumar, and S. Bhosekar. 1987. Studies on transvection at the bithorax complex in Drosophila melanogaster. Mol. Gen. Genet. 210:557-563. [DOI] [PubMed] [Google Scholar]
- 6.Badenhorst, P., M. Voas, I. Rebay, and C. Wu. 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16:3186-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badenhorst, P., H. Xiao, L. Cherbas, S. Y. Kwon, M. Voas, I. Rebay, P. Cherbas, and C. Wu. 2005. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 19:2540-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barges, S., J. Mihaly, M. Galloni, K. Hagstrom, M. Muller, G. Shanower, P. Schedl, H. Gyurkovics, and F. Karch. 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127:779-790. [DOI] [PubMed] [Google Scholar]
- 9.Bartkuhn, M., T. Straub, M. Herold, M. Herrmann, C. Rathke, H. Saumweber, G. D. Gilfillan, P. B. Becker, and R. Renkawitz. 2009. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 28:877-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 11.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 12.Belozerov, V. E., P. Majumder, P. Shen, and H. N. Cai. 2003. A novel boundary element may facilitate independent gene regulation in the Antennapedia Complex of Drosophila. EMBO J. 22:3113-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bi, X., and J. R. Broach. 1999. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 13:1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brehm, A., G. Langst, J. Kehle, C. R. Clapier, A. Imhof, A. Eberharter, J. Muller, and P. B. Becker. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19:4332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bushey, A. M., E. R. Dorman, and V. G. Corces. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell 32:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bushey, A. M., E. Ramos, and V. G. Corces. 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 23:1338-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai, H. N. 2006. Function and mechanism of chromatin boundaries. In J. Ma (ed.), Gene expression and regulation. Higher Education Press, Beijing, China.
- 18.Cai, H. N., and M. Levine. 1997. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 16:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai, H. N., and P. Shen. 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291:493-495. [DOI] [PubMed] [Google Scholar]
- 20.Celniker, S. E., and R. A. Drewell. 2007. Chromatin looping mediates boundary element promoter interactions. Bioessays 29:7-10. [DOI] [PubMed] [Google Scholar]
- 21.Clemens, J. C., C. A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B. A. Hemmings, and J. E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. U. S. A. 97:6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corona, D. F., G. Langst, C. R. Clapier, E. J. Bonte, S. Ferrari, J. W. Tamkun, and P. B. Becker. 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3:239-245. [DOI] [PubMed] [Google Scholar]
- 23.Deuring, R., L. Fanti, J. A. Armstrong, M. Sarte, O. Papoulas, M. Prestel, G. Daubresse, M. Verardo, S. L. Moseley, M. Berloco, T. Tsukiyama, C. Wu, S. Pimpinelli, and J. W. Tamkun. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5:355-365. [DOI] [PubMed] [Google Scholar]
- 24.Dunn, K. L., H. Zhao, and J. R. Davie. 2003. The insulator binding protein CTCF associates with the nuclear matrix. Exp. Cell Res. 288:218-223. [DOI] [PubMed] [Google Scholar]
- 25.Elfring, L. K., R. Deuring, C. M. McCallum, C. L. Peterson, and J. W. Tamkun. 1994. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol. Cell. Biol. 14:2225-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farkas, G., B. A. Leibovitch, and S. C. Elgin. 2000. Chromatin organization and transcriptional control of gene expression in Drosophila. Gene 253:117-136. [DOI] [PubMed] [Google Scholar]
- 27.Ferreira, R., A. Eberharter, T. Bonaldi, M. Chioda, A. Imhof, and P. B. Becker. 2007. Site-specific acetylation of ISWI by GCN5. BMC Mol. Biol. 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu, Y., M. Sinha, C. L. Peterson, and Z. Weng. 2008. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 4:e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galloni, M., H. Gyurkovics, P. Schedl, and F. Karch. 1993. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 12:1087-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaszner, M., and G. Felsenfeld. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7:703-713. [DOI] [PubMed] [Google Scholar]
- 31.Gaszner, M., J. Vazquez, and P. Schedl. 1999. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 13:2098-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerasimova, T. I., K. Byrd, and V. G. Corces. 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6:1025-1035. [DOI] [PubMed] [Google Scholar]
- 33.Geyer, P. K., and I. Clark. 2002. Protecting against promiscuity: the regulatory role of insulators. Cell. Mol. Life Sci. 59:2112-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geyer, P. K., and V. G. Corces. 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6:1865-1873. [DOI] [PubMed] [Google Scholar]
- 35.Gruzdeva, N., O. Kyrchanova, A. Parshikov, A. Kullyev, and P. Georgiev. 2005. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 25:3682-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gyurkovics, H., J. Gausz, J. Kummer, and F. Karch. 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9:2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamiche, A., R. Sandaltzopoulos, D. A. Gdula, and C. Wu. 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97:833-842. [DOI] [PubMed] [Google Scholar]
- 38.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 39.Hogga, I., and F. Karch. 2002. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development 129:4915-4922. [DOI] [PubMed] [Google Scholar]
- 40.Holohan, E. E., C. Kwong, B. Adryan, M. Bartkuhn, M. Herold, R. Renkawitz, S. Russell, and R. White. 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 3:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson, C. A., A. L. Barlow, and B. M. Turner. 1998. Molecular cloning of Drosophila melanogaster cDNAs that encode a novel histone deacetylase dHDAC3. Gene 221:127-134. [DOI] [PubMed] [Google Scholar]
- 42.Kehle, J., D. Beuchle, S. Treuheit, B. Christen, J. A. Kennison, M. Bienz, and J. Muller. 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282:1897-1900. [DOI] [PubMed] [Google Scholar]
- 43.Kellum, R., and P. Schedl. 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64:941-950. [DOI] [PubMed] [Google Scholar]
- 44.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 45.Kunert, N., and A. Brehm. 2008. Mass production of Drosophila embryos and chromatographic purification of native protein complexes. Methods Mol. Biol. 420:359-371. [DOI] [PubMed] [Google Scholar]
- 46.Kunert, N., and A. Brehm. 2009. Novel Mi-2 related ATP-dependent chromatin remodelers. Epigenetics 4:209-211. [DOI] [PubMed] [Google Scholar]
- 47.Kunert, N., E. Wagner, M. Murawska, H. Klinker, E. Kremmer, and A. Brehm. 2009. dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. EMBO J. 28:533-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kyrchanova, O., S. Toshchakov, A. Parshikov, and P. Georgiev. 2007. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol. Cell. Biol. 27:3035-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanctot, C., T. Cheutin, M. Cremer, G. Cavalli, and T. Cremer. 2007. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8:104-115. [DOI] [PubMed] [Google Scholar]
- 50.Langst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 51.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34-39. [DOI] [PubMed] [Google Scholar]
- 52.Li, M., V. E. Belozerov, and H. N. Cai. 2008. Analysis of chromatin boundary activity in Drosophila cells. BMC Mol. Biol. 9:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin, Q., Q. Chen, L. Lin, S. Smith, and J. Zhou. 2007. Promoter targeting sequence mediates enhancer interference in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 104:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lutz, M., L. J. Burke, G. Barreto, F. Goeman, H. Greb, R. Arnold, H. Schultheiss, A. Brehm, T. Kouzarides, V. Lobanenkov, and R. Renkawitz. 2000. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic Acids Res. 28:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahmoudi, T., K. R. Katsani, and C. P. Verrijzer. 2002. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 21:1775-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maier, V. K., M. Chioda, and P. B. Becker. 2008. ATP-dependent chromatosome remodeling. Biol. Chem. 389:345-352. [DOI] [PubMed] [Google Scholar]
- 57.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 58.Melnikova, L., F. Juge, N. Gruzdeva, A. Mazur, G. Cavalli, and P. Georgiev. 2004. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 101:14806-14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mishra, R. K., J. Mihaly, S. Barges, A. Spierer, F. Karch, K. Hagstrom, S. E. Schweinsberg, and P. Schedl. 2001. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and Pleiohomeotic for Silencing. Mol. Cell. Biol. 21:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mlynarova, L., A. Loonen, E. Mietkiewska, R. C. Jansen, and J. P. Nap. 2002. Assembly of two transgenes in an artificial chromatin domain gives highly coordinated expression in tobacco. Genetics 160:727-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moon, H., G. Filippova, D. Loukinov, E. Pugacheva, Q. Chen, S. T. Smith, A. Munhall, B. Grewe, M. Bartkuhn, R. Arnold, L. J. Burke, R. Renkawitz-Pohl, R. Ohlsson, J. Zhou, R. Renkawitz, and V. Lobanenkov. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohtsuki, S., and M. Levine. 1998. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 12:3325-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okada, M., and S. Hirose. 1998. Chromatin remodeling mediated by Drosophila GAGA factor and ISWI activates fushi tarazu gene transcription in vitro. Mol. Cell. Biol. 18:2455-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petesch, S. J., and J. T. Lis. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134:74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schweinsberg, S., K. Hagstrom, D. Gohl, P. Schedl, R. P. Kumar, R. Mishra, and F. Karch. 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168:1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spana, C., D. A. Harrison, and V. G. Corces. 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2:1414-1423. [DOI] [PubMed] [Google Scholar]
- 67.Tautz, D., and C. Pfeifle. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81-85. [DOI] [PubMed] [Google Scholar]
- 68.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917-921. [DOI] [PubMed] [Google Scholar]
- 69.Tsukiyama, T., P. B. Becker, and C. Wu. 1994. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature 367:525-532. [DOI] [PubMed] [Google Scholar]
- 70.Tsukiyama, T., C. Daniel, J. Tamkun, and C. Wu. 1995. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83:1021-1026. [DOI] [PubMed] [Google Scholar]
- 71.Varga-Weisz, P. D., T. A. Blank, and P. B. Becker. 1995. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J. 14:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.West, A. G., S. Huang, M. Gaszner, M. D. Litt, and G. Felsenfeld. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453-463. [DOI] [PubMed] [Google Scholar]
- 73.Xiao, H., R. Sandaltzopoulos, H. M. Wang, A. Hamiche, R. Ranallo, K. M. Lee, D. Fu, and C. Wu. 2001. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell 8:531-543. [DOI] [PubMed] [Google Scholar]
- 74.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 75.Yasui, D., M. Miyano, S. Cai, P. Varga-Weisz, and T. Kohwi-Shigematsu. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419:641-645. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 77.Zhao, K., C. M. Hart, and U. K. Laemmli. 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81:879-889. [DOI] [PubMed] [Google Scholar]
- 78.Zhou, J., H. Ashe, C. Burks, and M. Levine. 1999. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development 126:3057-3065. [DOI] [PubMed] [Google Scholar]