Abstract
Oscillatory synthesis and secretion of the gonadotropins, follicle-stimulating hormone (FSH) and luteinizing hormone (LH), under the control of pulsatile hypothalamic gonadotropin-releasing hormone (GnRH), is essential for normal reproductive development and fertility. The molecular mechanisms by which various patterns of pulsatile GnRH regulate gonadotrope responsiveness remain poorly understood. In contrast to the α and LHβ subunit genes, FSHβ subunit transcription is preferentially stimulated at low rather than high frequencies of pulsatile GnRH. In this study, mutation of a cyclic AMP response element (CRE) within the FSHβ promoter resulted in the loss of preferential GnRH stimulation at low pulse frequencies. We hypothesized that high GnRH pulse frequencies might stimulate a transcriptional repressor(s) to attenuate the action of CRE binding protein (CREB) and show that inducible cAMP early repressor (ICER) fulfills such a role. ICER was not detected under basal conditions, but pulsatile GnRH stimulated ICER to a greater extent at high than at low pulse frequencies. ICER binds to the FSHβ CRE site to reduce CREB occupation and abrogates both maximal GnRH stimulation and GnRH pulse frequency-dependent effects on FSHβ transcription. These data suggest that ICER production antagonizes the stimulatory action of CREB to attenuate FSHβ transcription at high GnRH pulse frequencies, thereby playing a critical role in regulating cyclic reproductive function.
The maintenance of normal reproductive function in all vertebrate species is dependent on the regulation of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) synthesis and release by pituitary gonadotropes. These hormones are released in a pulsatile manner to regulate gametogenesis and gonadal hormone synthesis (2, 11, 17). The intermittent synthesis and secretion of LH and FSH by pituitary gonadotropes are tightly regulated, as evidenced by predictable and reproducible changes in circulating levels throughout the menstrual or estrous cycle. Although the synthesis and release of pituitary gonadotropins are affected by a number of endocrine, paracrine, and autocrine factors, the most important influence appears to be that of the hypothalamic decapeptide, gonadotropin-releasing hormone (GnRH). The tight inter-relationship between GnRH release and gonadotropin production is evidenced in patients with Kallmann's syndrome, in which GnRH deficiency results in low gonadotropin levels, absence of pubertal maturation, and infertility (42). Thus, GnRH is an essential coordinator of reproductive function.
Regulation of gonadotropin biosynthesis and secretion by GnRH is critically dependent on GnRH delivery to the anterior pituitary. Pulsatile GnRH results in the stimulation of gonadotropin subunit mRNA levels and of LH and FSH secretion, whereas continuous exposure to GnRH downregulates mRNA levels and secretion (2, 45). Furthermore, the frequency and amplitude of GnRH pulses varies temporally and developmentally, for example, during different phases of the menstrual or estrous cycle, and determines, in part, the relative proportions of LH and FSH synthesis and secretion (34). Increased frequency of pulsatile hypothalamic GnRH release favors LHβ gene transcription over FSHβ and increases the ratio of secreted LH to FSH (1, 2, 15, 19, 34, 45). Conversely, a decreased GnRH pulse frequency, characteristic of the luteal and early follicular phases of the ovulatory cycle, favors FSHβ, allowing for increased pituitary FSH secretion essential for the recruitment and selection of the maturing ovum (1, 2, 15, 19, 34, 45).
The response of gonadotropes to GnRH in terms of relative FSH and LH production is thus exquisitely sensitive to the pattern of GnRH stimulation. This is exemplified in polycystic ovarian syndrome (PCOS), the most common cause of infertility in women of reproductive age, affecting up to 10% of this population (13). This disorder, which is becoming increasingly prevalent, is often associated with obesity, insulin resistance, and metabolic and cardiovascular abnormalities similar to those of the metabolic syndrome (23). The pathogenesis of this disorder remains unclear, but one hallmark of PCOS is that of disrupted reproductive cycles as a consequence of elevated serum LH and depressed FSH levels, leading to an increase in androgen production by ovarian thecal cells (3, 12, 23). This change in gonadotropin dynamics reflects increased hypothalamic GnRH neuronal activity which manifests itself in predominantly high frequency GnRH pulsatility (3, 12, 23). In the present study, we propose a mechanism by which changes in GnRH pulse frequency cause differential pituitary FSHβ gene expression.
We (8) and others (10, 44) have characterized a major GnRH responsive element within the proximal FSHβ promoter, which contains a partial cyclic AMP (cAMP) response element (CRE) that, in the rat, is predominantly bound by CRE binding protein (CREB) (8). Since this GnRH responsive element is 100% conserved in humans (44), it may be of clinical relevance, with protein-DNA interactions at this site a potential focus for therapeutic intervention. GnRH stimulates rat (r)FSHβ transcription by inducing phosphorylation of CREB bound to this site, leading to the recruitment of the histone acetyltransferase CREB binding protein (CBP) (8). We show here that a mutation of this GnRH responsive element abolishes the preferential stimulation of FSHβ transcription by low frequencies of pulsatile GnRH, leading us to hypothesize that high GnRH pulse frequencies stimulate a transcriptional repressor(s) to attenuate the action of CREB. We provide evidence that functional antagonism between CREB and a transcriptional repressor, inducible cAMP early repressor (ICER), exists in the gonadotrope to mediate the differential regulation of FSHβ transcription by various patterns of pulsatile GnRH. Given the central role of FSH in the control of gametogenesis, we provide here a context for the design of novel therapeutic approaches to contraception and the treatment of infertility, PCOS, and other reproductive disorders.
MATERIALS AND METHODS
Reporter plasmids, shRNA constructs, and expression vectors.
−140/+15 rFSHβLuc was generated as previously described by fusing the −140/+15 portion of the rFSHβ gene promoter upstream of the luciferase reporter gene in pXP2 (48). A 6-bp mutation was introduced into the CRE site of −140/+15 rFSHβLuc (5′-GGTCACGTT-3′ to 5′-GcggccgTT-3′; ΔCRE) by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's suggested protocol as previously reported (8). For an internal standard, an expression vector encoding β-galactosidase driven by the simian virus 40 early promoter was used in all luciferase studies (SV40βGal; Promega, Madison, WI). Short hairpin RNA (shRNA) constructs containing either an shICER or a control shCtrl scrambled sequence were purchased from Superarray Biosciences Corp. (Frederick, MD). The expression vectors for ICER I, Iγ, II, and IIγ in pcDNA3 were a generous gift from Kelly Mayo (Northwestern University, Chicago, IL), and the expression vector for CREB in pCMX was kindly provided by J. Larry Jameson (Northwestern University, Chicago, IL).
Cell culture and transfection.
The murine gonadotrope-derived LβT2 cell line was kindly provided by Pamela L. Mellon (University of California San Diego, San Diego, CA) and were grown and maintained in high-glucose Dulbecco modified Eagle medium (DMEM; HyClone, Logan, UT) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Omega, Tarzana, CA), 100 U of penicillin/ml, and 100 μg of streptomycin sulfate (Invitrogen, Carlsbad, CA)/ml in 5% CO2 humidified air at 37°C. HEK293 cells were cultured in low-glucose DMEM (HyClone) with the same supplements outlined above and transiently transfected by the calcium phosphate coprecipitation method. Transient transfections in LβT2 cells were performed by electroporation as described previously (8). Briefly, LβT2 cells were transiently transfected by electroporation with the indicated vectors. The cells were suspended in 0.5 ml of Dulbecco phosphate-buffered saline (PBS) supplemented with 5 mM glucose containing the plasmid DNA to be transfected. The LβT2 cells were exposed to a single electrical pulse of 0.24V with a total capacitance of 960 μF and allowed to recover in PBS supplemented with 5 mM glucose and 20% FBS before plating. Static cultures of LβT2 were stimulated by 100 nM GnRHAg (des-Gly10,[d-Ala6]-LHRH ethylamide; Sigma, St. Louis, MO) or vehicle.
Perifusion studies.
Perifusion studies were conducted as previously reported (1). Briefly, LβT2 cells were plated in perifusion chambers coated with Matrigel (Becton Dickinson Labware, Bedford, MA) and incubated for 24 h in static culture. The chambers were subsequently mounted in the perifusion system and continuously perifused with high-glucose DMEM supplemented with 1% FBS and 1% penicillin-streptomycin at a constant flow rate (0.25 ml/min). During perifusion, groups of three chambers were treated for 20 h with either medium alone or 10 nM GnRH (Sigma) pulses at high (every 30 min) or low (every 120 min) frequency. GnRH pulse frequencies were delivered by peristaltic pumps controlled by a time controller (ChronTrol XT, San Diego, CA). At 20 min after the last GnRH pulse, the chambers were disconnected, and the cells were harvested for analyses.
mRNA and protein quantification.
Total RNA from LβT2 cells was extracted by using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's protocol, including DNase treatment. One microgram of RNA was reverse transcribed using Superscript III cDNA synthesis kit (Invitrogen). Semiquantitative reverse transcription-PCR analyses were performed in a PTC-100 thermal cycler (MJ Research, Waltham, MA) with Taq polymerase (Promega) containing the addition of 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates spiked with 1 μCi of [α-32P]dCTP (Perkin-Elmer, Waltham, MA), 2 μl of cDNA, and specific primers. The primer sequences for each respective gene were as follows: ICER sense (5′-ATGGCTGTAACTGGAGATGAAACT-3′) and antisense (5′-CTAATCTGTTTTGGGAGAGCAAATGTC-3′), FSHβ sense (5′-AGACAGCTGACTGCACAGGA-3′) and antisense (5′-CCGAGCTGGGTCCTTATACA-3′), and L19 sense (5′-CTGAAGGTCAAAGGGAATGTG-3′) and antisense (5′-GGACAGAGTCTTGATGATCTC-3′). The amplification cycling profile used for all genes was 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. To ensure linearity of amplification for each primer set, different cycling conditions were tested, and the final PCR assays were performed using the optimal, nonsaturating conditions attained for each gene. Amplified samples were analyzed on a 5% native acrylamide gel and subsequently dried and scanned in a phosphorimager.
Total protein was purified in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 0.1 μM aprotinin, and 1 μM pepstatin to a final pH of 7.4). Portions (50 μg) of protein were subjected to SDS-PAGE separation and transferred onto a nitrocellulose membrane (Whatman, Waltham, MA) for Western blot analyses. The membranes were then blocked with 5% nonfat dry milk in TBST (Tris-buffered saline plus Tween 20) for 1 h at room temperature with shaking and then incubated with an ICER (kindly provided by Carlos Molina, New Jersey Medical School, Newark, NJ) antibody at 4°C for 12 h with gentle shaking. Blots were subsequently rinsed with TBST and incubated with secondary antibody (donkey anti-rabbit antibody conjugated to horseradish peroxidase; Santa Cruz); after rinsing again with TBST, the antibody-antigen complexes were visualized by using an enhanced chemiluminescence reagent (Perkin-Elmer). The blots were stripped (100 mM β-mercaptoethanol, 2% SDS, 62.5 mM Tris [pH 6.8]) and reprobed with a β-actin antibody (Sigma) for normalization purposes. The signal intensities for each band, either cDNA or protein, were quantified by using ImageJ (National Institute of Health [www.rsb.info.nih.gov/nih-image/]). Values were normalized by each respective housekeeping gene and expressed as arbitrary units.
Electrophoretic mobility shift assay (EMSA).
The rFSHβ probes containing the wild-type (WT) (5′-TGGTATTGGTCACGTTAACACCCAGTAAAT-3′) and mutant (Δ) (5′-TGGTATTGGCGGCCGTTAACACCCAGTAAAT-3′) CRE sites were annealed with complementary antisense oligonucleotides in annealing buffer (100 nM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]) and purified by PAGE. The probes were 5′ end labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and extracted by using a quick spin column (Roche, Nutley, NJ). Nuclear extracts were purified from HEK293 or LβT2 cells as previously described (8). Then, 2 μg of HEK293 or 10 μg of LβT2 nuclear extract per sample was used with 100,000 cpm of probe in binding buffer (0.01 μg of salmon sperm/μl, 2.15 mM phenylmethylsulfonyl fluoride, 5 mM dithiothreitol, 20 mM HEPES [pH 7.9], 60 mM KCl, 5 mM MgCl2, 1 mg of bovine serum albumin/ml, and 5% [vol/vol] glycerol). Gels were dried and scanned in a phosphorimager. An ICER antibody (kindly provided by Kelly Mayo, Northwestern University, Chicago, IL) was used to supershift protein-DNA complexes, while rabbit IgG (Santa-Cruz) was used as a negative control.
ChIP and semiquantitative and quantitative real-time PCR.
Chromatin immunoprecipitation (ChIP) analyses were performed as previously described (8). Briefly, LβT2 cells were transfected by electroporation with either WT or ΔCRE −140/+15 rFSHβLuc and either a fixed or increasing amounts of an ICER II expression vector as indicated. Cells were incubated for 48 h after transfection, stimulated with 100 nM GnRHAg for 1 h immediately before collection, cross-linked with 1% formaldehyde at room temperature for 15 min, and rinsed twice in ice-cold PBS. Cross-linked DNA was sonicated to fragments ranging from 200 to 500 bp in length. After sonication, the chromatin solutions were precleared with protein A-agarose (Upstate, Charlottesville, VA) and salmon sperm DNA and subjected to immunoprecipitation by incubation with CREB or ICER antibodies, followed by incubation with protein A-agarose-salmon sperm DNA. Preimmune IgG was used as a negative control. After sequential washes with 2× low salt (20 mM Tris [pH 8.0], 150 mM NaCl, 2 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% SDS), 2× high salt (20 mM Tris [pH 8.0], 500 mM NaCl, 2 mM EDTA [pH 8.0], 1% Triton X-100, 0.1% SDS), 2× LiCl (0.25 M LiCl, 1% NP-40, 1% sodium desoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and 4× Tris-EDTA buffer, precipitated chromatin was eluted, and cross-linking was reversed with 0.3 M NaCl. After protein and RNA removal with proteinase K and RNase (Roche Applied Science), respectively, chromatin was purified by phenol-chloroform extraction and ethanol precipitation. Semiquantitative PCR was performed by using a sense primer directed against the rFSHβ (−117/−93) promoter sequence (5′-TGTCTAAACAATGATTCCCTTTCA-3′) and an antisense primer directed against the pXP2 vector (5′-CTTTCTTTATGTTTTTGGCGTCTT-3′), which produced a product encompassing the CRE in the rFSHβ promoter. For ChIP studies on the endogenous mouse FSHβ promoter, LβT2 cells were transfected with 12 μg of an ICER II expression vector or the empty control vector and were treated with 100 nM GnRHAg for 1 h immediately before harvest for ChIP assay as outlined above. Previously published oligonucleotides (5′-GGTGTGCTGCCATATCAGATTCGG-3′) and (5′-GCATCAAGTGCTGCTACTCACCTGTG-3′) spanning a 280-bp region of the mouse FSHβ gene from −223 to +57 that encompasses the partial CRE/AP1 site (10) were used to amplify immunoprecipitated DNA. Since the ICER II expression vector used in this experiment encodes a hemagglutinin (HA) sequence (6), an HA antibody (Santa Cruz) was incubated with cell lysates to determine ICER binding, whereas the CREB antibody was used to determine CREB binding.
DNA was amplified in a PTC-100 thermal cycler (MJ Research) for 24 to 28 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 30 s). Samples were then analyzed on a 5% native acrylamide gel and subsequently dried and scanned on a phosphorimager, which measured incorporated [α-32P]dCTP. Cyclophilin A (5′-CGAGCTCTGAGCACTGGAGA-3′) and (5′-TGGCGTGTAAAGTCACCACC-3′) was used as a control to verify specificity of the immunoprecipitation. Quantitative real-time PCR assays were performed on an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) using primers described above and SYBR green mix (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. The results were analyzed using ABI Prism 7000 SDS software (Applied Biosystems), and data presented as either a fold change from LβT2 cells transfected with empty pcDNA3 vector or as absolute values as quantified by a standard curve generated through serial dilution of the −140/+15 rFSHβLuc construct. PCRs were subsequently subjected to electrophoresis on an agarose gel to verify that only a single band was amplified.
Statistical analyses.
All graphs were drawn and statistical analyses were performed in Prism 3.00 for Windows (GraphPad Software, San Diego, CA). Details of statistical analyses for each individual experiment can be found in each respective figure legend. P values of <0.05 were considered statistically significant.
RESULTS
Mutation of a major GnRH responsive element within the FSHβ promoter results in loss of preferential GnRH stimulation at low pulse frequencies.
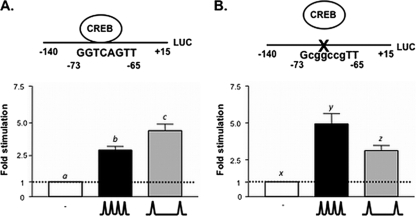
We recently identified a major GnRH responsive element within the rFSHβ promoter (positions −80/−51 relative to the transcription initiation start site), which contains a partial CRE site (5′-GGTCA-3′) (8). This sequence is 100% conserved in both human (44) and mouse (10) and is bound in vivo by the bZIP transcription factor, CREB. In static cultures of the murine gonadotrope LβT2 cell line, mutation of this element reduces both basal and GnRH stimulation of FSHβ gene expression (8). To determine the role of this CRE site in mediating the response of FSHβ transcription to pulsatile GnRH, LβT2 cells were transfected with a luciferase reporter construct driven by either −140/+15 of the rFSHβ gene promoter containing the wild-type CRE site (5′-GGTCACGTT-3′) or the corresponding construct harboring a mutation in this CRE (5′-GcggccgTT-3′). Twelve hours after transfection, the cells were perifused for 20 h and exposed to pulsatile GnRH at either high (every 30 min) or low (every 120 min) pulse frequencies or with medium alone, as previously described (1). Here and in all subsequent experiments, GnRH pulses were administered every 30 min to reflect the high-pulse-frequency state and every 2 h to reflect the low-pulse-frequency state. These pulse frequencies were chosen based on previous studies, indicating that these frequencies were optimal for LHβ gene expression and LH secretion and FSHβ gene expression and FSH secretion, respectively (1, 4, 26). In keeping with our previous studies, LβT2 cells harboring the wild-type FSHβ promoter construct show preferential stimulation of FSHβ transcription at the low GnRH pulse frequency (Fig. 1A) (1). However, mutation of the CRE site resulted in a loss of this preferential GnRH stimulation at the low pulse frequency (Fig. 1B). These results indicate that the FSHβ partial CRE plays a critical role in GnRH pulse frequency-dependent regulation of FSHβ gene expression. We hypothesized that a transcriptional repressor may be stimulated in the gonadotrope at high GnRH pulse frequencies to attenuate FSHβ transcription by antagonizing the formation of a multiprotein complex composed of CREB, CBP, and elements of the transcriptional machinery.
FIG. 1.
Mutation of a CRE site in the FSHβ promoter results in loss of preferential GnRH stimulation at low pulse frequencies. LβT2 cells were transfected with (A) wild-type −140/+15 rFSHβLuc reporter or (B) mutant CRE −140/+15 rFSHβLuc reporter. The cells were stimulated with pulsatile GnRH at a low (∧__∧; one pulse/120 min) or high (∧∧∧; one pulse/30 min) frequency. Bar graphs show means ± the standard errors of the mean (SEM) fold stimulation for each experimental condition. The data presented are a pool of four to five independent experiments, each performed in triplicate. Significant differences (P < 0.05), measured by one-way analysis of variance (ANOVA) with a post hoc Tukey-Kramer multiple-comparison test, are indicated by different letters.
GnRH induces ICER expression in a gonadotrope cell line.
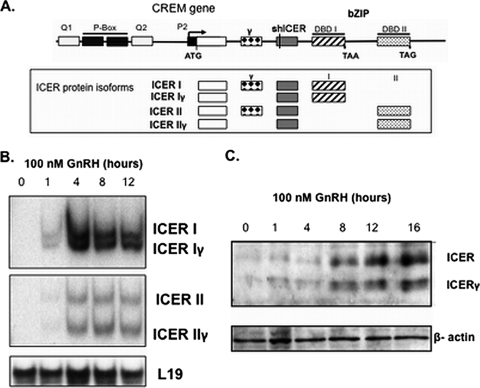
Proteins that inhibit the recruitment of CBP to the promoter of CREB responsive genes will cause transcriptional repression. There are bZIP repressor proteins that are endogenous antagonists of CREB, including a well-characterized class of repressor isoforms known as inducible cAMP early repressor (ICER) (28, 36). As a consequence of alternative splicing events, ICER is expressed as a family of four isoforms. ICER I and II contain distinct bZIP domains; in addition, both ICER I and ICER II can be deficient in exon γ and are accordingly named ICER Iγ and ICER IIγ (Fig. 2A). These isoforms have the ability to bind to CRE or CRE-like elements but, since they lack both kinase-inducible and transactivation domains, they are unable to recruit CBP to increase transcription. Instead, they act as transcriptional repressors by abrogating CREB action. To establish whether these ICER isoforms play a functional role in controlling FSHβ transcription, we first determined whether ICER is expressed in the LβT2 gonadotrope-derived cell line, under basal or GnRH-stimulated conditions. As measured by PCR, and in keeping with the dogma that ICER isoforms are inducible products of the CREM gene, there was no detectable ICER mRNA expression under basal conditions in static LβT2 cell cultures (Fig. 2B). However, GnRH exposure stimulated expression of transcripts of all four ICER isoforms (Fig. 2B). ICER protein levels were similarly induced by GnRH (Fig. 2C). The expression of ICER isoforms following GnRH stimulation supports a potential role for ICER in the GnRH control of the FSHβ gene.
FIG. 2.
GnRH stimulates inducible cAMP early repressor (ICER) in LβT2 cells. (A) Structure of the CREM gene that encodes ICER from an internal promoter (P2). Alternative splicing events give rise to four distinct ICER protein isoforms as depicted in the boxed field. DBD I and II, DNA-binding domain I and II; ATG, translational start site; TAA and TAG, translational stop codons; bZIP, basic leucine zipper motif. Regions encoding glutamine rich (Q1 and Q2) and kinase-inducible (P-Box) domains of the CREM gene, absent in ICER isoforms, are also depicted. The shICER sequence used in later experiments is common to all ICER isoforms. (B) ICER isoform mRNA levels in LβT2 cells at the indicated time points after stimulation with 100 nM GnRHAg. L19 mRNA levels are included as a control. (C) ICER isoform protein levels at the indicated time points after stimulation with 100 nM GnRHAg. β-Actin protein levels are shown as a loading control.
ICER is induced to a greater extent at high GnRH pulse frequencies.
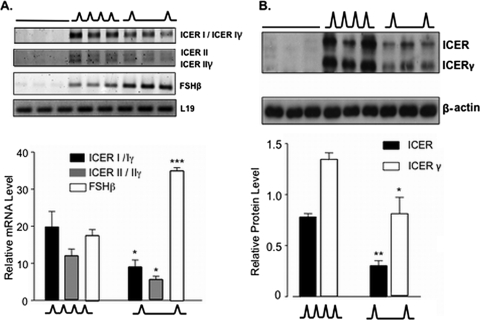
Based on the evidence that GnRH can induce ICER production in LβT2 cells, we hypothesized that ICER may be preferentially stimulated at high GnRH pulse frequencies to attenuate FSHβ transcription. LβT2 cells were perifused for 20 h with either high (every 30 min) or low (every 120 min) frequencies of pulsatile GnRH or with medium alone and subsequently harvested for mRNA or protein quantification. Confirming our initial study, ICER was undetectable under basal conditions at both the mRNA (Fig. 3A) and protein (Fig. 3B) levels but was induced by pulsatile GnRH. Furthermore, ICER mRNA (Fig. 3A) and protein (Fig. 3B) were significantly more abundant in LβT2 cells exposed to high, rather than low, GnRH pulse frequencies. In contrast, FSHβ mRNA was significantly more abundant at low, rather than high, GnRH pulse frequencies (Fig. 3A), complementing previous studies (1, 4, 26) and further supporting a potential inhibitory role for ICER in the differential control of FSHβ transcription by pulsatile GnRH.
FIG. 3.
ICER mRNA and protein isoforms are preferentially stimulated at high GnRH pulse frequencies. (A) ICER, FSHβ, and L19 mRNA levels were quantified after 20 h of perifusion and stimulation with high (∧∧∧; every 30 min) or low (∧__∧; every 120 min) frequencies of pulsatile GnRH or with medium alone (___). (B) ICER and β-actin protein levels were quantified by Western blot analysis under the same conditions as in panel A. In both panels A and B, the upper panels show the results of a representative experiment. The lower panels show the means ± the SEM of three independent experiments, each performed in triplicate. *, P < 0.05; **, P < 0.005; ***, P < 0.001 (compared to the corresponding mRNA or protein level at the high GnRH pulse frequency [Student unpaired t test]).
ICER isoforms attenuate maximal GnRH stimulation of FSHβ transcription.
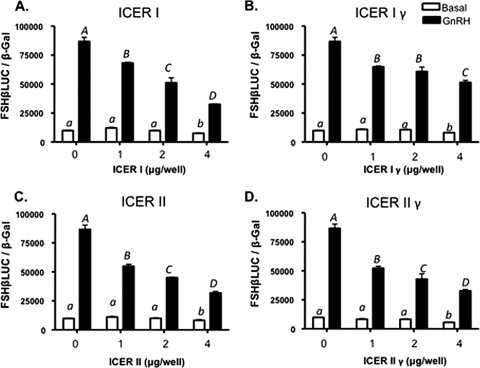
Cotransfection studies were performed to determine whether ICER is able to downregulate GnRH/CREB stimulation of FSHβ transcription. LβT2 cells were transfected with increasing amounts of expression vectors encoding each of the four ICER isoforms, together with an expression vector encoding CREB as well as −140/+15 rFSHβLuc. At 44 h after transfection, cells were treated with 100 nM GnRHAg or vehicle for an additional 4 h prior to harvest for subsequent luciferase and β-galactosidase assays. All four ICER isoforms significantly attenuated maximal GnRH stimulation of the FSHβ promoter in a dose-dependent fashion (Fig. 4).
FIG. 4.
Overexpression of ICER I (A), ICER Iγ (B), ICER II (C), and ICER IIγ (D) attenuates the maximal GnRH transcriptional response of the rFSHβ gene in a dose-dependent manner. LβT2 cells were transfected with 2 μg of −140/+15 rFSHβLuc and increasing amounts of the indicated ICER expression vector, together with 1 μg of a CREB expression vector. Cells were treated 44 h after transfection with 100 nM GnRHAg or vehicle for 4 h prior to harvest. Bar graphs show the luciferase activity (means ± the SEM from triplicate samples, normalized to β-galactosidase activity) from a representative experiment, repeated on three separate occasions with comparable results. Significant differences (P < 0.05), as measured by one-way ANOVA with post hoc Tukey-Kramer multiple-comparison test, are denoted by different letters.
ICER can bind to the FSHβ CRE site.
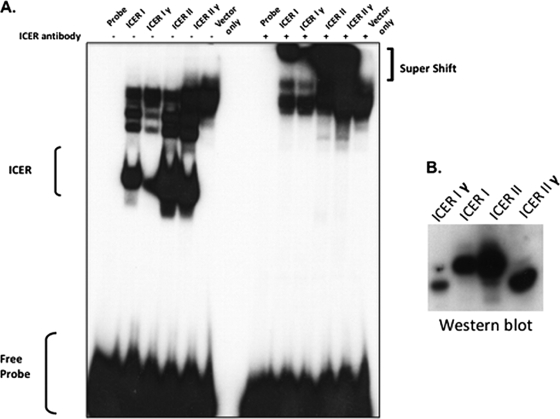
One possible mechanism by which ICER may attenuate GnRH-stimulated FSHβ transcription is by antagonizing the binding of CREB to the FSHβ CRE site. Since both CREB and ICER are members of the bZIP family of transcription factors, they share high homology at the protein level, particularly within their DNA-binding domains (DBD) and the adjacent leucine zipper motif, which is responsible for the dimerization of bZIP members (36). Therefore, the occupancy of a CRE site is dependent on the relative abundance of these dimers as well as their affinity to DNA. At high GnRH pulse frequencies, elevated ICER protein levels could antagonize CREB-induced transcription of the FSHβ gene by forming either ICER homodimers or CREB-ICER heterodimers that can bind to the FSHβ CRE site. Since ICER I/Iγ and ICER II/IIγ have different DBDs, it is conceivable that ICER isoforms may have various affinities to the FSHβ CRE site. This hypothesis is supported by previous findings that suggest DBD II has a consistently higher affinity for noncanonical CRE sites than DBD I (28). To determine which ICER isoforms have the ability to bind to the noncanonical FSHβ CRE site, ICER isoform expression vectors (ICER I, Iγ, II, and IIγ) were transfected into HEK293 cells, and nuclear extracts were prepared from the transfected cells and used in an EMSA with a radiolabeled probe encompassing the FSHβ CRE site. These studies demonstrated that all four ICER isoforms have the ability to bind to the FSHβ CRE site (Fig. 5A). The identities of these complexes were verified by further retardation of DNA-protein complexes after incubation with an ICER-specific antibody (Fig. 5A). In agreement with previous observations (28), ICER II isoforms appear to have a higher affinity for the noncanonical FSHβ CRE site than ICER I isoforms, as reflected by the stronger band intensities in the EMSA study. These differences in band intensities do not appear to be due to differences in protein expression levels between ICER I and ICER II, with the exception of ICER Iγ, which was poorly expressed in the transfected HEK293 cells (Fig. 5B). It has been shown previously that the rate of degradation of ICER isoforms can vary (16), which may explain the relatively poor ICER Iγ expression in this experiment.
FIG. 5.
ICER isoforms can bind to the FSHβ CRE site. ICER expression vectors (10 μg) (ICER I, ICER Iγ, ICER II, and ICER IIγ) were transfected into HEK293 cells and nuclear extracts were prepared and used in EMSA. (A) A protein-DNA complex (labeled ICER) was detected only in the presence of transfected ICER isoforms but not in control empty vector-transfected cells. Supershifted complexes were detected with incubation of an ICER-specific antibody. (B) Western blot indicating relative amounts of each respective ICER protein present in nuclear extract samples from transfected HEK293 cells.
ICER-induced suppression of GnRH-stimulated FSHβ transcription is dependent on an intact CRE site.
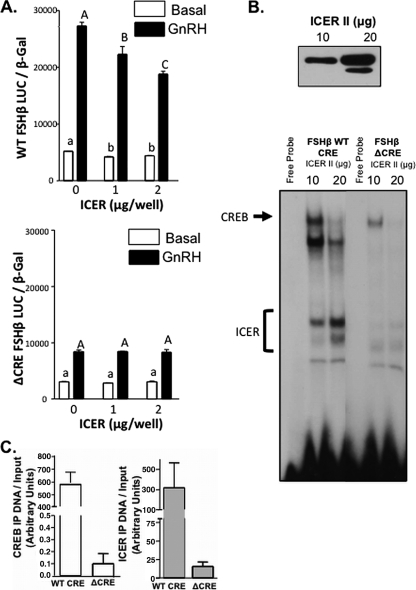
If the observed ICER suppression of GnRH stimulation of FSHβ transcription is mediated through the CRE site within the FSHβ promoter, then mutation of this CRE site would be expected to abolish the repressive effects of ICER. To test this hypothesis, LβT2 cells were transfected with increasing amounts of an expression vector encoding ICER II, together with an expression vector encoding CREB and a luciferase reporter linked to the rFSHβ(−140/+15) promoter containing the mutant CRE site as used in previous perifusion experiments (Fig. 1B), and then stimulated with 100 nM GnRHAg or vehicle in static culture. No repression of GnRH-stimulated luciferase activity following ICER II overexpression was observed (Fig. 6A), and this was associated with a marked reduction of ICER II binding to the mutated (Δ) FSHβ CRE as observed by EMSA (Fig. 6B) and ChIP (Fig. 6C). In addition, increasing ICER II expression resulted in the reduction of protein-DNA complexes formed on the WT CRE probe (Fig. 6B), complexes previously characterized to include CREB and USF (8). The reduction in these other protein-DNA complexes likely include CREB, since the binding of CREB to the ΔCRE FSHβ promoter was reduced compared to the WT FSHβ promoter as quantified by ChIP (Fig. 6C). Collectively, these results suggest that ICER mediates its inhibitory effects on FSHβ transcription through a mechanism that requires DNA binding to the intact FSHβ CRE site and may involve CREB displacement.
FIG. 6.
ICER-induced suppression of FSHβ transcription is dependent on an intact CRE site. (A) LβT2 cells were cotransfected with either a WT or mutant (Δ) CRE −140/+15 rFSHβLuc reporter and increasing amounts of an ICER II expression vector as indicated, together with 1 μg of a CREB expression vector. LβT2 cells were treated with either 100 nM GnRHAg or medium alone for 4 h. Bar graphs show luciferase activity (means ± the SEM from triplicate samples, normalized to β-galactosidase activity) from a representative experiment, repeated on three separate occasions with comparable results. Significant differences are denoted as different letters as tested by one-way ANOVA with a post hoc Tukey-Kramer multiple-comparison test. (B) Nuclear extracts from LβT2 cells transfected with increasing amounts of an ICER II expression construct (10 and 20 μg per plate) were initially subjected to Western blot to verify increasing ICER II expression (top panel), and nuclear extracts were subsequently used in EMSA studies (bottom panel) with either FSHβ WT CRE or ΔCRE probes. Protein-DNA complexes containing ICER or CREB are indicated. (C) LβT2 cells were transfected with either WT or mutant (Δ) CRE −140/+15 rFSHβLuc and an ICER II expression vector. After 48 h, and after 100 nM GnRHAg treatment for 1 h, the cells were harvested for a ChIP assay with immunoprecipitation with CREB, ICER, or IgG antibodies. Immunoprecipitated DNA was quantified by real-time qPCR with reference to a standard curve generated by serial dilutions of a known concentration of plasmid (as outlined in the ABI prism user manual) and normalized to respective input amounts. The results for CREB (left panel) and ICER (right panel) are represented in the bar graphs (means ± the SEM).
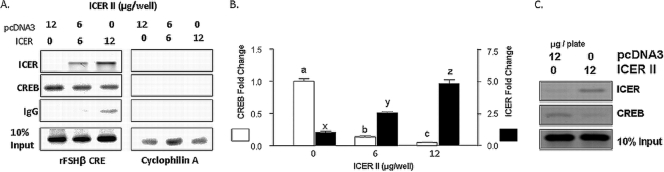
ICER reduces CREB binding to the FSHβ promoter in vivo.
The ability of ICER to bind to the noncanonical FSHβ CRE site (Fig. 5A, 6B, and 6C) suggests that ICER may antagonize CREB function by direct competition for the occupation of the FSHβ promoter (Fig. 6B). To test this hypothesis and determine whether ICER displaces CREB binding to the FSHβ CRE site in an in vivo context, ChIP was performed in LβT2 cells transfected with increasing amounts of an ICER II expression vector, together with −140/+15 rFSHβLuc. Oligonucleotides encompassing the CRE site were used to amplify DNA immunoprecipitated with ICER or CREB antibodies. Overexpression of ICER resulted in a reduction in CREB occupation of the FSHβ promoter in a dose dependent manner (Fig. 7A and B). Accompanying this finding, there was a corresponding increase in ICER occupation of the FSHβ promoter, as evidenced by immunoprecipitation with an ICER antibody (Fig. 7A and 7B). This suggests that the in vivo reduction in CREB binding to the FSHβ promoter is a direct consequence of increased ICER occupation. The observed binding of CREB and ICER to the FSHβ promoter was specific, as determined both by immunoprecipitation with nonspecific IgG antibody and by PCR amplification of immunoprecipitated DNA using mouse cyclophilin A oligonucleotides (Fig. 7A).
FIG. 7.
ICER reduces CREB binding to the FSHβ promoter through competition for the FSHβ CRE site. (A) LβT2 cells were cotransfected with −140/+15 rFSHβLuc and increasing amounts of an ICER II expression vector. ChIP analysis was performed by specific PCR amplification of rFSHβ or cyclophilin after immunoprecipitation with ICER, CREB, or IgG antibodies as indicated. Input samples (10-fold diluted) were subjected to PCR as positive controls. Representation of a radiolabeled PCR scanned in a phosphorimager depicting the amount of FSHβ CRE immunoprecipitation with the respective antibodies is shown. (B) Results of real-time qPCR of FSHβ DNA immunoprecipitated with CREB or ICER antibodies. The bar graph depicts the means ± the SEM of triplicate samples from a representative experiment, repeated on three separate occasions with comparable results. Significant differences (P < 0.05), as measured by one-way ANOVA with a post hoc Tukey-Kramer multiple-comparison test, are denoted by different letters. (C) LβT2 cells were transfected with either an HA-tagged ICER II or control empty expression vector and cultured for 48 h. After 100 nM GnRHAg treatment for 1 h, cells were harvested, and ChIP analysis was performed by PCR amplification of the endogenous murine FSHβ promoter on samples immunoprecipitated with either an HA antibody to specifically measure ICER binding or a CREB antibody to determine CREB binding. Input samples (10-fold diluted) were subjected to PCR as positive controls; PCR image shown is a representative radiolabeled PCR scanned in a phosphorimager.
CREB and ICER were also shown to bind to the endogenous mouse FSHβ promoter in LβT2 cells. LβT2 cells were transfected with an HA-tagged ICER II expression vector or empty control vector and oligonucleotides encompassing the partial CRE/AP1 site in the endogenous mouse FSHβ promoter were used to amplify DNA immunoprecipitated with HA or CREB antibodies from cell lysates. LβT2 cells transfected with empty vector showed that CREB can bind to the −223/+57 portion of the endogenous FSHβ promoter (Fig. 7C). As is the case with the rFSHβ CRE site, overexpression of ICER reduces CREB binding to the endogenous mouse FSHβ promoter (Fig. 7C), suggesting that ICER can also antagonize CREB binding to the murine FSHβ promoter. Indeed, immunoprecipitation of ICER using an HA antibody confirmed that ICER can also bind to the endogenous mouse FSHβ promoter (Fig. 7C).
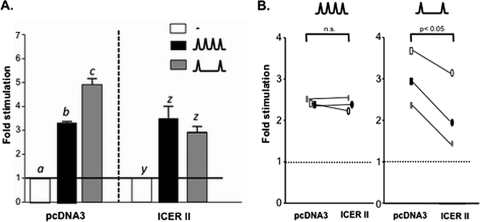
ICER abrogates GnRH pulse frequency-dependent effects on FSHβ transcription.
Since ICER has the ability to attenuate GnRH-stimulated FSHβ transcription in static LβT2 cultures, we hypothesized that overexpression of ICER could abrogate the preferential stimulation of FSHβ gene expression at low GnRH pulse frequencies. LβT2 cells were cotransfected with −140/+15 rFSHβLuc and an expression vector encoding ICER II and exposed to pulsatile GnRH at either high or low pulse frequencies or medium alone. As usual, FSHβ gene promoter activity was preferentially stimulated at the low, rather than high, GnRH pulse frequency (Fig. 8A, left panel). In contrast, overexpression of ICER resulted in a loss of the preferential stimulation of FSHβ transcription at the low GnRH pulse frequency, such that there were no longer GnRH pulse frequency-dependent differential effects on GnRH stimulation of FSHβ (Fig. 8A, right panel). Taking into account that endogenous ICER is preferentially induced at high GnRH pulse frequencies (Fig. 3), it is plausible that overexpression of ICER in LβT2 cells exposed to high GnRH pulse frequencies would show no further attenuation of FSHβ transcription. In contrast, ICER overexpression would be expected to attenuate FSHβ transcription in LβT2 cells treated with low GnRH pulse frequencies. To test this hypothesis, LβT2 cells were again cotransfected with a luciferase reporter linked to the rFSHβ(−140/+15) gene promoter and either an ICER II-expressing or empty pcDNA3 control vector and then exposed to high-frequency GnRH pulses or medium alone. Under this experimental paradigm, there was no inhibition of FSHβ luciferase activity (Fig. 8B, left panel), suggesting that endogenous ICER is produced to a sufficient level to cause maximal repression of GnRH-stimulated FSHβ transcription. In contrast, exposing the transfected cells to low-frequency GnRH pulses revealed a significant reduction in GnRH-stimulated FSHβ promoter activity in the cells overexpressing ICER (Fig. 8B, right panel). Thus, overexpression of ICER selectively attenuated stimulation of FSHβ promoter activity by GnRH at the low pulse frequency, with no effect at the high frequency, resulting in the loss of GnRH pulse frequency-dependent differential regulation of FSHβ expression.
FIG. 8.
ICER overexpression abrogates GnRH pulse frequency-dependent effects on FSHβ transcription. (A) LβT2 cells were transfected with −140/+15 rFSHβLuc (4 μg) and with 2 μg of an expression vector encoding ICER II or with empty vector (pcDNA3), followed by perifusion and stimulation with low (∧__∧; every 120 min) or high (∧∧∧; every 30 min) GnRH pulse frequencies. Bar graphs show the fold stimulation (means ± the SEM) relative to unstimulated levels. Significant differences (P < 0.05), as measured by one-way ANOVA with a post hoc Tukey-Kramer multiple-comparison test, are denoted by different letters. (B) LβT2 cells were transfected with the ICER II expression vector and exposed to either high or low GnRH pulse frequencies in two separate experiments, repeated on three separate occasions with each individual experiment done in triplicate. Paired t tests, performed on experimental data with connected data points denoting paired experiments, revealed a significant effect at low, but not high, GnRH pulse frequencies.
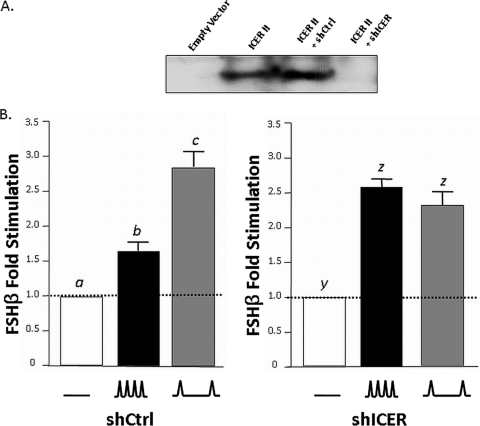
It would follow that if ICER overexpression disrupts the GnRH pulse frequency-dependent response of FSHβ transcription, then knockdown of endogenously expressed ICER would have comparable results. To test this hypothesis, LβT2 cells were transfected with −140/+15 rFSHβLuc and a shRNA construct containing either a shICER sequence or a scrambled, nonspecific shCtrl sequence. The shICER construct is directed to a sequence that is common to all ICER isoforms (Fig. 2A). The ability of the shICER construct to knock down ICER was verified by Western blot analysis (Fig. 9A). Transfected LβT2 cells were then perifused with either high or low GnRH pulse frequencies or medium alone, as outlined above. Complementing our ICER overexpression data, knockdown of endogenous ICER with a shICER construct causes a disruption to the GnRH pulse frequency-dependent response of FSHβ transcription (Fig. 9B, right panel). This effect was not seen in LβT2 cells transfected with a shCtrl construct, which retained the ability to respond preferentially to low GnRH pulse frequency (Fig. 9B, left panel). Collectively, these studies support our hypothesis that increased expression of ICER is specifically mediating the observed attenuation of FSHβ promoter activity in LβT2 cells exposed to high GnRH pulse frequencies.
FIG. 9.
ICER shRNA abrogates GnRH pulse frequency-dependent effects on FSHβ transcription. (A) Representative Western blot analysis using LβT2 cell lysates transfected with empty vector (pcDNA3) or an ICER II expression vector with either an shCtrl construct containing a nonspecific scrambled sequence or an shICER construct containing a sequence directed against ICER. (B) LβT2 cells were transfected with −140/+15 rFSHβLuc (4 μg) and with 2 μg of shCtrl (left panel) or shICER (right panel) constructs, followed by perifusion and stimulation with low (∧__∧; every 120 min) or high (∧∧∧; every 30 min) GnRH pulse frequencies. Bar graphs show the fold stimulation (means ± the SEM) relative to unstimulated levels. Significant differences (P < 0.05), as measured by one-way ANOVA with a post hoc Tukey-Kramer multiple comparison test, are denoted by different letters.
DISCUSSION
Ovarian dynamics during the ovulatory cycle are dependent on changes in systemic gonadotropin levels, making episodic release of these pituitary hormones, LH and FSH, fundamental to the maintenance of cyclic reproductive function (11). Befitting their important roles in endocrine physiology, the synthesis and secretion of LH and FSH are controlled by a complex interplay of both inhibitory and stimulatory endocrine components that include feedback from gonad-derived factors and stimulation by the hypothalamic decapeptide, GnRH (2, 11, 34). GnRH binds to specific high-affinity cell surface receptors (GnRHR) on pituitary gonadotropes to activate signal transduction cascades that ultimately modulate the biosynthesis of gonadotropin subunits (α, LHβ, and FSHβ) and secretion of biologically active hormones (LH and FSH) (24).
GnRH can cause differential effects on the rate of gonadotropin subunit gene transcription and on LH and FSH secretion. How this occurs likely rests on the ability of the gonadotrope to decipher different GnRH input patterns (15, 41); this is possible because GnRH secretion from hypothalamic neurons is not continuous. Rather, GnRH is released in a regulated pulsatile manner that varies in frequency and amplitude under normal physiological conditions such as at the onset of puberty and during the ovulatory cycle (34). Interestingly, these variations in GnRH pulse pattern are associated with divergent LH and FSH secretion, allowing for a mechanism by which a single hypothalamic neuropeptide can induce differential changes on two distinct hormones released from the same pituitary cell type. High GnRH pulse frequencies, a characteristic of the late follicular phase of the ovulatory cycle, leads to greater LH secretion; conversely, low GnRH frequencies, associated with the luteal phase, lead to increased FSH release (34). These frequency-dependent effects have been shown to occur at the transcriptional level (1, 15, 19, 26). Disruption of the correct pulsatile GnRH pattern and, hence, gonadotropin synthesis and secretion, manifests itself in a number of clinical disorders that cause infertility. In women, hypothalamic amenorrhea is predominantly associated with low GnRH pulse frequencies and abnormal serum gonadotropin levels (40); conversely, PCOS is associated with both high GnRH pulse frequencies and circulating LH levels but reduced circulating FSH levels (3, 12, 13).
With the use of the gonadotrope-derived LβT2 and αT3-1 cell lines, progress has been made in elucidating the molecular mechanisms underlying tissue-specific and static GnRH-stimulated expression of the gonadotropin genes (8-10, 22, 29-31, 44, 46, 48, 49). In contrast, relatively few studies have addressed the physiologically relevant effects of GnRH pulse frequency on gonadotropin transcription (1, 4, 5, 25-27, 29). Recent studies suggest that GnRH mediates stimulatory transcriptional effects on both LHβ and FSHβ genes through modifying the histone acetylation status of the gonadotropin gene promoters. It appears that GnRH-mediated repression of histone modifying enzymes, the histone deacetylases (HDACs), is a hallmark of gonadotrope development (30). In the immature αT3-1 gonadotrope cell line, HDACs occupy both FSHβ and LHβ promoters under basal conditions to cause transcriptional silencing (30). Other studies in the more mature LβT2 gonadotrope model have implicated the histone acetyltransferases (HATs), CBP and/or its paralogue P300, in mediating the GnRH stimulation of both gonadotropin β-subunit promoter genes (8, 37). Collectively, these observations suggest that histone modification through acetylation is an important aspect in GnRH control of gonadotropin β-subunit gene expression.
The synthesis of FSHβ is the rate-limiting step in FSH production (14, 35). Therefore, a greater perspective of how FSHβ transcription is regulated is key to understanding the control of FSH release and hence the development and maintenance of reproductive function. In the context of the rFSHβ promoter, GnRH stimulates transcription by increasing bound histone-modifying enzyme CBP (8). This increase in CBP is mediated in turn through the bZIP transcription factor CREB, bound to the FSHβ CRE site (8). In the present study, we have shown that mutation of this CRE site abolishes preferential FSHβ transcription at low GnRH pulse frequency, implicating this site as an important mediator of GnRH pulse frequency-dependent FSHβ gene expression (Fig. 1B). Our subsequent investigations have elucidated a potential mechanism by which patterns of GnRH pulsatility cause differential effects on FSHβ transcription by orchestrating changes in the occupancy of this CRE site by two bZIP family members that have contrasting functions, CREB and ICER.
Pulsatile hormone synthesis and secretion are characteristic features of various oscillatory systems. The present study further strengthens and expands the view that ICER is a major integrative player in the maintenance of such biological rhythms. For example, through LH-dependent signaling within the ovary, ICER has been implicated in controlling the oscillatory synthesis of the inhibin α-subunit gene during the ovulatory cycle by antagonizing stimulatory CREB function induced through FSH receptor-dependent signaling (6, 38). Furthermore, ICER is an essential component of molecular mechanisms involved in rhythmic production of pineal gland melatonin and, under hypothalamic control, participates in a transcriptional autoregulatory loop which ultimately mediates oscillations of serotonin N-acetyltransferase, the rate-limiting enzyme for melatonin synthesis (33, 39, 43). However, it remains unclear whether this hypothalamic stimulation of the pineal gland is pulsatile or tonic in nature. Here, we have extended our understanding of ICER biology and show that differential activation of a single receptor in a pulsatile manner can induce disparate ICER expression (Fig. 3) that ultimately reduces promoter bound CREB (Fig. 7). In this way, ICER acts as an essential and multifunctional regulator of oscillatory hormonal levels.
An intriguing question is what causes the preferential ICER stimulation at high GnRH pulse frequencies that ultimately reduces FSHβ transcription. The answer may rest in either the differential activation of signaling systems within the gonadotrope and/or synthesis, modification, and degradation of either transcription factors or regulatory proteins. It is well established that members of the mitogen activated protein kinase (MAPK) family of signaling proteins are stimulated by GnRH in vivo (50), in cultured rat pituitary fragments (22) and in LβT2 cells (9, 31) to contribute to regulation of gonadotropin subunit gene expression. In particular, activation of phosphorylated extracellular signal-regulated kinases (pERK) 1/2 is more rapid and more sustained in LβT2 cells perifused with low, rather than high, GnRH pulse frequencies, suggesting that pERK is important in the preferential increase of FSHβ transcription observed at low GnRH pulse frequency (27). It is conceivable that pERK may mediate stimulatory effects on FSHβ gene expression, at least in part, through an ICER-dependent mechanism. In support of this view, pERK1/2 have been found to physically interact and phosphorylate ICER proteins. The phosphorylation of ICER at position serine 41 marks the protein for ubiquitination and subsequent proteasomal degradation (47). This pERK-dependent degradation of ICER proteins may explain the reduction in ICER levels at low GnRH pulse frequencies. This model is in agreement with the emerging concept that differential effects mediated by changes in GnRH pulse frequency are associated with stability or degradation of important regulatory proteins and transcription factors (15, 29). In this way, the regulation of protein stability appears to be a generic mechanism used by oscillatory systems. For example, the production of cryptochrome proteins, essential components of the circadian clock, are regulated in part by ubiquitination and subsequent protein degradation (7).
It is conceivable that Ca2+-mediated signaling is preferentially elevated at high GnRH pulse frequencies, which may explain the observed increase in ICER mRNAs (Fig. 3A). In support of this view, high-frequency Ca2+ stimulation has an inhibitory effect on FSHβ mRNA (21). This suggests that GnRH, at least in part, signals through a calcium-dependent mechanism, which at high GnRH pulse frequencies might attenuate FSHβ transcription. Recently, in cultured primary rat neurons, Ca2+ signaling through calcium/calmodulin-dependent kinase II (CaMKII) has been found to be important in inducing immediate-early gene expression, including CREM/ICER (32). Furthermore, in primary rat pituitary cultures, CaMKII has been implicated as an essential mediator of GnRH-dependent gonadotropin gene expression (18, 20, 21). In this way, Ca2+ signaling, potentially through CaMKII, may explain elevated ICER gene expression at high GnRH pulse frequencies and suggests that ICER transcription and subsequent ICER protein stability are controlled by two independent mechanisms which have the potential to be functionally significant.
The data presented here provide evidence that ICER causes transcriptional repression through protein-DNA interactions. ICER isoforms are able to bind to the intact FSHβ CRE site (Fig. 5B and 7), which is essential for transcriptional repression of the gonadotropin subunit gene since ICER is unable to bind or cause transcriptional repression when this CRE site is mutated (Fig. 5A and B). In this way, it appears that ICER binding to the FSHβ promoter is the primary mechanism of FSHβ subunit transcriptional repression. Furthermore, our current data help to elucidate how a single neuropeptide, through different patterns of GnRH pulsatility, can have differential effects on FSHβ subunit transcription. An important aspect in the GnRH pulse frequency-mediated transcriptional control of the FSHβ subunit appears to be the stimulation of inducible members of the CREM gene, ICER isoforms, to a greater extent at high GnRH pulse frequencies (Fig. 3A and B), which may account for decreased FSHβ transcription by inhibiting recruitment of CBP to the GnRH responsive element within the FSHβ promoter. Insights into the mechanisms by which changes in GnRH pulse frequency cause differential pituitary FSHβ gene expression as provided by the present study will contribute to our understanding of abnormal gonadotropin secretion in disorders such as hypothalamic amenorrhea and PCOS and provide a context for the design of novel therapeutic approaches.
Acknowledgments
This research was supported in part by NIH grants R01 HD33001 (U.B.K.) and R01 HD19938 (U.B.K.) and by NICHD/NIH through cooperative agreement U54 HD28138 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (U.B.K.).
We thank Kelly Mayo, J. Larry Jameson, and Carlos Molina for generous gifts of antibodies and expression constructs and Yujiang Shi for stimulating discussion.
Footnotes
Published ahead of print on 14 December 2009.
REFERENCES
- 1.Bedecarrats, G. Y., and U. B. Kaiser. 2003. Differential regulation of gonadotropin subunit gene promoter activity by pulsatile gonadotropin-releasing hormone (GnRH) in perifused L beta T2 cells: role of GnRH receptor concentration. Endocrinology 144:1802-1811. [DOI] [PubMed] [Google Scholar]
- 2.Belchetz, P. E., T. M. Plant, Y. Nakai, E. J. Keogh, and E. Knobil. 1978. Hypophysial responses to continuous and intermittent delivery of hypothalamic gonadotropin-releasing hormone. Science 10:631-633. [DOI] [PubMed] [Google Scholar]
- 3.Blank, S. K., C. R. McCartney, K. D. Helm, and J. C. Marshall. 2007. Neuroendocrine effects of androgens in adult polycystic ovary syndrome and female puberty. Semin. Reprod. Med. 25:352-359. [DOI] [PubMed] [Google Scholar]
- 4.Burger, L. L., A. C. Dalkin, K. W. Aylor, D. J. Haisenleder, and J. C. Marshall. 2002. GnRH pulse frequency modulation of gonadotropin subunit gene transcription in normal gonadotropes: assessment by primary transcript assay provides evidence for roles of GnRH and follistatin. Endocrinology 143:3243-3249. [DOI] [PubMed] [Google Scholar]
- 5.Burger, L. L., D. J. Haisenleder, K. W. Aylor, and J. C. Marshall. 2008. Regulation of intracellular signaling cascades by GNRH pulse frequency in the rat pituitary: roles for CaMK II, ERK, and JNK activation. Biol. Reprod. 79:947-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkart, A. D., A. Mukherjee, and K. E. Mayo. 2006. Mechanism of repression of the inhibin alpha-subunit gene by inducible 3′,5′-cyclic adenosine monophosphate early repressor. Mol. Endocrinol. 20:584-597. [DOI] [PubMed] [Google Scholar]
- 7.Busino, L., F. Bassermann, A. Maiolica, C. Lee, P. M. Nolan, S. I. Godinho, G. F. Draetta, and M. Pagano. 2007. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science 316:900-904. [DOI] [PubMed] [Google Scholar]
- 8.Ciccone, N. A., C. T. Lacza, M. Y. Hou, S. J. Gregory, K. Y. Kam, S. Xu, and U. B. Kaiser. 2008. A composite element that binds basic helix loop helix and basic leucine zipper transcription factors is important for gonadotropin-releasing hormone regulation of the follicle-stimulating hormone beta gene. Mol. Endocrinol. 22:1908-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coss, D., C. M. Hand, K. K. Yaphockun, H. A. Ely, and P. L. Mellon. 2007. p38 mitogen-activated protein kinase is critical for synergistic induction of the FSH(beta) gene by gonadotropin-releasing hormone and activin through augmentation of c-Fos induction and Smad phosphorylation. Mol. Endocrinol. 21:3071-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coss, D., S. B. Jacobs, C. E. Bender, and P. L. Mellon. 2004. A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone beta gene by gonadotropin-releasing hormone. J. Biol. Chem. 279:152-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley, W. F., M. Filicori, D. I. Spratt, and N. F. Santoro. 1985. The physiology of gonadotropin-releasing hormone (GnRH) secretion in men and women. Recent Prog. Horm. Res. 41:473-531. [DOI] [PubMed] [Google Scholar]
- 12.Dunaif, A. 1997. Insulin resistance and the polycystic ovary syndrome: mechanism and implications for pathogenesis. Endocrinol. Rev. 18:774-800. [DOI] [PubMed] [Google Scholar]
- 13.Ehrmann, D. A. 2005. Polycystic ovary syndrome. N. Engl. J. Med. 352:1223-1236. [DOI] [PubMed] [Google Scholar]
- 14.Farnworth, P. G. 1995. Gonadotrophin secretion revisited. How many ways can FSH leave a gonadotroph? J. Endocrinol. 145:387-395. [DOI] [PubMed] [Google Scholar]
- 15.Ferris, H. A., and M. A. Shupnik. 2006. Mechanisms for pulsatile regulation of the gonadotropin subunit genes by GNRH1. Biol. Reprod. 74:993-998. [DOI] [PubMed] [Google Scholar]
- 16.Folco, E. J., and G. Koren. 1997. Degradation of the inducible cAMP early repressor (ICER) by the ubiquitin-proteasome pathway. Biochem. J. 328:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gharib, S. D., M. E. Wierman, M. A. Shupnik, and W. W. Chin. 1990. Molecular biology of the pituitary gonadotropins. Endocrinol. Rev. 11:177-199. [DOI] [PubMed] [Google Scholar]
- 18.Haisenleder, D. J., L. L. Burger, K. W. Aylor, A. C. Dalkin, and J. C. Marshall. 2003. Gonadotropin-releasing hormone stimulation of gonadotropin subunit transcription: evidence for the involvement of calcium/calmodulin-dependent kinase II (Ca/CAMK II) activation in rat pituitaries. Endocrinology 144:2768-2774. [DOI] [PubMed] [Google Scholar]
- 19.Haisenleder, D. J., A. C. Dalkin, G. A. Ortolano, J. C. Marshall, and M. A. Shupnik. 1991. A pulsatile gonadotropin-releasing hormone stimulus is required to increase transcription of the gonadotropin subunit genes: evidence for differential regulation of transcription by pulse frequency in vivo. Endocrinology 128:509-517. [DOI] [PubMed] [Google Scholar]
- 20.Haisenleder, D. J., H. A. Ferris, and M. A. Shupnik. 2003. The calcium component of gonadotropin-releasing hormone-stimulated luteinizing hormone subunit gene transcription is mediated by calcium/calmodulin-dependent protein kinase type II. Endocrinology 144:2409-2416. [DOI] [PubMed] [Google Scholar]
- 21.Haisenleder, D. J., L. J. Workman, L. L. Burger, K. W. Aylor, A. C. Dalkin, and J. C. Marshall. 2001. Gonadotropin subunit transcriptional responses to calcium signals in the rat: evidence for regulation by pulse frequency. Biol. Reprod. 65:1789-1793. [DOI] [PubMed] [Google Scholar]
- 22.Harris, D., D. Chuderland, D. Bonfil, S. Kraus, R. Seger, and Z. Naor. 2003. Extracellular signal-regulated kinase and c-Src, but not Jun N-terminal kinase, are involved in basal and gonadotropin-releasing hormone-stimulated activity of the glycoprotein hormone alpha-subunit promoter. Endocrinology 144:612-622. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman, L. K., and D. A. Ehrmann. 2008. Cardiometabolic features of polycystic ovary syndrome. Nat. Clin. Pract. Endocrinol. Metab. 4:215-222. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser, U. B., P. M. Conn, and W. W. Chin. 1997. Studies of gonadotropin-releasing hormone (GnRH) action using GnRH receptor-expressing pituitary cell lines. Endocrinol. Rev. 18:46-70. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser, U. B., L. M. Halvorson, and M. T. Chen. 2000. Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (egr-1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-beta gene promoter: an integral role for SF-1. Mol. Endocrinol. 14:1235-1245. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser, U. B., A. Jakubowiak, A. Steinberger, and W. W. Chin. 1997. Differential effects of gonadotropin-releasing hormone (GnRH) pulse frequency on gonadotropin subunit and GnRH receptor messenger ribonucleic acid levels in vitro. Endocrinology 138:1224-1231. [DOI] [PubMed] [Google Scholar]
- 27.Kanasaki, H., G. Y. Bedecarrats, K. Y. Kam, S. Xu, and U. B. Kaiser. 2005. Gonadotropin-releasing hormone pulse frequency-dependent activation of extracellular signal-regulated kinase pathways in perifused LβT2 cells. Endocrinology 146:5503-5513. [DOI] [PubMed] [Google Scholar]
- 28.Laoide, B. M., N. S. Foulkes, F. Schlotter, and P. Sassone-Corsi. 1993. The functional versatility of CREM is determined by its modular structure. EMBO. J. 12:1179-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawson, M. A., R. Tsutsumi, H. Zhang, I. Talukdar, B. K. Butler, S. J. Santos, P. L. Mellon, and N. J. Webster. 2007. Pulse sensitivity of the luteinizing hormone beta promoter is determined by a negative feedback loop Involving early growth response-1 and Ngfi-A binding protein 1 and 2. Mol. Endocrinol. 21:1175-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim, S., M. Luo, M. Koh, M. Yang, M. N. Bin Abdul Kadir, J. H. Tan, Z. Ye, W. Wang, and P. Melamed. 2007. Distinct mechanisms involving diverse histone deacetylases repress expression of the two gonadotropin beta-subunit genes in immature gonadotropes, and their actions are overcome by gonadotropin-releasing hormone. Mol. Cell. Biol. 27:4105-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, F., D. A. Austin, P. L. Mellon, J. M. Olefsky, and N. J. Webster. 2002. GnRH activates ERK1/2 leading to the induction of c-fos and LHβ protein expression in LbetaT2 cells. Mol. Endocrinol. 16:419-434. [DOI] [PubMed] [Google Scholar]
- 32.Machado, H. B., L. J. Vician, and H. R. Herschman. 2008. The MAPK pathway is required for depolarization-induced “promiscuous” immediate-early gene expression but not for depolarization-restricted immediate-early gene expression in neurons. J. Neurosci. Res. 86:593-602. [DOI] [PubMed] [Google Scholar]
- 33.Maronde, E., M. Pfeffer, J. Olcese, C. A. Molina, F. Schlotter, F. Dehghani, H. W. Korf, and J. H. Stehle. 1999. Transcription factors in neuroendocrine regulation: rhythmic changes in pCREB and ICER levels frame melatonin synthesis. J. Neurosci. 19:3326-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marshall, J. C., A. C. Dalkin, D. J. Haisenleder, S. J. Paul, G. A. Ortolano, and R. P. Kelch. 1991. Gonadotropin-releasing hormone pulses: regulators of gonadotropin synthesis and ovulatory cycles. Recent Prog. Horm. Res. 47:155-187. [DOI] [PubMed] [Google Scholar]
- 35.McNeilly, J. R., P. Brown, A. J. Clark, and A. S. McNeilly. 1991. Gonadotropin-releasing hormone modulation of gonadotrophins in the ewe: evidence for differential effects on gene expression and hormone secretion. J. Mol. Endocrinol. 7:35-43. [DOI] [PubMed] [Google Scholar]
- 36.Mioduszewska, B., J. Jaworski, and L. Kaczmarek. 2003. Inducible cAMP early repressor (ICER) in the nervous system: a transcriptional regulator of neuronal plasticity and programmed cell death. J. Neurochem. 87:1313-1320. [DOI] [PubMed] [Google Scholar]
- 37.Mouillet, J. F., C. Sonnenberg-Hirche, X. Yan, and Y. Sadovsky. 2004. p300 regulates the synergy of steroidogenic factor-1 and early growth response-1 in activating luteinizing hormone-beta subunit gene. J. Biol. Chem. 279:7832-7839. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee, A., J. Urban, P. Sassone-Corsi, and K. E. Mayo. 1998. Gonadotropins regulate inducible cyclic adenosine 3′,5′-monophosphate early repressor in the rat ovary: implications for inhibin alpha subunit gene expression. Mol. Endocrinol. 12:785-800. [DOI] [PubMed] [Google Scholar]
- 39.Pfeffer, M., E. Maronde, C. A. Molina, H. W. Korf, and J. H. Stehle. 1999. Inducible cyclic AMP early repressor protein in rat pinealocytes: a highly sensitive natural reporter for regulated gene transcription. Mol. Pharmacol. 56:279-289. [DOI] [PubMed] [Google Scholar]
- 40.Reame, N. E., S. E. Sauder, G. D. Case, R. P. Kelch, and J. C. Marshall. 1985. Pulsatile gonadotropin secretion in women with hypothalamic amenorrhea: evidence that reduced frequency of gonadotropin-releasing hormone secretion is the mechanism of persistent anovulation. J. Clin. Endocrinol. Metab. 61:851-858. [DOI] [PubMed] [Google Scholar]
- 41.Ruf, F., M. J. Park, F. Hayot, G. Lin, B. Roysam, Y. Ge, and S. C. Sealfon. 2006. Mixed analog/digital gonadotrope biosynthetic response to gonadotropin-releasing hormone. J. Biol. Chem. 281:30967-30978. [DOI] [PubMed] [Google Scholar]
- 42.Seminara, S. B., F. J. Hayes, and W. F. Crowley, Jr. 1998. Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann's syndrome): pathophysiological and genetic considerations. Endocrinol. Rev. 19:521-539. [DOI] [PubMed] [Google Scholar]
- 43.Stehle, J. H., N. S. Foulkes, C. A. Molina, V. Simonneaux, P. Pevet, and P. Sassone-Corsi. 1993. Adrenergic signals direct rhythmic expression of transcriptional repressor CREM in the pineal gland. Nature 365:314-320. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Y., J. Fortin, P. Lamba, M. Bonomi, L. Persani, M. S. Roberson, and D. J. Bernard. 2008. Activator protein-1 and smad proteins synergistically regulate human follicle-stimulating hormone beta-promoter activity. Endocrinology 149:5577-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss, J., J. L. Jameson, J. M. Burrin, and W. F. Crowley, Jr. 1990. Divergent responses of gonadotropin subunit messenger RNAs to continuous versus pulsatile gonadotropin-releasing hormone in vitro. Mol. Endocrinol. 4:557-564. [DOI] [PubMed] [Google Scholar]
- 46.Xie, J., S. P. Bliss, T. M. Nett, B. J. Ebersole, S. C. Sealfon, and M. S. Roberson. 2005. Transcript profiling of immediate-early genes reveals a unique role for activating transcription factor 3 in mediating activation of the glycoprotein hormone alpha-subunit promoter by gonadotropin-releasing hormone. Mol. Endocrinol. 19:2624-2638. [DOI] [PubMed] [Google Scholar]
- 47.Yehia, G., F. Schlotter, R. Razavi, A. Alessandrini, and C. A. Molina. 2001. Mitogen-activated protein kinase phosphorylates and targets inducible cAMP early repressor to ubiquitin-mediated destruction. J. Biol. Chem. 276:35272-35279. [DOI] [PubMed] [Google Scholar]
- 48.Zakaria, M. M., K. H. Jeong, C. Lacza, and U. B. Kaiser. 2002. Pituitary homeobox 1 activates the rat FSHβ (rFSHβ) gene through both direct and indirect interactions with the rFSHβ gene promoter. Mol. Endocrinol. 16:1840-1852. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, H., J. S. Bailey, D. Coss, B. Lin, R. Tsutsumi, M. A. Lawson, P. L. Mellon, and N. J. Webster. 2006. Activin modulates the transcriptional response of LbetaT2 cells to gonadotropin-releasing hormone and alters cellular proliferation. Mol. Endocrinol. 20:2909-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, T., and M. S. Roberson. 2006. Role of MAP kinase phosphatases in GnRH-dependent activation of MAP kinases. J. Mol. Endocrinol. 36:41-50. [DOI] [PubMed] [Google Scholar]