Abstract
Terminally differentiated cell types are needed to live and function in a postmitotic state for a lifetime. Cellular senescence is another type of permanent arrest that blocks the proliferation of cells in response to genotoxic stress. Here we show that the retinoblastoma protein (pRB) uses a mechanism to block DNA replication in senescence that is distinct from its role in permanent cell cycle exit associated with terminal differentiation. Our work demonstrates that a subtle mutation in pRB that cripples its ability to interact with chromatin regulators impairs heterochromatinization and repression of E2F-responsive promoters during senescence. In contrast, terminally differentiated nerve and muscle cells bearing the same mutation fully exit the cell cycle and block E2F-responsive gene expression by a different mechanism. Remarkably, this reveals that pRB recruits chromatin regulators primarily to engage a stress-responsive G1 arrest program.
Terminal differentiation is fundamental to the development of a multicellular organism (9). Of particular importance is the commitment to permanently exit the cell cycle. Many cells enter a postmitotic state early in life and must remain viable and nonproliferative throughout the life span of the organism. Cellular senescence is another form of proliferative control that can be induced as a natural consequence of aging or prematurely in response to stimuli such as DNA damage (12). The physiological differences between terminal differentiation and senescence suggest that there may be differences in their mechanisms of growth control; however, the robust control of cell cycle entry is an obvious similarity. Comparisons between the two are rare in the current literature.
Coupling cell cycle exit with terminal differentiation requires the coordinated activities of the retinoblastoma (RB) family of proteins and cyclin-dependent kinase (CDK) inhibitors (9). Studies with organisms such as Drosophila and Caenorhabditis elegans support a general model in which cell cycle exit requires simultaneous regulation of E2F transcription by RB family proteins and cyclin/CDK activity by their inhibitors (4, 10, 22). While it is unclear how this regulation is coordinated, it has been speculated that changes in chromatin structure could offer an explanation. Under this interpretation, heterochromatinization of cell cycle promoters blocks cyclin/CDKs from activating transcription through E2Fs; likewise, promiscuous E2F activity is unable to induce expression of cyclins. For these reasons, much attention has been focused on chromatin regulation in transcriptional control by pRB, and this function has been reviewed extensively (8, 14, 29). Cell cycle exit during terminal differentiation of neurons and skeletal muscle requires pRB function (13, 18, 21, 26, 35, 57). In addition, deposition of heterochromatin at E2F-responsive cell cycle promoters is reported to be pRB dependent during the differentiation of these same cell types (3, 45). Unfortunately, efforts to uncouple cell cycle exit from differentiation through loss of pRB have been complicated because this often leads to cell death, particularly in muscle development (11, 26, 56). This raises the question of whether chromatin regulation by pRB is the cause of cell cycle exit or a consequence of differentiation.
Cell cycle exit in senescence also involves the coordinated action of CDK inhibitors and RB family proteins (46, 51). However, the frequent participation of p53 in the induction of senescence distinguishes it from cell cycle exit in differentiation (12, 16). In this cell cycle arrest paradigm, pRB has a central role in the generation of senescence-associated heterochromatic foci (SAHF) (42). SAHF are single chromosomes compacted into microscopically visible heterochromatin bodies (24, 58). This compressed genomic structure ensures efficient silencing of E2F-regulated cell cycle genes. Thus, pRB function is critical to establishing one of the features of senescence that best define its permanence. However, not all senescent human fibroblasts form SAHF (24). Fibroblasts from knockout mice have been used extensively to genetically dissect the pathway that induces senescence, and this analysis has demonstrated that it requires RB family proteins (17, 46, 50). Interestingly, the presence of SAHF in senescent mouse cells remains in question because pericentromeric heterochromatin bodies are present under all growth conditions. Because not all senescent cells contain SAHF, it is unclear whether pRB regulates chromatin structure in senescence in their absence.
Despite these gaps in our knowledge, regulation of chromatin structure by pRB is frequently linked with its function in cell cycle control (8, 14). Many reports have shown that chromatin-regulating enzymes such as Brg1 (20), Brm (53), HDAC1 (36), DNMT1 (48), and Suv39h1 (43), among others, use a peptide motif called LXCXE to interact with the pocket domain of pRB (14, 40). Through the simultaneous interaction with E2F transcription factors, this complex is recruited to E2F target genes to block transcription and arrest the cell cycle in G1 (8). In this way, a one-size-fits-all model of pRB has emerged in which this E2F-pRB-chromatin-regulating repressor module is activated under all G1 cell cycle arrest circumstances to remodel chromatin and block proliferation. However, it is noteworthy that studies investigating the myriad of chromatin regulators that interact with pRB have largely been carried out using cell culture assays and this has prevented us from truly understanding the biological significance of chromatin remodeling by pRB. It is unclear if induction of senescence or terminal differentiation invokes the same pRB functions, even though they both can lead to a permanent G1 arrest that is frequently characterized by changes in chromatin structure.
To investigate how the recruitment of chromatin-regulating activities by pRB influences mammalian development and disease, we have generated a gene-targeted mouse strain in which mutations in pRB disrupt only LXCXE-dependent interactions (28). We have validated that this mutation (called Rb1ΔL) disrupts numerous interactions between chromatin regulators and pRB but leaves interactions with E2Fs intact (28). Importantly, the Rb1ΔL allele expresses pRB at levels equivalent to those of the wild type and the expression of the related RB family proteins p107 and p130 is unchanged (28). This suggests that defects in Rb1ΔL/ΔL are not suppressed by overexpression of other family members, as is the case for Rb1−/− mutant mice (27, 41). Despite the interactions that are disrupted, Rb1ΔL/ΔL knock-in mice are viable (28), raising the question of what physiological circumstances require pRB to use chromatin regulation in cell cycle control.
In this study, we compared the cell cycle exit properties of skeletal muscle and retinal neurons, two long-lived cell types, with those of senescent cells derived from Rb1ΔL/ΔL mutant mice. Our work shows that there is defective inhibition of DNA replication in senescent Rb1ΔL/ΔL mutant cells, but not in permanent cell cycle exit during development. This indicates that one of the primary functions of chromatin regulation by pRB is an arrest checkpoint that is used during senescence. The defect in senescence is a failure to create a repressive chromatin structure at E2F-responsive genes and is characterized by a deficiency in H3K9me3. Conversely, chromatin immunoprecipitation (ChIP) analysis of the same promoters in Rb1ΔL/ΔL mutant muscle reveals a different transcriptional silencing pathway characterized by a combination of H3K27me3 and H3K9me3 modifications that are present in normal abundance in Rb1ΔL/ΔL mutants. Unexpectedly, this reveals that pRB possesses a stress-responsive growth control mechanism that is distinct from cell cycle exit in terminal differentiation during development.
MATERIALS AND METHODS
Mice.
The generation of Rb1ΔL/ΔL mutant mice has been described previously (28). Rb1−/− mice were obtained from MMHCC. Mice bearing Rb1f/f alleles and the α-crystallin-Cre transgene were generated as described before (13). All animals were housed and handled according to Canadian Council on Animal Care regulations.
Cell culture.
Mouse embryonic fibroblasts (MEFs) were generated from day 13.5 embryos using standard procedures and cultured as previously described (28). Retroviral transduction with pBABE-H-RasV12 was as reported by Serrano et al. (51), and viruses were packaged in Bosc-23 cells. Cells infected with viruses encoding Ras were selected in 4 μg/ml puromycin for at least 3 days before processing for further experiments using flow cytometry, microscopy, or extract preparation. Senescent cells prepared by this method were allowed to senescence for at least 10 days following retroviral infection. Cells induced to senesce with gamma irradiation were exposed to 15 Gys. Senescence-associated β-galactosidase (SA β-Gal) staining was performed as described previously (51). Infections with adenovirus (Ad)-E2F1 were done according to standard methods, and cells were cultured for an additional 48 h before labeling with bromodeoxyuridine (BrdU) for 16 h or preparing extracts. Myogenic differentiation was carried out by infecting MEFs with a pBABE-MyoD-based retrovirus as described above and following the differentiation protocol of Novitch et al. (44). Following selection, cells were differentiated and restimulated with 15% serum and labeled with BrdU for 24 h (6 days total). 3T3 culture assays were carried out following previously reported methods (55), as modified by Classon et al. (15).
Histology and fluorescence microscopy.
Hematoxylin-and-eosin (H&E)- and Ki67-stained tissues were fixed in formalin, embedded, and stained using standard procedures. All other tissues were fixed in optimum cutting temperature compound and embedded for cryosectioning. Staining of retinal sections was carried out as described by Chen et al. (13), and anti-BrdU staining was done as recommended by the manufacturer (BD, San Jose, CA). Cell cultures were fixed and permeabilized in alcohol, blocked, and stained for BrdU or protein markers as previously described (28). Antibodies against the major histocompatibility complex (MHC) were obtained from the Developmental Studies Hybridoma Bank, University of Iowa.
Quantitation of DNA, protein, and mRNA.
DNA content and BrdU incorporation were measured by flow cytometry (see Fig. 1A and C) as described by Isaac et al. (28). All other measurements of BrdU incorporation were generated from in situ staining and microscopic evaluation as described above. Flow cytometry measurements of hepatocyte nuclear DNA content were done as described by Mayhew et al. (38). Protein expression levels were detected by Western blotting using antibodies against E2F1 (KH95), p107 (C-18), PCNA (pc10), p130 (C-20), and MCM7 (141.2) from Santa Cruz. Actin (A2066; Sigma) or lamin A/C (MAB3211; Chemicon) levels were detected as loading controls. Message levels for Pcna, Ccne1, Ccna2, Tyms, and Rbl1 were detected using the Quantigene Plex 2.0 reagent system from Panomics (Fremont, CA) and quantified by comparison with the message for acidic ribosomal phosphoprotein P0 (Rplp0) using a BioPlex200 multiplex analysis system according to the Panomics instructions. ChIP assays were performed as described previously, using anti-H3K9me3 and -H3K27me3 antibodies (Upstate) and 2 × 107 cells per immunoprecipitation (2). DNA released from precipitated complexes was amplified by PCR using primers specific to the promoter regions of Airn (AGG GTG AAA AGC TGC ACA AG and CCC TGA TCA CAG AAC CCT TC) (47), Pcna (CTG CGC GAG GTC ATG ACG CCA and CTT CCG TGG CGC GGA AAC TTC C), Ccne1 (TGA GGG GCT CGC AGC CCT CG and CCC GGC TTC GAG CGG GAC AT), Mcm3 (GAA TGC AGT GCT TCC TAG CC and CGG AAG TTT ATG GTG GAG GA) (3), Mcm5 (AAC CAA TAG GAG CGC AGA GA and AAG CCC GAC ATG ACT GTA CC) (3). Hoxd10 (GCT GAA AAC CTC CCC ATC TT and CCT ACT TGG CGC ATT TTC TC), and Ccna2 (ATC CAC TGA GCA GCA GAG AT and TTG TAG TTC AAG TAG CCC GCG).
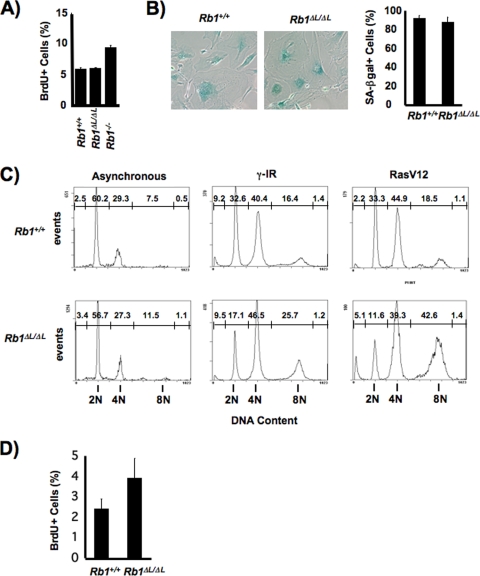
FIG. 1.
Defective arrest of DNA synthesis in Rb1 mutant cells during senescence. (A) MEFs of the indicated genotypes were serum deprived for 72 h, and their resulting DNA synthesis was measured by BrdU incorporation. (B) Ten days following retroviral transduction of MEFs with oncogenic Ras, senescent cell morphology and SA β-Gal activity were examined by light microscopy. The percentage of SA β-Gal-positive cells of each genotype was determined and is displayed in the graph to the right. (C) DNA content of wild-type and mutant MEFs was examined by propidium iodide staining and flow cytometry 5 days following irradiation or viral infection to express RasV12. The values above the peaks are the percentages of cells with this DNA content. (D) DNA synthesis in senescent MEFs was measured by BrdU incorporation over 8 h at 10 days following viral infection. Error bars in all graphs indicate 1 standard deviation from the mean of at least three replicates. The P value for a t test comparing mean measurements in panel D is 0.04.
RESULTS
The Rb1ΔL mutation causes defects in a senescent cell cycle arrest.
Based on pRB's well-known role in controlling G1-to-S-phase progression, we surveyed the ability of fibroblast cells from Rb1ΔL/ΔL mutant mice to respond to DNA damage agents, activated oncogenes, and other stimuli that are known to impinge upon proliferative control by pRB. Consistent with the discrete nature of the knock-in mutation, some growth arrest mechanisms worked normally, including serum deprivation over periods ranging from 3 to 5 days (Fig. 1A and data not shown). However, a number of senescence-inducing stimuli, like gamma irradiation and oncogenic Ras, were unable to generate a complete cell cycle exit in Rb1ΔL/ΔL mutant fibroblast cells, despite the fact that the cells ceased to divide and assumed a senescent morphology characterized by SA β-Gal staining (Fig. 1B). Rb1ΔL/ΔL mutant MEFs showed a normal response to DNA damage during the first 48 h following gamma irradiation and largely ceased to incorporate BrdU (data not shown). However, even at this early time point, reduced accumulation of cells in G1 became apparent (data not shown). A similar analysis of DNA content 5 days posttreatment revealed a striking failure of mutant cells to collect in G1, with many cells exhibiting abnormally high DNA content at 8N and beyond, indicative of endoreduplication (Fig. 1C). This occurred regardless of whether the arrest was induced by gamma irradiation or activated Ras. Furthermore, 10 days following the induction of senescence by RasV12, mutant MEFs still had elevated levels of BrdU incorporation relative to wild-type controls (Fig. 1D). This suggests that persistent but low levels of DNA synthesis lead to the elevated DNA content found in Rb1ΔL/ΔL mutant MEFs following the induction of senescence. We interpret this phenotype to mean that Rb1ΔL/ΔL mutant cells are capable of entering a senescent state based on morphology, the presence of SA β-Gal staining, and the inability to undergo mitosis. However, the mutation in pRB prevents an irreversible withdrawal from the cell cycle that allows endoreduplication. For these reasons, we will refer to the state of these cells as defective, or incomplete, senescence throughout this report.
Permanent cell cycle exit during development is normal in Rb1ΔL/ΔL mutant mice.
Because cellular senescence is thought to be an irreversible arrest, we decided to examine cell cycle exit and differentiation in long-lived cell types that remain growth arrested throughout life. A number of tissues possessing permanently arrested cells that fit this description are also known to require pRB for cell cycle exit during terminal differentiation. In particular, pRB has a well-recognized role in cell cycle control of muscle (18, 56), as well as the retina (13, 35, 57), and we have examined the effects of the Rb1ΔL mutation in these contexts.
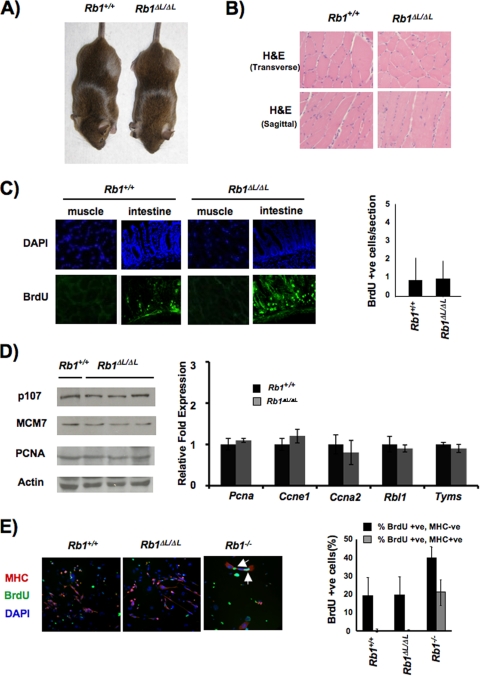
When the placental defects of Rb1−/− mutant mice are complemented, knockout animals die at birth with defects in myogenesis that are characterized by gross histological abnormalities and numerous apoptotic cells (18). The fact that Rb1ΔL/ΔL mutant mice are viable and appear normal suggests that pRB's role in muscle differentiation is complemented by the Rb1ΔL allele (Fig. 2A). Indeed, histological analysis of skeletal muscle stained with H&E from Rb1ΔL/ΔL mutant mice reveals that they are indistinguishable from wild-type controls (Fig. 2B). Beyond the ability of the Rb1ΔL mutant to function in the differentiation of muscle, we also investigated the permanence of cell cycle exit in this tissue. Anti-BrdU staining demonstrates infrequent proliferation in cross-sections of wild-type and Rb1ΔL/ΔL mutant muscle fibers, less than one per microscopic field of view (Fig. 2C). The quantity of rare, positively stained nuclei is consistent with proliferation of myosatellite cells that repair postmitotic muscle fibers. From this analysis, ectopic DNA replication in myotubes appears to be absent. As a control for our ability to sensitively detect DNA replication, we also stained highly proliferative cells from intestinal crypts in the same mice to confirm that our labeling and staining robustly detect DNA replication (Fig. 2C). This analysis of cell proliferation in the muscle of Rb1ΔL/ΔL mutant mice indicates that cells exit the cell cycle and remain postmitotic in a manner comparable to that of the wild type. In order to test if the transcriptional silencing function of pRB is intact in differentiated muscle of Rb1ΔL/ΔL mutant mice, we studied the expression of E2F target genes like Pcna, Ccne1 (cyclin E), Rbl1 (p107), Ccna2 (cyclin A), and Tyms (thymidylate synthase). We found equal expression of these genes between wild-type and mutant muscles. Western blots also showed similar levels of protein expression among E2F targets across the two genotypes, further suggesting that control of gene expression is properly maintained in Rb1ΔL/ΔL mutant muscle. Importantly, this also reveals that expression of the related pRB family protein p107 remains normal under these conditions. This indicates that myogenesis in Rb1ΔL/ΔL mutants is likely not the result of compensation by other pRB family members.
FIG. 2.
Normal cell cycle exit and differentiation of muscle. (A) Rb1ΔL/ΔL mutant animals are viable and appear indistinguishable from wild-type littermates. (B) Anterior tibialis muscle tissue from 8- to 10-week-old animals was stained with H&E to examine the gross morphology of the wild type and Rb1ΔL/ΔL mutants. Transverse and sagittal sections are shown. (C) Cell proliferation in muscle was examined by BrdU staining, the number of BrdU-positive cells per microscopic field was quantified, and the average is displayed in the graph. As a control for detection of BrdU in mature muscle fibers, we also stained cryosections of intestinal epithelia prepared from the same mice. DAPI, 4′,6-diamidino-2-phenylindole. (D) mRNA and protein were extracted from the muscle of 6-week-old wild-type and Rb1ΔL/ΔL mutant mice. Western blot assays show the expression of known E2F target genes, and the graph to the right displays the relative abundance of the specified transcripts. Message levels of acidic ribosomal phosphoprotein P0 (Rplp0) and actin protein levels were used as controls. (E) The indicated genotypes of MEFs were infected with MyoD-expressing retroviruses and induced to differentiate into myocytes under low-serum conditions. Cells were then restimulated with 15% fetal bovine serum and pulse-labeled with BrdU (24 h) to detect DNA synthesis. Myocytes were identified by MHC staining (red), DNA synthesis was detected by BrdU staining (green), and DNA was counterstained with DAPI (blue). The percentage of myocytes (MHC positive) and surrounding fibroblasts (MHC negative) that incorporated BrdU in response to serum is shown in the graph to the right. The error bars in all of the graphs indicate 1 standard deviation from the mean of at least three replicates.
To complement the in situ analysis of muscle proliferation described above, we also analyzed the permanence of cell cycle exit in a cell culture-based assay of muscle differentiation. This allows us to directly compare the Rb1ΔL/ΔL, Rb1−/−, and Rb1+/+ genotypes, since Rb1−/− mutant myoblasts do not form muscle fibers (26). We infected MEFs with a MyoD-expressing retrovirus to induce the formation of myocytes and stimulated differentiation in low serum as described previously (44). Prior reports have revealed that Rb1−/− mutant myocytes generated by this methodology are susceptible to cell cycle reentry upon serum stimulation (44). The Rb1ΔL allele readily supports a growth factor-resistant cell cycle exit that is indistinguishable from that of the wild-type control (Fig. 2E). These data suggest that the Rb1ΔL mutant is capable of supporting a permanent cell cycle exit in terminal differentiation that is just as robust as that of the wild type. This result stands in stark contrast to the incomplete senescent arrest described in Fig. 1 because these cell cycle arrest assays start with the same fibroblast cells.
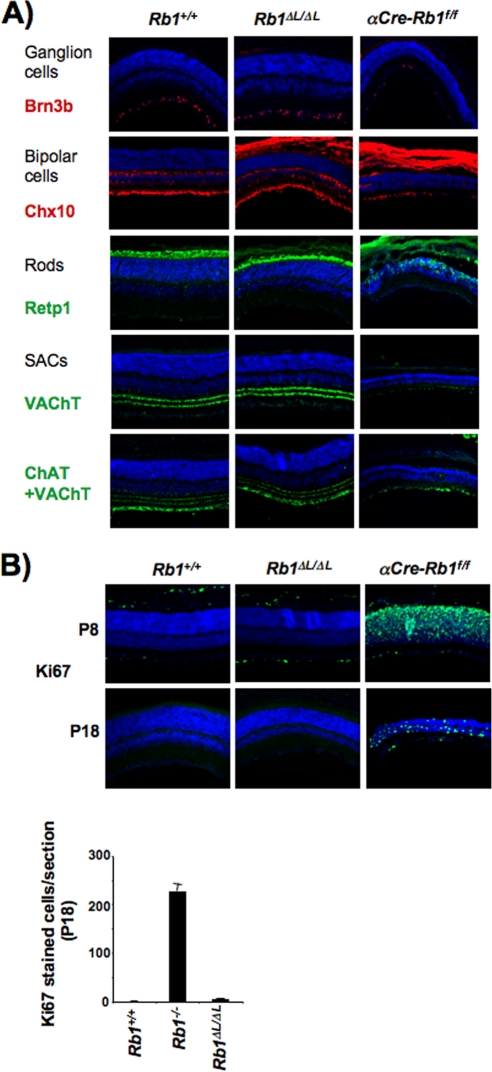
Similarly, it is known that conditional deletion of Rb1 in the retina causes cell death of ganglions, as well as bipolar and rod cells (13). In addition, Rb1 deficiency causes differentiation defects in starburst amacrine cells (SACs). Our analysis of Rb1ΔL/ΔL mutants reveals that all of these cell types are specified normally and at the same developmental time as the wild type (Fig. 3A). Cell types that are unaffected by conditional deletion of pRB are also normal in the Rb1ΔL/ΔL mutant (data not shown), indicating that retinal cells are correctly specified in Rb1ΔL/ΔL mutant mice. We also investigated the proliferative status of Rb1ΔL/ΔL mutant retinas in both 8- and 18-day-old mice. As shown in Fig. 3B (and data not shown), proliferation has ceased in Rb1ΔL/ΔL mutant retinas at 8 days of age and remains absent at 18 days of age. In contrast, proliferation persists in conditional Rb1 knockouts at both time points, as indicated by Ki67 staining (Fig. 3B). This reveals that the Rb1ΔL mutant is capable of mediating normal cell cycle exit during retinal development, further emphasizing that the cell cycle exit and terminal differentiation of long-lived cell types is essentially normal in Rb1ΔL/ΔL mutant mice.
FIG. 3.
Developmental cell cycle arrest is normal in Rb1ΔL/ΔL mutant retinas. (A) Cross-sections of retinas were used to examine morphology and cellular composition in 8-day-old newborn mice. Ganglion, bipolar, and rod cells and SACs were stained for the protein marker indicated to the left of each panel (red or green), and nuclei were counterstained with DAPI (blue). The effect of α-crystallin-Cre deletion of Rb1f/f in the retina on the development of these cells was included as a control. (B) Cell proliferation in differentiated retinal cells was examined by Ki67 staining (green) and counterstaining with DAPI (blue) in 8- and 18-day-old (P8 and P18, respectively) mice. Deregulation of proliferation by α-crystallin-Cre deletion of Rb1f/f in the retina was included as a control. The number of Ki67-positive cells per tissue section was determined for each genotype and is shown to the right. The error bars in all of the graphs indicate 1 standard deviation from the mean of at least three replicates.
Defective senescence in Rb1ΔL/ΔL mutant fibroblasts contributes to immortalization.
In contrast to the cell cycle exit that occurs normally during development in Rb1ΔL/ΔL mutant mice, we investigated whether the defective senescent arrest allows cells to escape and resume proliferating. Using a 3T3 culture protocol, we passaged wild-type and mutant fibroblasts to determine if they have similar proliferative potentials by measuring the passage at which they enter senescence. Figure 4A shows that they enter senescence at an equivalent passage. DNA replication was measured in successive passages of senescent cultures by BrdU incorporation, and levels were found to be elevated in Rb1ΔL/ΔL mutants (data not shown). This indicates that Rb1ΔL/ΔL mutant MEFs respond similarly to a 3T3 culture protocol as they do to other senescence-inducing stimuli (Fig. 1) that produce an incomplete arrest. To detect early escape from this defective senescence, we continued to culture these fibroblasts and counted the first passage at which they resumed doubling as escape (Fig. 4A). Based on this criterion, Rb1ΔL/ΔL mutant cultures become immortal significantly earlier than wild-type controls do (P < 0.05). This suggests that not only do cells from Rb1ΔL/ΔL mutant mice enter into an incomplete senescent state, but this allows them to escape and resume proliferating more readily.
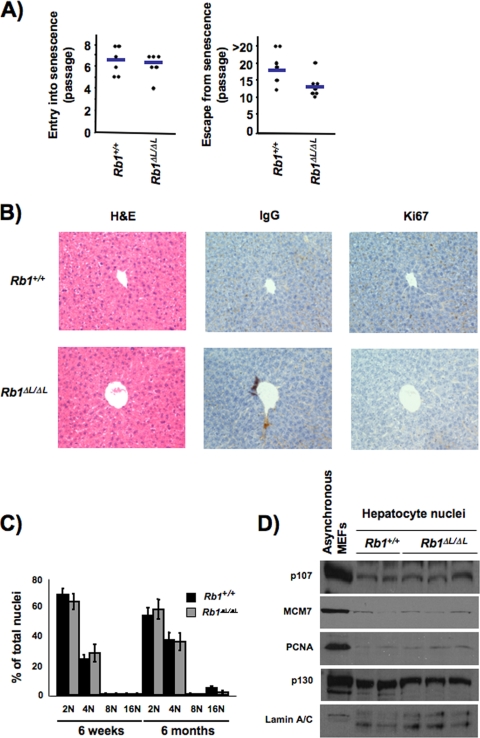
FIG. 4.
Defective senescence of Rb1ΔL/ΔL mutants contributes to immortalization. (A) MEFs were subjected to a 3T3 culture protocol to induce entry into senescence. We measured the number of passages that it took the cells to senesce and the number of passages it took them to become immortalized. Senescence was defined as the first passage without a population doubling, and immortalization was the next passage where cells resumed doubling and continued to double each passage thereafter. Scatterplots showing the passage where each wild-type or Rb1ΔL/ΔL mutant culture ceased to proliferate are shown at the left. Plots that reveal when cultures resumed proliferating are shown at the right. Horizontal bars represent the mean of each measurement. P values are 0.66 (for entry) and 0.04 (for escape). (B) H&E and immunohistochemical staining of liver sections from wild-type and Rb1ΔL/ΔL mutant mice stained with Ki67 antibodies (or immunoglobulin G [IgG] control). Each field of view is centered on a portal duct to ensure equivalent orientation of the tissue. (C) DNA content of nuclei extracted from livers was analyzed by propidium iodide staining and flow cytometry. Each ploidy content category is expressed as a percentage of the total number of nuclei analyzed. Error bars indicate 1 standard deviation from the mean of at least three replicates. (D) Protein expression of known E2F target genes, as well as other pRB family proteins, is shown for nuclear extracts prepared from hepatocytes.
We also sought a developmental comparison for the rapid escape from senescence that we observed in Rb1ΔL/ΔL mutant 3T3 cells. Unfortunately, none of the experiments characterizing the cell cycle arrest of whole tissues in Rb1ΔL/ΔL mutant mice in Fig. 2 or 3 are capable of detecting rare cells that undergo sporadic DNA replication. Thus, to search for rare DNA replication events, we analyzed the DNA content of hepatocyte nuclei. While hepatocytes retain proliferative potential for regeneration that separates them from muscle and retinal cells, they become extensively growth arrested in adult mice (54). As mice age, ectopic DNA replication occurs in hepatocytes at a low level; however, many of these cells fail to undergo a subsequent cell division, resulting in endoreduplication (38). Thus, rare replication events that accumulate over time are identifiable by increased nuclear DNA content. Importantly, conditional deletion of pRB in hepatocytes is known to exacerbate this age-dependent endoreduplication effect (38). Our analysis of Rb1ΔL/ΔL mutant livers showed that they appear histologically normal by H&E staining of tissue sections (Fig. 4B). We also did not detect any proliferating cells, as measured by Ki67 staining, in either wild-type or mutant liver sections (Fig. 4B), suggesting that the growth arrest is normal in Rb1ΔL/ΔL mutant livers. As an internal control for our staining efficiency, we also stained other mouse tissues in parallel and could detect cells positive for Ki67 staining (data not shown). We also found that DNA content increases uniformly with age in wild-type and Rb1ΔL/ΔL mutant animals, indicative of normal control of DNA replication (Fig. 4C). In addition, expression levels of E2F targets and other RB family proteins remain normal under these circumstances, further suggesting that compensation by related proteins does not underlie the maintenance of cell cycle arrest in Rb1ΔL/ΔL mutant hepatocytes (Fig. 4D). Because hepatocytes undergo sporadic DNA replication as part of a normal aging process, this analysis shows that even the most sensitive measurements of DNA replication support the conclusion that cell cycle exit in development is as robust in Rb1ΔL/ΔL mutant mice as in wild-type controls.
Incomplete senescence in Rb1ΔL/ΔL mutant cells is characterized by defective transcriptional repression.
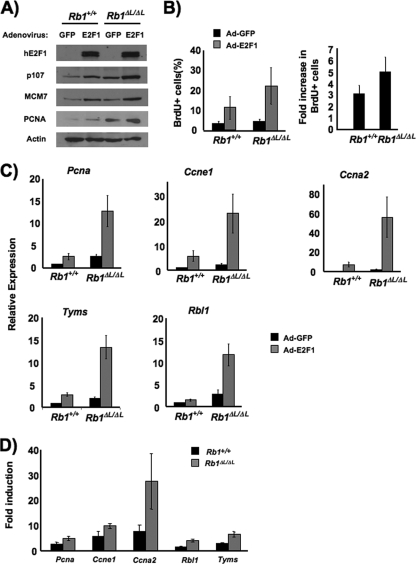
Our initial experiments have revealed that cells from Rb1ΔL/ΔL mutant mice are defective in their senescent cell cycle arrest. To explore the cause of this defect further, we investigated the effects of the Rb1ΔL mutation on the transcriptional silencing of E2F target genes. In order to generate an opportunity to manipulate E2F-dependent gene transcription in a senescent environment, we induced senescence by using oncogenic Ras and ectopically expressed human E2F1 by subsequent adenoviral infection. In this way, we were able to probe the accessibility and potential for transcriptional activation of E2F-responsive promoters. As shown in Fig. 5A, E2F1 was expressed equally in wild-type and mutant cells. In addition, expression levels of three E2F-responsive targets, p107, MCM7, and PCNA, were increased in the incompletely senescent Rb1ΔL/ΔL mutant MEFs and are further elevated by E2F1 expression. More importantly, E2F1 induced higher levels of BrdU incorporation into Rb1ΔL/ΔL mutant cells than into wild-type cells (Fig. 5B). Because basal levels of BrdU incorporation into Rb1ΔL/ΔL mutant cells are slightly higher than those of the wild type under these conditions (Fig. 1D), we also calculated the fold induction of BrdU incorporation in response to E2F1 (Fig. 5B), and this was also significantly higher in mutant cells (P < 0.05). Thus, by using ectopic E2F1 expression, we have demonstrated that the incomplete senescent cell cycle arrest in Rb1ΔL/ΔL mutant cells is more susceptible to being overridden by proliferative signals that activate E2F-dependent transcription.
FIG. 5.
Defective repression of E2F target genes in senescent Rb1ΔL/ΔL mutant MEFs. (A) Ten days following retroviral transduction with oncogenic Ras, senescent MEFs were infected with recombinant Ad expressing either green fluorescent protein (GFP) or human E2F1 at a multiplicity of infection of 100 PFU/cell. The expression level of ectopic E2F1 was measured by Western blotting with a human-specific anti-E2F1 antibody (KH95) after 48 h. The protein expression levels of three known E2F target genes are also shown. Western blotting for actin served as a loading control. (B) Synthesis of DNA in response to E2F1 expression was measured by BrdU incorporation. Two days following Ad-E2F1 infection, cells were pulse-labeled with BrdU for 16 h and positive cells were identified by immunofluorescence microscopy. The fold increase in BrdU incorporation between control and E2F1-infected cells was calculated and is shown in the graph on the right. The mean fold increase was compared by a t test (P < 0.05). (C) The relative abundance of mRNA corresponding to five E2F target genes is shown. To facilitate comparisons, the expression level in uninfected wild-type cells is designated as a relative abundance of 1. Expression of acidic ribosomal phosphoprotein P0 (Rplp0) was used as an internal control. (D) The fold increase in mRNA abundance in E2F1-expressing Rb1ΔL/ΔL mutant cells relative to that of a wild-type control is shown for each E2F target gene (the P value is <0.05 for each gene). Error bars indicate 1 standard deviation from the mean of at least three replicates.
To examine the transcriptional effects of E2F1 expression more closely, we compared mRNA levels from five well-characterized E2F-responsive genes, Pcna, Ccne1, Ccna2, Rbl1, and Tyms (thymidylate synthase). In each case, the expression level was higher in senescent Rb1ΔL/ΔL mutant cells than in the wild-type controls (Fig. 5C). Upon E2F1 expression, these target genes were also more readily transcribed as they accumulated to higher levels in the Rb1ΔL/ΔL mutants. Furthermore, as determined by measuring the fold induction of each E2F target gene, the ability of E2F1 to activate transcription in Rb1ΔL/ΔL mutant cells was again significantly higher than in the wild type (Fig. 5D). This demonstrates that cell cycle-regulated, E2F-responsive promoters are more readily activated in defective senescence. This suggests that transcriptional silencing is likely altered in senescent Rb1ΔL/ΔL mutant cells. Importantly, this difference allows E2F1 expression to stimulate senescent Rb1ΔL/ΔL mutant cells to synthesize DNA more readily.
Rb1ΔL/ΔL mutant cells fail to heterochromatinize E2F target gene promoters in senescence.
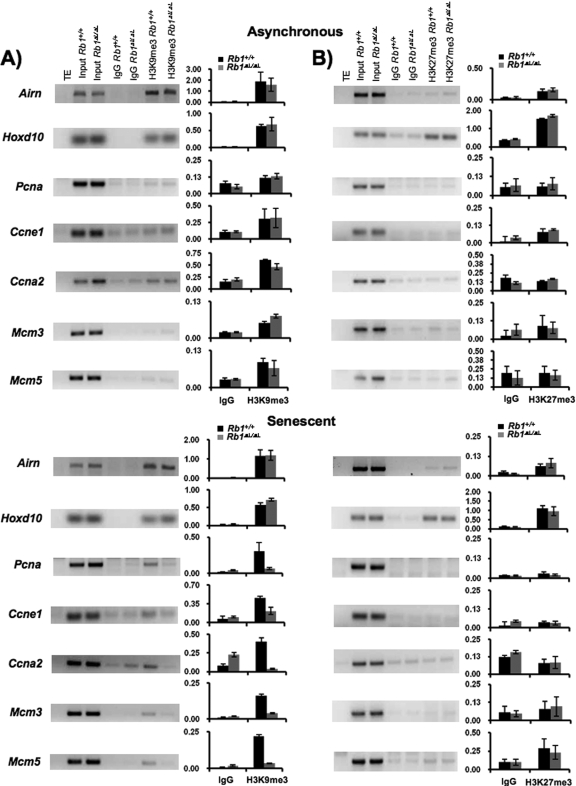
Given recent studies that have demonstrated the role of chromatin regulation in reorganizing the genome during senescence (42), we wondered if the Rb1ΔL mutation affects this process. We sought to investigate repressive histone tail modifications to determine if they are also altered or absent. In particular, we were interested in H3K9me3 status because one of the histone methyltransferases responsible for adding this modification, Suv39h1, is required for oncogene-induced senescence (6) and is reported to interact with pRB through its LXCXE binding cleft (43).
Chromatin from proliferating and Ras-induced senescent cells was immunoprecipitated to determine the relative abundance of H3K9me3 at E2F-responsive promoters. As a control for our immunoprecipitation experiments, we amplified sequences from the imprinted Airn promoter. Because of its allele-specific expression, we are able to detect H3K9me3 that originates from the silenced allele under all growth conditions (47). In cells that have been induced to senesce with oncogenic Ras, H3K9me3 becomes enriched at E2F-responsive promoters (Fig. 6A). Importantly, H3K9me3 is not enriched at E2F promoters in Rb1ΔL/ΔL mutant cells. This analysis reveals that LXCXE interactions with pRB are crucial for assembling heterochromatin in senescence. In addition to the increase in H3K9me3, it has also been proposed that repressive marks like H3K27me3 play an important role in silencing cell cycle genes such as Ink4a in an RB family-dependent manner (5, 30). For these reasons, we also investigated H3K27me3 histone tail modifications at the same E2F-responsive promoters in senescence (Fig. 6B). We also amplified sequences from the Hoxd10 homeobox gene promoter, which has been shown previously to be enriched for this mark, as an additional control for our immunoprecipitations (3). This analysis demonstrates that some E2F target genes also increase their abundance of H3K27me3 in senescence compared with asynchronously growing cells. Interestingly, deposition of this histone tail modification is not dependent on pRB-LXCXE interactions. Given the ability of ectopic E2F1 to activate genes like Ccne1 in incompletely senescent Rb1ΔL/ΔL mutant cells and the fact that only H3K9me3 is added at this promoter in a pRB-LXCXE-dependent manner, we suggest that H3K9me3 is a key repressive modification that silences gene expression under senescent growth arrest conditions.
FIG. 6.
Disrupted heterochromatin structure in incompletely senescent Rb1ΔL/ΔL mutant cells. (A and B) ChIPs were performed on extracts from asynchronous or Ras-induced senescent MEFs. Sheared chromatin was precipitated with rabbit immunoglobulin G (IgG) control, anti-H3K9me3, or anti-H3K27me3 antibodies. An input control PCR was performed on 0.5% of the chromatin used for each ChIP. Precipitated DNA fragments were amplified by PCR using primers specific for the promoter regions of Airn, Hoxd10, Pcna, Ccne1, Ccna2, Mcm3, and Mcm5. Band intensities were quantified using image quantification software from Bio-Rad and are presented as graphs. Error bars indicate standard deviation from the mean value generated from multiple trials of ChIPs. TE, Tris-EDTA buffer.
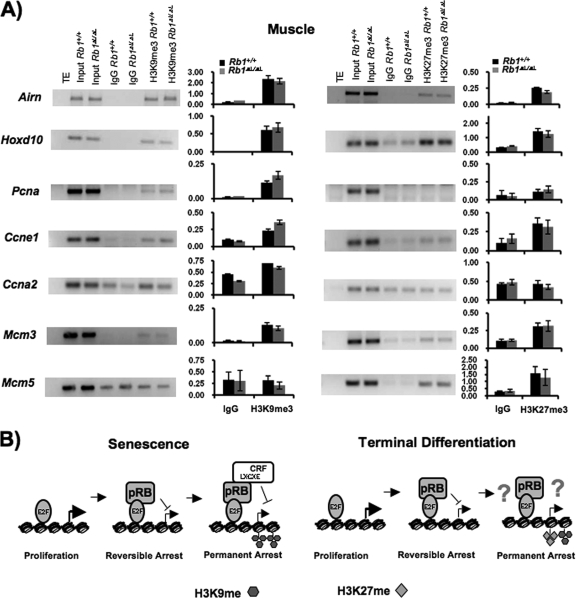
Assembly of repressive heterochromatin has also been implicated in the establishment of a stable cell cycle exit in terminal differentiation (1, 3, 45). We next wanted to determine if the epigenetic landscape of the same E2F-responsive genes was similar in differentiated muscle and whether it differs between wild-type and mutant mice. In agreement with previous work, we found that the H3K9me3 mark can be detected at the promoters of some E2F-responsive cell cycle genes in muscle (Fig. 7A). We observed that some promoters, like the Ccne1 and Mcm3 promoters, are enriched for both H3K9me3 and H3K27me3. In the case of genes like Mcm5, there is a significant enrichment of H3K27me3 whereas we were unable to detect H3K9me3 levels above the background, suggesting that H3K27me3 has a prominent role in the repression of this gene. Conversely, Ccna2 and Pcna are enriched for H3K9me3, but not H3K27me3, in muscle. Thus, comparing the epigenetic landscape of these E2F targets in senescence and muscle indicates that senescence is highly dependent on H3K9me3 while gene silencing in muscle is likely a varied combination of both H3K9me3 and H3K27me3. This strongly suggests that there is a fundamental difference in the growth arrest pathways that silence E2F target genes under these different arrest conditions. Furthermore, neither the deposition of H3K27me3 nor that of H3K9me3 is different between wild-type and mutant muscle tissues. Providing additional evidence that a different mechanism governs the silencing of E2F-responsive genes in terminal differentiation. Previous reports demonstrate that H3K9me3 and H3K27me3 deposition at E2F target genes occurs in response to pRB-dependent myogenesis (3). For these reasons, we interpret our results to mean that pRB has multiple growth arrest mechanisms at its disposal and that the pathways used in cell cycle arrest during senescence and terminal differentiation are fundamentally distinct.
FIG. 7.
Heterochromatin regulation during terminal differentiation is distinct from senescence. (A) Muscle tissue from 6-week-old wild-type and mutant mice were used for ChIP with rabbit immunoglobulin G (IgG) control, anti-H3K9me3, or anti-H3K27me3 antibodies as described in the legend to Fig. 6. Band intensities were quantified in the same way as for Fig. 6. (B) Model of cell cycle exit regulation in senescence and terminal differentiation. During the induction of a senescent arrest, pRB-E2F interactions regulate proliferation sufficiently to induce a reversible arrest state. Incomplete senescence of Rb1ΔL/ΔL mutant cells appears to reach this state where they remain susceptible to rereplication of their DNA. Establishment of a heterochromatin barrier that can block inappropriate cell cycle reentry is dependent on pRB-LXCXE interactions and H3-K9me3 histone tail modifications. Cell cycle exit associated with terminal differentiation requires pRB regulation of E2Fs. Through unknown mechanisms, the initial withdrawal from the cell cycle becomes permanent. E2F target promoters become heterochromatinized with H3-K27me3 and H3-K9me3 modifications in a manner that is independent of pRB-LXCXE interactions. TE, Tris-EDTA buffer.
DISCUSSION
Our work reveals the surprising finding that pRB possesses the ability to block DNA replication in senescence using a fundamentally different mechanism from a permanent cell cycle arrest in development. In particular, pRB requires LXCXE-type interactions to regulate chromatin structure and silence E2F-responsive genes in senescence. This is an important distinction because it demonstrates that pRB uses more than just a single growth-suppressive mechanism to block proliferation. It reveals that specific growth arrest signals like DNA damage elicit different functions from pRB than the development programs that govern myogenesis and neurogenesis. It also suggests that different external signals (as conveyed by expression of MyoD versus oncogenic Ras) activate different functions of pRB. Although both are growth restrictive, there is clearly an important distinction between these two types of stimuli that activate distinct functions of pRB. An important distinction may be that expression of MyoD under growth-restrictive conditions further signals cells to exit the cell cycle and differentiate. Alternatively, expression of oncogenic Ras under high-serum conditions activates conflicting signals by driving rapid proliferation on the one hand and activating growth arrest signals by inducing DNA damage on the other. We think it is in the context of persistent, conflicting signals that the LXCXE-dependent functions of pRB are activated or required.
This study emphasizes that cell cycle arrest in senescence requires a repressor module containing E2F-pRB and a chromatin-regulatory component (Fig. 7A). Rowland et al. have previously shown that expression of a pRB binding-deficient mutant of E2F3 can disrupt pRB-E2F function in senescence, demonstrating the need for E2F to recruit this complex to promoters (49). The response of Rb1ΔL/ΔL mutant cells to DNA damage indicates that the initial steps in cell cycle arrest take place normally, allowing these cells to reach a reversible arrest. This suggests that pRB-E2F interactions are sufficient to mediate this initial step (Fig. 7B). The low level of DNA synthesis that persists over time in Rb1ΔL/ΔL mutants suggests that the true role of chromatin regulation by pRB in senescence is to function as a failsafe mechanism in cell cycle arrest that establishes permanence. Because complete cell cycle exit in senescence is dependent on chromatin remodeling, we describe pRB's role at this step as a checkpoint.
The discovery that pRB-LXCXE interactions are dispensable for a terminal differentiation-related cell cycle arrest is very surprising. As stated earlier, an E2F-pRB-chromatin regulatory complex such as that shown for a senescent arrest in Fig. 7B is highlighted in many reviews of pRB function as controlling cell cycle exit in a ubiquitous arrest scenario that includes terminal differentiation (7, 8, 14, 25, 29). We offer the following explanations, as well as our own data in support of the model of terminal differentiation shown in Fig. 7B, where LXCXE-dependent chromatin regulation is dispensable. We think that pRB's role in a developmentally induced cell cycle exit may be accomplished largely through negative regulation of activator E2F activity. This interpretation is supported by the fact that a number of differentiation defects caused by complete loss of pRB can be rescued by crossing to null alleles of activator E2Fs. In the murine retina, it is known that conditional deletion of Rb1 triggers ectopic division and death of ganglion, bipolar, and rod cells (13). These defects in terminal differentiation are reversed by E2f1 deficiency. Furthermore, ablation of Rb1 in the telencephalon has been reported to dissociate proliferative control from the initiation of neuronal differentiation (21). Ectopic cell division in the intermediate zone and cortical plate regions of Rb1−/− mutant mouse brain tissue can be suppressed by E2f1 or E2f3 deficiency (39). Beyond E2F regulation, a number of reports have also shown pRB-dependent effects on chromatin in terminal differentiation of muscle that may, on the surface, seem to contradict our model. Ablation of Rb1 in skeletal muscle progenitors has been demonstrated to lead to complete failure of myogenesis (26), and recent experiments using RNA interference to deplete pRB expression in myotubes indicate that cell cycle reentry is triggered in its absence (3). For these reasons, formation of myotubes and resulting chromatin changes are clearly pRB dependent. However, pRB is also able to influence the activity of differentiation-inducing factors like ID2 and MyoD, and through molecules like these, it may regulate chromatin in differentiation indirectly (8). For these reasons, we suggest that chromatin regulation in terminal differentiation of muscle that is pRB dependent is an indirect consequence of cell cycle exit, is independent of LXCXE interactions with pRB, or is induced indirectly through prodifferentiation factors (Fig. 7B).
In addition to senescence, we have also determined that pRB-LXCXE interactions are critical to transforming growth factor β (TGF-β) regulation of continuously proliferating mammary epithelial cells (23). While this is a different growth-regulatory paradigm, the ability of TGF-β to induce senescence through chronic stimulation further suggests that pRB-LXCXE interactions can be implicated in a broad, stress-responsive growth control program (33). It is tempting to speculate that the pRB-LXCXE-dependent arrest pathway plays a key role in pRB's tumor suppressor function. We have not detected spontaneous tumors in our Rb1ΔL/ΔL mutants (data not shown). However, it is noteworthy that the Rb1ΔL mutation does not abrogate senescence completely but uncouples its permanence from the initial arrest. Other genetically modified strains of mice whose lesions completely abrogate this senescence arrest pathway, such as Ink4a−/− mutant mice, already have surprisingly low rates of spontaneous tumorigenesis themselves (31, 52). Future work to determine the importance of heterochromatin at E2F-responsive targets in senescence will require crosses to transgenic mice with defined oncogenic lesions. In this way, we will be able to directly relate the chromatin assembly step in senescence to cancer progression.
Intriguingly, our data reveal an unexpected parallel between pRB and p53 in mammalian physiology. Like our Rb1ΔL/ΔL mutant mice, Trp53−/− mutant mice are relatively normal developmentally (19). While, p53's role in responding to cancer-causing insults like DNA damage is well known, only recently has it been demonstrated that p53's role in stellate cell senescence is essential for the liver to respond appropriately to chemical toxicity and avoid fibrosis (32). The unique role for pRB-LXCXE regulation of chromatin in senescence that we describe offers a similar glimpse at a fundamental stress response mechanism. Indeed, other reports have suggested a role for pRB in stress responses (34, 37). In particular, lung epithelium appears to use pRB in a very specific role in controlling proliferation following injury, but not in development (37). Thus, it seems that pRB plays a unique role in this growth control paradigm that developed to respond to stressful exogenous stimuli, including DNA damage or the release of TGF-β in response to tissue trauma, or as a protective response to chemical toxicity. Such responses, which are largely independent of cell cycle control during development, imply that a stress-responsive growth control program is a pervasive and important aspect of mammalian physiology. It is difficult to know to what degree evolution has selected for anticancer functions in the genes that code for p53 and pRB; however, the involvement of these master regulators in a stress-specific growth arrest reveals an important biological feature of proliferative control. Genes involved in a checkpoint that is stress responsive, as opposed to ones that are largely regulated by developmental cues, may offer a starting point for growth control mechanisms that, in the present day, offer antioncogenic properties as genetic damage accumulates in response to environmental pressures.
Acknowledgments
We have no conflict of interest with the publication of this research.
We are grateful to A. Chicas and S. Lowe for freely discussing unpublished work. We thank many colleagues at the LRCP and CHRI for advice during the course of this work, especially Jai Ablack for providing Ad-GFP. The Ad-E2F1 virus was a kind gift of E. Knudsen (Kimmel, Philadelphia, PA). We are grateful for technical assistance by the CHRI histology, LHSC histology, and LHSC flow cytometry core facilities.
S.T. is supported by an ERA award. C.E.I., S.A.H., and S.M.F. have all been members of the CIHR strategic training program in cancer research. S.M.F. acknowledges fellowship support from CBCF Ontario and OGSST, and S.A.H. thanks CBCF Ontario for postdoctoral fellowship support. F.A.D. is a research scientist of the Canadian Cancer Society. This work was supported by operating grants from the CIHR to R.B. (MOP-64243) and to F.A.D. (MOP-64253).
Footnotes
Published ahead of print on 14 December 2009.
REFERENCES
- 1.Ait-Si-Ali, S., V. Guasconi, L. Fritsch, R. Yahi, I. Sekhri, I. Naguibneva, P. Robin, F. Cabon, A. Polesskaya, and A. Harel-Bellan. 2004. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not cycling cells. EMBO J. 23:605-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio, O., J. V. Geisberg, E. Sekinger, E. Yang, Z. Moqtaderi, and K. Struhl. 2005. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo, p. 21.3.1-21.3.33. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. E. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates & Wiley-Interscience, New York, NY. [DOI] [PubMed]
- 3.Blais, A., C. J. van Oevelen, R. Margueron, D. Acosta-Alvear, and B. D. Dynlacht. 2007. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J. Cell Biol. 179:1399-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boxem, M., and S. van den Heuvel. 2001. lin-35 and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development 128:4349-4359. [DOI] [PubMed] [Google Scholar]
- 5.Bracken, A. P., D. Kleine-Kohlbrecher, N. Dietrich, D. Pasini, G. Gargiulo, C. Beekmean, K. Theilgaard-Monch, S. Minucci, B. T. Porse, J. C. Marine, K. H. Hansen, and K. Helin. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braig, M., S. Lee, C. Loddenkemper, C. Rudolph, A. H. Peters, B. Schlegelberger, H. Stein, B. Dorken, T. Jenuwein, and C. A. Schmitt. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436:660-665. [DOI] [PubMed] [Google Scholar]
- 7.Brehm, A., and T. Kouzarides. 1999. Retinoblastoma protein meets chromatin. Trends Biochem. Sci. 24:142-145. [DOI] [PubMed] [Google Scholar]
- 8.Burkhart, D. L., and J. Sage. 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8:671-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buttitta, L., and B. Edgar. 2007. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buttitta, L., A. Katzaroff, C. Perez, A. de la Cruz, and B. Edgar. 2007. A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell 12:631-643. [DOI] [PubMed] [Google Scholar]
- 11.Camarda, G., F. Siepi, D. Pajalunga, C. Bernardini, R. Rossi, A. Montecucco, E. Meccia, and M. Crescenzi. 2004. A pRb-independent mechanism preserves the postmitotic state in terminally differentiated skeletal muscle cells. J. Cell Biol. 167:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campisi, J., and F. d'Adda di Fagagna. 2007. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8:729-740. [DOI] [PubMed] [Google Scholar]
- 13.Chen, D., I. Livne-bar, J. Vanderluit, R. S. Slack, M. Agochiya, and R. Bremner. 2004. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5:539-551. [DOI] [PubMed] [Google Scholar]
- 14.Classon, M., and E. Harlow. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910-917. [DOI] [PubMed] [Google Scholar]
- 15.Classon, M., S. R. Salama, C. Gorka, R. Mulloy, P. Braun, and E. E. Harlow. 2000. Combinatorial roles for pRB, p107 and p130 in E2F-mediated cell cycle control. Proc. Natl. Acad. Sci. U. S. A. 97:10820-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collado, M., M. Blasco, and M. Serrano. 2007. Cellular senescence in cancer and aging. Cell 130:223-233. [DOI] [PubMed] [Google Scholar]
- 17.Dannenberg, J.-H., A. van Rossum, L. Schuijff, and H. te Riele. 2000. Ablation of the Retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruin, A., L. Wu, H. I. Saavedra, P. Wilson, Y. Yang, T. J. Rosol, M. Weinstein, M. L. Robinson, and G. Leone. 2003. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 100:6546-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donehower, L., M. Harvey, B. Slagle, M. McArthur, C. Montgomery, J. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 20.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson, K., J. Vanderluit, J. Hébert, W. McIntosh, E. Tibbo, J. MacLaurin, D. Park, V. Wallace, M. Vooijs, S. McConnell, and R. Slack. 2002. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 21:3337-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firth, L., and N. Baker. 2005. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 8:541-551. [DOI] [PubMed] [Google Scholar]
- 23.Francis, S. M., J. Bergseid, C. H. Coschi, C. E. Isaac, C. V. Hojilla, S. Chakrabarti, G. E. DiMattia, R. Khoka, J. Y. J. Wang, and F. A. Dick. 2009. A functional connection between pRB and TGFbeta in growth arrest and mammary gland development. Mol. Cell. Biol. 29:4455-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Funayama, R., M. Saito, H. Tanobe, and F. Ishikawa. 2006. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 175:869-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harbour, J. W., and D. C. Dean. 2000. Chromatin remodeling and Rb activity. Curr. Opin. Cell Biol. 12:685-689. [DOI] [PubMed] [Google Scholar]
- 26.Huh, M. S., M. H. Parker, A. Scime, R. Parks, and M. A. Rudnicki. 2004. Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J. Cell Biol. 166:865-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurford, R., D. Cobrinik, M.-H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 28.Isaac, C. E., S. M. Francis, A. L. Martens, L. M. Julian, L. A. Seifried, N. Erdmann, U. K. Binne, L. Harrington, P. Sicinski, N. J. Dyson, and F. A. Dick. 2006. The retinoblastoma protein regulates pericentric heterochromatin. Mol. Cell. Biol. 26:3659-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korenjak, M., and A. Brehm. 2005. E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr. Opin. Genet. Dev. 15:520-527. [DOI] [PubMed] [Google Scholar]
- 30.Kotake, Y., R. Cao, P. Viatour, J. Sage, Y. Zhang, and Y. Xiong. 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4A tumor suppressor gene. Genes Dev. 21:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krimpenfort, P., K. C. Quon, W. J. Mooi, A. Loonstra, and A. Berns. 2001. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413:83-86. [DOI] [PubMed] [Google Scholar]
- 32.Krizhanovsky, V., M. Yon, R. Dickins, S. Hearn, J. Simon, C. Miething, H. Yee, L. Zender, and S. Lowe. 2008. Senescence of activated stellate cells limits liver fibrosis. Cell 134:657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, H. K., S. Bergmann, and P. P. Pandolfi. 2004. Cytoplasmic PML function in TGF-beta signalling. Nature 431:205-211. [DOI] [PubMed] [Google Scholar]
- 34.MacLeod, K. 2008. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat. Rev. Cancer 8:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacPherson, D., J. Sage, T. Kim, D. Ho, M. E. McLaughlin, and T. Jacks. 2004. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 18:1681-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-604. [DOI] [PubMed] [Google Scholar]
- 37.Mason-Richie, N. A., M. J. Mistry, C. A. Gettler, A. Elayyadi, and K. A. Wikenheiser-Brokamp. 2008. Retinoblastoma function is essential for establishing lung epithelial quiescence after injury. Cancer Res. 68:4068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayhew, C. N., E. E. Bosco, S. R. Fox, T. Okaya, P. Tarapore, S. J. Schwemberger, G. F. Babcock, A. B. Lentsch, K. Fukasawa, and E. S. Knudsen. 2005. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 65:4568-4577. [DOI] [PubMed] [Google Scholar]
- 39.McClellan, K., V. Ruzhynsky, D. Douda, J. Vanderluit, K. Ferguson, D. Chen, R. Bremner, D. Park, G. Leone, and R. Slack. 2007. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol. Cell. Biol. 27:4825-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClellan, K., and R. S. Slack. 2007. Specific in vivo roles for E2Fs in differentiation and development. Cell Cycle 6:2917-2927. [DOI] [PubMed] [Google Scholar]
- 41.Mulligan, G. J., J. Wong, and T. Jacks. 1998. p130 is dispensable in peripheral T lymphocytes: evidence for functional compensation by p107 and p130. Mol. Cell. Biol. 18:206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narita, M., S. Nunez, E. Heard, M. Narita, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 44.Novitch, B. G., G. J. Mulligan, T. Jacks, and A. B. Lassar. 1996. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 135:441-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panteleeva, I., S. Boutillier, V. See, D. Spiller, C. Rouaux, G. Almouzni, D. Bailly, C. Maison, H. Lai, J. Loeffler, and A. Boutillier. 2007. HP1alpha guides neuronal fate by timing E2F-targeted genes silencing during terminal differentiation. EMBO J. 26:3616-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peeper, D., J. Dannenberg, S. Douma, H. teRiele, and R. Bernards. 2001. Escape from premature senescence is not sufficient for oncogenic transformation by ras. Nat. Cell Biol. 3:198-203. [DOI] [PubMed] [Google Scholar]
- 47.Regha, K., M. A. Sloane, R. Huang, F. M. Pauler, K. E. Warczok, B. Melikant, M. Radolf, J. H. Martens, G. Schotta, T. Jenuwein, and D. P. Barlow. 2007. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell 27:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 49.Rowland, B. D., S. G. Denissov, S. Douma, H. G. Stunnenberg, R. Bernards, and D. S. Peeper. 2002. E2F transcriptional repressor complexes are critical downstream targets of p19arf/p53-induced proliferative arrest. Cancer Cell 2:55-65. [DOI] [PubMed] [Google Scholar]
- 50.Sage, J., G. Mulligan, L. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature senescence associated with accumulation of p53 and p16. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 52.Sharpless, N. E., N. Bardeesy, K. H. Lee, D. Carrasco, D. H. Castrillon, A. J. Aguirre, E. A. Wu, J. W. Horner, and R. A. DePinho. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86-91. [DOI] [PubMed] [Google Scholar]
- 53.Singh, P., J. Coe, and W. Hong. 1995. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature 374:562-565. [DOI] [PubMed] [Google Scholar]
- 54.Steer, C. J. 1995. Liver regeneration. FASEB J. 9:1396-1400. [DOI] [PubMed] [Google Scholar]
- 55.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zacksenhaus, E., Z. Jiang, D. Chung, J. D. Marth, R. A. Phillips, and B. L. Gallie. 1996. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 10:3051-3064. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, J., J. Gray, L. Wu, G. Leone, S. Rowan, C. L. Cepko, X. Zhu, C. M. Craft, and M. A. Dyer. 2004. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat. Genet. 36:351-360. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, R., W. Chen, and P. Adams. 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27:2343-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]