Abstract
Autophagy is a bulk proteolytic process that is indispensable for cell survival during starvation. Autophagy is induced by nutrient deprivation via inactivation of the rapamycin-sensitive Tor complex1 (TORC1), a protein kinase complex regulating cell growth in response to nutrient conditions. However, the mechanism by which TORC1 controls autophagy and the direct target of TORC1 activity remain unclear. Atg13 is an essential regulatory component of autophagy upstream of the Atg1 kinase complex, and here we show that yeast TORC1 directly phosphorylates Atg13 at multiple Ser residues. Additionally, expression of an unphosphorylatable Atg13 mutant bypasses the TORC1 pathway to induce autophagy through activation of Atg1 in cells growing under nutrient-rich conditions. Our findings suggest that the direct control of the Atg1 complex by TORC1 induces autophagy.
Autophagy is a highly conserved cellular process that leads to the degradation of cytoplasmic contents and organelles (25, 32, 44). Cellular material is sequestered into the autophagosome, a unique double-membrane enclosed compartment, and transported to the vacuole/lysosome for degradation (1). Eighteen different essential autophagy-related (ATG) genes have been identified: ATG1 to ATG10, ATG12 to ATG14, ATG16 to ATG18, ATG29, and ATG31 (32, 47).
In most eukaryotes, autophagy is controlled by cellular nutrient status, and this process is indispensable for cell survival during starvation (44, 45). Among the proteins involved in autophagy, Tor (Target of rapamycin) is a protein kinase that regulates cell growth and autophagy in response to changes in cellular nutrient conditions, and rapamycin, a Tor inhibitor, induces autophagy even under nutrient-rich conditions (34). Tor forms two distinct complexes, TORC1 and TORC2, within cells, but only the function of TORC1 is sensitive to rapamycin (7, 12, 26), suggesting that TORC1, but not TORC2, is responsible for controlling autophagy. Several studies have characterized the biochemical properties of different Atg proteins under TORC1-inactivating, autophagy-inducing conditions (32). In particular, most Atg proteins assemble at the preautophagosomal structure (PAS), a putative site for autophagosome formation (42, 43), and Atg9 cycles between the PAS and its cytosolic pool (9). Additionally, phosphatidylinositol 3-kinase plays a role in regulating autophagy (35). Atg8 is upregulated at the transcriptional level (23), and it undergoes phosphatidylethanolamine conjugation (Atg8-PE) mediated by the Atg12-Atg5 complex (6). However, it remains unclear which Atg protein(s) is directly downstream of TORC1 signaling to induce autophagy.
ATG1 encodes a protein kinase, and the catalytic activity of Atg1 is essential for autophagy (19, 28). Atg1 forms a complex with several proteins, including Atg11, Atg13, Atg17, Atg29, and Atg31 (5, 16, 19, 21, 22). Atg13 is highly phosphorylated in a TORC1-dependent manner under nutrient-rich conditions, and phosphorylated Atg13 does not appreciably bind to Atg1. This, in turn, reduces the kinase activity of Atg1. However, immediately upon starvation or rapamycin treatment, Atg13 is rapidly dephosphorylated, and binding of dephosphorylated Atg13 by Atg1 leads to Atg1 activation (19). Thus, the association of Atg1 with Atg13 and subsequent Atg1 activation are regulated by the phosphorylation state of Atg13. The dephosphorylation of Atg13 is thought to be one of the initial steps in autophagy; it remains unresolved whether Atg13 is a direct target of TORC1 or whether Atg13 dephosphorylation alone is sufficient for autophagy induction (24). In this study, we examined the relationship between TORC1 activity, Atg13 dephosphorylation, and autophagy induction, and we show that dephosphorylation of Atg13 acts as an initiating trigger for autophagy.
MATERIALS AND METHODS
Strains, plasmids, media, and genetic methods.
The yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Standard techniques were used for yeast manipulation (30). The replacement of Ser residues with Ala to generate the ATG13-SA alleles was performed by PCR-mediated mutagenesis using QuikChange (Stratagene, La Jolla, CA). To induce Atg13, yeast cells were grown to early logarithmic phase (optical density at 600 nm [OD600] of ca. 1) in synthetic medium with 2% raffinose (SCRaf) at 30°C. Cells were then incubated in SCRaf plus 2% galactose (SCRafGal) for the indicated times (1 to 6 h) to activate the GAL1 promoter.
TABLE 1.
Yeast strains used in this study
| Strain name | Genotype | Source or reference |
|---|---|---|
| W303-1B | MATaade2 his3 leu2 trp1 ura3 can1 | Laboratory stock |
| BJ2168 | MATaleu2 trp1 ura3 pep4-3 prb1-1122 prc1-407 | Laboratory stock |
| BY4741 | MATahis3 leu2 met15 ura3 | Laboratory stock |
| OND88 | W303-1B pho8::TDH3p::pho8Δ60 | 20 |
| YYK742 | OND88 atg17Δ::KanMX | This study |
| YYK744 | OND88 atg29Δ::KanMX | This study |
| YYK757 | OND88 atg2Δ::KanMX | This study |
| YYK762 | OND88 atg11Δ::LEU2 | 20 |
| YYK797 | OND88 atg1Δ::URA3 | This study |
| YYK334 | W303-1B tor1Δ::KanMX | This study |
| YYK798 | YYK334 pRS313[HA-TOR1] pRS316vector | This study |
| YYK799 | YYK334 pRS313[HA-TOR1] pRS316[Flag-KOG1] | This study |
| YYK800 | YYK334 pRS313[HA-tor1-D2294E] pRS316[Flag-KOG1] | This study |
| YYK412 | BJ2168 HA-ATG1 | 15 |
| TMK625 | BY4741 atg11Δ::LEU2 ATG17-GFP::KanMX | 20 |
TABLE 2.
Plasmids used in this study
| Plasmid name | Description | Source or reference |
|---|---|---|
| YEp352 | URA3 2μm ARS Ampr | Laboratory stock |
| p416GAL1 | URA3 CEN6-ARSH4 AmprGAL1 promoter | Laboratory stock |
| pRS313 | HIS3 CEN6-ARSH4 Ampr | Laboratory stock |
| pRS314 | TRP1 CEN6-ARSH4 Ampr | Laboratory stock |
| pRS316 | URA3 CEN6-ARSH4 Ampr | Laboratory stock |
| YEp352[ATG13] | YEp352 ATG13 | 5 |
| YEp352[ATG13-8SA] | YEp352 ATG13 S348A S437A S438A S496A S535A S541A S646A S649A | This study |
| p416GAL1[ATG13] | p416GAL1 ATG13orf | This study |
| p416GAL1[ATG13-2SA] | p416GAL1 ATG13orf S348A S496A | This study |
| p416GAL1[ATG13-3SAa] | p416GAL1 ATG13orf S437A S438A S496A | This study |
| p416GAL1[ATG13-3SAb] | p416GAL1 ATG13orf S496A S535A S541A | This study |
| p416GAL1[ATG13-3SAc] | p416GAL1 ATG13orf S496A S646A S649A | This study |
| p416GAL1[ATG13-4SAa] | p416GAL1 ATG13orf S437A S438A S646A S649A | This study |
| p416GAL1[ATG13-4SAb] | p416GAL1 ATG13orf S348A S496A S535A S541A | This study |
| p416GAL1[ATG13-5SA] | p416GAL1 ATG13orf S437A S438A S496A S646A S649A | This study |
| p416GAL1[ATG13-6SA] | p416GAL1 ATG13orf S348A S437A S438A S496A S646A S649A | This study |
| p416GAL1[ATG13-7SAa] | p416GAL1 ATG13orf S437A S438A S496A S535A S541A S646A S649A | This study |
| p416GAL1[ATG13-7SAb] | p416GAL1 ATG13orf S348A S437A S438A S535A S541A S646A S649A | This study |
| p416GAL1[ATG13-8SA] | p416GAL1 ATG13-8SAorf | This study |
| pRS313[HATOR1] | pRS313 3×HA-TOR1 | 30 |
| pRS313[HATOR1-KD] | pRS313 3×HA-TOR1-D2294E | 30 |
| pRS316[KOG1] | pRS316 KOG1 | 30 |
| pRS316[FlagKOG1] | pRS316 Flag-KOG1 | 30 |
| pRS316[ATG1] | pRS316 ATG1 | 28 |
| pRS316[atg1D211A] | pRS316 atg1-D211A | 28 |
| pRS314[atg1K54A] | pRS316 atg1-K54A | 19 |
Preparation of recombinant Atg13.
To prepare recombinant Atg13 protein from Escherichia coli, the pCold TF vector (TaKaRa) was used. Expression and purification of His6-tagged trigger factor (TF)-fused Atg13 proteins by use of nitrilotriacetic acid (NTA)-agarose beads were performed as described previously (31).
To purify His6-Atg13 from yeast, cells (BJ2168) harboring p416GAL1[His6-ATG13] were grown in YEPRaf (3-liter culture) and incubated in 2% galactose for 6 h to induce Atg13 expression. The culture was converted to spheroplasts with Zymolyase 100T (Seikagaku Kogyo), and the resulting spheroplasts were disrupted by resuspension in lysis buffer (1× phosphate-buffered saline [PBS], 2 mM Na3VO4, 50 mM KF, 15 mM Na2H2P2O7, 15 mM p-nitrophenylphosphate [pNPP], 1% Tween 20, 20 μg/ml leupeptin, 20 μg/ml benzamidine, 10 μg/ml pepstatin A, 40 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride [PMSF]). Cell lysates were cleared by centrifugation for 10 min at 10,000 × g. The His6-Atg13 protein was concentrated using an NTA-agarose column and purified by immunoprecipitation with anti-Atg13. The resulting precipitant was subjected to SDS-PAGE for mass spectrometry (MS) analyses.
Mass spectrometry.
The protein bands in SDS-PAGE gels were excised; incubated with 10 mM EDTA, 10 mM dithiothreitol, and 100 mM ammonium bicarbonate for 1 h at 50°C; and alkylated by treating with 10 mM EDTA, 40 mM iodoacetamide, and 100 mM ammonium bicarbonate for 30 min at room temperature. The proteins were in-gel digested with lysyl endopeptidase from Achromobacter lyticus (Wako Pure Chemical Industries) in 100 mM Tris-HCl (pH 8.9) for 15 h at 37°C. The peptide fragments were extracted from the gels and concentrated in vacuo. After concentration and desalting with ZipTipC18 (Millipore), peptide fragments in 30% acetonitrile and 0.1% formic acid were subjected to mass spectrometric analysis. Positive-ion mass spectra were acquired by direct infusion analysis on a Micromass Q-Tof2 hybrid quadrupole time-of-flight mass spectrometer equipped with a nano-electrospray ionization (ESI) source. Mass spectrometry/mass spectrometry (MS/MS) was performed by collision-induced dissociation using argon as the collision gas.
Immunoprecipitation and protein kinase assay.
Immunoprecipitation and kinase assay for TORC1 were performed as described previously with some modifications (18, 30).
Autophagy assays.
Green fluorescent protein (GFP)-Atg17 and the accumulation of autophagic bodies were observed by fluorescence microscopy and phase-contrast microscopy, respectively, as described previously (20, 44). Electron microscopy was performed as described previously (31). The alkaline phosphatase (ALP) assay used to measure autophagic activity was performed using α-naphthyl phosphate as described previously (33).
RESULTS
Determination of phosphorylation sites of Atg13.
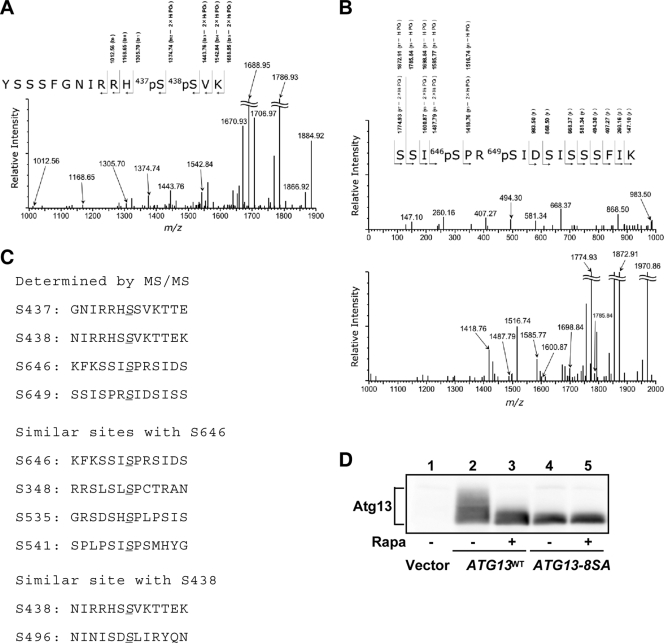
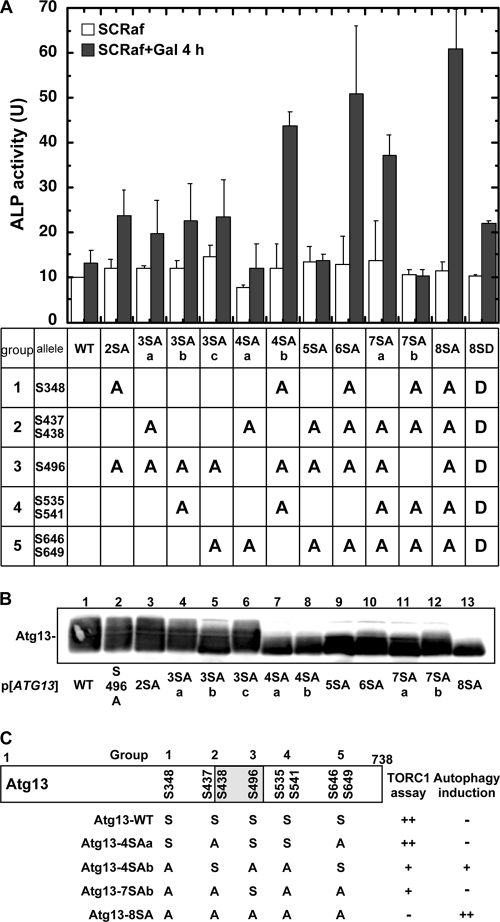
Atg13 is phosphorylated in a Tor-dependent manner, and when isolated from cells under nutrient-rich conditions, Atg13 migrates more slowly by SDS-PAGE, suggesting that it is multiply phosphorylated (19). To identify the sites of Atg13 phosphorylation, we purified His6-tagged Atg13 from yeast cells, and analyzed the isolated proteins by mass spectrometry/mass spectrometry (MS/MS) (Fig. 1A and B). The MS analysis covered approximately 60% of the total Atg13 protein (738 amino acids [aa]), and it indicated that 11 Ser/Thr sites were phosphorylated (data not shown). Among these sites, we clearly identified four phosphorylation sites: S437, S438, S646, and S649 (Fig. 1C). Several phosphorylated peptides were also detected, but the precise phosphorylated residues could not be determined. For example, peptide 483-504, which is located within the Atg1-binding domain, was phosphorylated (data not shown). We next scanned the entire Atg13 sequence to identify Ser/Thr residues that are putative phosphorylation sites by the following criteria: homology to the phosphorylation sites determined as described above, homology to known (m)TORC1 substrates such as 4E-BP1, or conservation among Saccharomyces species. Using these criteria, we chose four Ser residues (S348, S496, S535, and S541) in addition to the identified serines (Fig. 1C). S348, S535, and S541 were chosen due to their homology with S646. These residues have a conserved motif (S-X-S*-P) that is highly homologous to the phosphorylation sites of 4E-BP1 (S-T-T*-P), an mTORC1 substrate (2). S496 was chosen for further analysis because it is homologous to S438, and a peptide including S496 (peptide 483-504) was phosphorylated. These eight residues are well conserved among Saccharomyces species. We generated Ser-to-Ala substitution mutants for all of these residues (S348A, S437A, S438A, S496A, S535A, S541A, S646A, and S649A), and we analyzed the resultant Atg13-8SA mutant. As previously reported, wild-type Atg13 (Atg13WT) isolated from growing cells resolved as an indistinct smear by immunoblotting, but this smear became a single band following rapamycin treatment (Fig. 1D). In contrast, mutant Atg13-8SA resolved as a dephosphorylated, quickly migrating band, suggesting that TORC1-dependent phosphorylation of Atg13-8SA in vivo was largely eliminated (Fig. 1D).
FIG. 1.
Determination and prediction of Atg13 phosphorylation sites. (A and B) Product ion mass spectra of phosphopeptides 426-YSSSFGNIRRHpSpSVK-440 (A) and 643-SSIpSPRpSIDSISSSFIK-659 (B), obtained from the His6-tagged Atg13 protein. In these spectra, multicharged ion peaks, [M + nH]n+, on ESI mass spectra were converted to single-charged ion peaks, [M + H]+ (protonated molecules). Only monoisotopic ion peaks are shown. Isotopic ion peaks are not shown. (C) Phosphorylation sites of Atg13 mapped by mass spectrometry. Four Ser residues determined by MS/MS analysis (top) and the putative phosphorylation sites similar to the determined Ser sites (bottom) are shown. (D) Immunoblot of Atg13. YEPD-grown cells (OND88) expressing the indicated Atg13 protein were treated with rapamycin (Rapa) (0.2 μg/ml) for 1 h. The phosphorylation state of Atg13 was assessed by immunoblotting.
TORC1 directly phosphorylates Atg13.
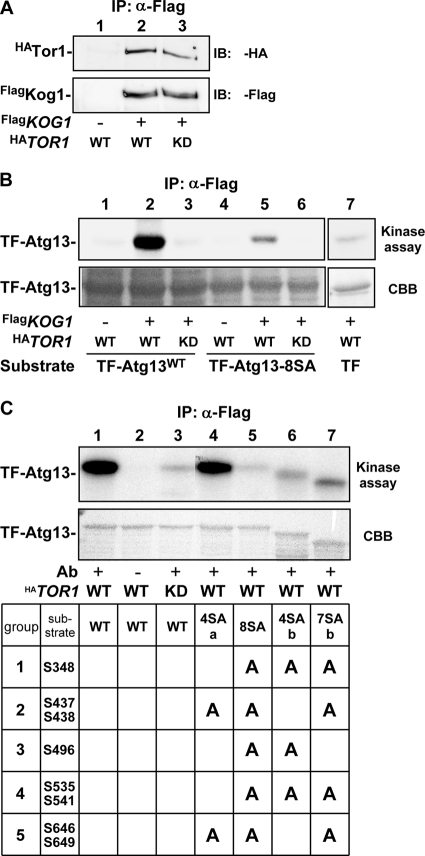
To determine whether TORC1 directly phosphorylates Atg13, we performed an in vitro TORC1 phosphorylation assay. Flag-tagged Kog1 (FlagKog1), a component of TORC1 (7, 26), was immunoprecipitated from yeast cell lysates, and the kinase activity of the coprecipitated Tor protein (monitored by hemagglutinin [HA]-tagged Tor1 [HATor1]) (Fig. 2A) was assayed using recombinant trigger factor (TF)-fused Atg13. Indeed, HATor1-containing TORC1 readily phosphorylated TF-Atg13 (Fig. 2B). However, TORC1 containing HATor1D2294E (a kinase-dead mutant) displayed only marginal kinase activity toward TF-Atg13, confirming the intrinsic activity of the Tor protein. In contrast, TF-Atg13-8SA was phosphorylated to a much lesser extent by TORC1 compared to TF-Atg13. In mammals, mTORC1 substrates such as S6K, 4E-BP1, and PRAS40 have a conserved TOS motif that is required for the recognition and binding to Raptor, a Kog1 ortholog (37, 38). However, Atg13 does not contain an obvious TOS motif, and we did not detect the coprecipitation of Atg1 or Atg13 with FlagKog1 (data not shown).
FIG. 2.
TORC1 directly phosphorylates Atg13 in vitro. (A) Detection of immunoprecipitated TORC1. FlagKog1 was immunoprecipitated, and wild-type HATor1 (WT) and a kinase-dead D2294E mutant (KD) contained in the immunocomplex were detected. Results of a control experiment using untagged Kog1 are shown in lane 1. α, anti. (B) In vitro TORC1 kinase assay. TORC1 isolated as for panel A was used in an in vitro kinase assay using 4 μg of recombinant trigger factor (TF)-fused Atg13 (wild type [lanes 1 to 3] or Atg13-8SA [lanes 4 to 6]) or TF (lane 7) as substrates. The results of autoradiography (kinase assay) and Coomassie blue staining (CBB) are shown. (C) TORC1 kinase assay using various Atg13 constructs. TORC1 assay was performed using the indicated TF-Atg13 proteins. TF-Atg13-4SAb and TF-Atg13-7SAb migrated faster than the wild type, presumably because they were C-terminally processed in E. coli cells.
We next examined whether the Ser residues identified above are phosphorylated by TORC1 in vitro. The eight Ser residues replaced in Atg13-8SA were divided into five groups (group 1, S348; group 2, S437 S438; group 3, S496; group 4, S535 S541; and group 5, S646 S649), and the alanine substitutions were reverted to serine. Atg13-4SAa (S437A, S438A, S646A, and S649A) was strongly phosphorylated by TORC1 in vitro, supporting our prediction that S348, S496, S535, and S541 may be targets of TORC1 (Fig. 2C). Additionally, Atg13-4SAb (S348A, S496A, S535A, and S541A) underwent phosphorylation in the in vitro assay, confirming our MS/MS analysis, but the degree of phosphorylation was less than that seen for Atg13-4SAa. Thus, TORC1 directly phosphorylates Atg13, and the identified Ser residues are candidate sites for TORC1 binding and modification.
Expression of Atg13-8SA directly induces autophagy.
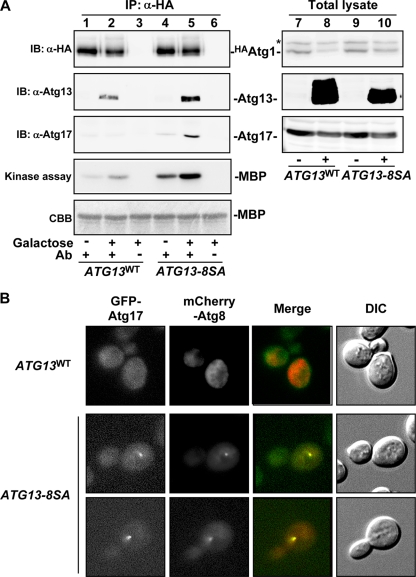
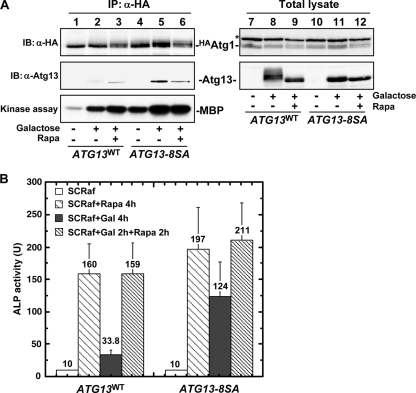
The Atg13-8SA mutant was not phosphorylated, and we hypothesized that it may act as a dephosphorylated form of Atg13 when expressed in vivo. Following TORC1 inhibition and subsequent Atg13 dephosphorylation, specific steps associated with Atg1 activation occur (15, 19, 20): the Atg1-Atg13 complex forms, it associates with the ternary complex of autophagy-specific proteins (Atg17, Atg29, and Atg31), and Atg1 kinase is activated (16, 17, 19-21). Thus, we tested whether Atg13-8SA induced these events in growing cells without TORC1 inhibition. HA-tagged Atg1 (HAAtg1) was immunoprecipitated from Atg13-expressing cells and analyzed. As shown in Fig. 3A, the association of Atg1 with Atg13 and Atg17 and Atg1 kinase activity were significantly enhanced following Atg13-8SA expression. The faster-migrating Atg13WT coprecipitated with Atg1 (Fig. 3A, lane 2), presumably because it exists as a mixture of phosphorylated forms (Fig. 3A, lane 8) (19). However, this low degree of phosphorylation might not be sufficient to stabilize Atg17 within the complex. Atg1 activity was slightly increased prior to Atg13-8SA induction, and this is likely due to the expression of undetectable levels of Atg13-8SA that were not able to induce Atg1 activation (Fig. 3A, lane 4).
FIG. 3.
Atg13-8SA promotes Atg1 complex formation, Atg1 activation, and PAS organization. (A) Formation of the Atg1 complex. Cells (YYK412) harboring the indicated p416GAL1[ATG13] plasmid grown in SCRaf were incubated in SCRafGal for 2 h to induce Atg13 protein. Atg13 and Atg17 coimmunoprecipitated with in HAAtg1 were detected by immunoblotting. The catalytic activity of Atg1 kinase was also examined using myelin basic protein (MBP) as a substrate. The asterisk denotes a nonspecific band in the total cell lysates that was recognized by the anti-HA ascites. α, anti. (B) PAS assembly of Atg proteins. Cells (TMK625, atg11Δ) grown in SCRaf were shifted to SCRafGal for 1 h to induce Atg13. GFP-Atg17 and mCherry-Atg8 were observed by fluorescence microscopy.
The recruitment of Atg proteins to the PAS requires Atg17, indicating that Atg17 plays a central role in PAS organization (43). Organization of the PAS in atg11Δ cells is tightly regulated by nutrient conditions, since Atg17 protein assembles at the PAS only under autophagy-inducing conditions (20, 40). Thus, localization of Atg17 in Atg13-8SA-expressing atg11Δ cells was examined by fluorescence microscopy. While GFP-tagged Atg17 was uniformly distributed throughout the cytosol in ATG13WT cells, it localized to the PAS in ATG13-8SA-expressing cells (Fig. 3B). Atg8 tagged with mCherry was also recruited to the PAS in ATG13-8SA cells. Together, these results suggest that dephosphorylation of Atg13 is sufficient to mediate the formation and activation of the Atg1 complex and assembly of Atg proteins at the PAS.
Atg13-8SA induces autophagy in normally growing cells.
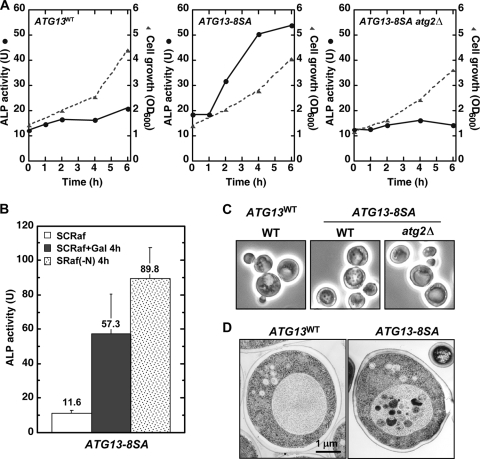
We next examined whether Atg13-8SA expression can induce autophagy. First, we performed an alkaline phosphatase (ALP) assay, an established method to monitor autophagy (33). ALP activity was significantly increased when expression of Atg13-8SA was controlled by the GAL1 promoter, demonstrating that autophagy is induced in ATG13-8SA cells (Fig. 4A). In contrast, ATG13WT and ATG13-8SA atg2Δ cells showed only a basal level of ALP activity. There was a low level of ALP activity in ATG13-8SA cells before Atg13-8SA induction, and this is likely related to the slight Atg1 kinase activity seen under these conditions (Fig. 3A). Autophagic activity was induced 4- to 6-fold after expression of Atg13-8SA for 4 h, and it reached 50 to 70% of that seen in cells subjected to nitrogen starvation or rapamycin treatment (Fig. 4B). Expression of Atg13-8SA had only a slight synergistic effect on rapamycin-induced autophagy, and rapamycin treatment had little effect on the formation of the Atg1 complex or Atg1 kinase activity in Atg13-8SA-expressing cells (Fig. 5A and B). These results suggest that dephosphorylation of Atg13 can mimic TORC1 inactivation to induce autophagy.
FIG. 4.
Atg13-8SA induces autophagy in normally grown cells. (A) Pho8Δ60-expressing (OND88) and YYK757 (atg2Δ) cells harboring the indicated p416GAL1[ATG13] plasmids were grown in SCRaf and shifted to SCRafGal. Alkaline phosphatase (ALP) activity (U, solid line) and cell growth (OD600, broken line) were monitored. (B) Pho8Δ60-expressing cells harboring the p306GAL1[ATG13-8SA] plasmid were subjected to the ALP assay before or after a 4-h incubation with SCRafGal or a 4-h incubation with nitrogen-depleted medium [SRaf(−N)]. The ALP activity of each sample is shown. Error bars represent the standard deviations [SD] from three independent experiments. (C) Accumulation of autophagic bodies. Cells used for panel A were incubated in SCRafGal in the presence of 1 mM PMSF (to inhibit degradation of autophagic bodies) for 6 h. Autophagic bodies were visualized by phase-contrast microscopy. (D) Electron microscopy of cells used for panel B. Bar, 1 μm.
FIG. 5.
Expression of Atg13-8SA mimics TORC1 inactivation to induce autophagy. (A) Cells grown in SCRaf were treated with 0.2 μg/ml rapamycin for 4 h, incubated in SCRafGal for 4 h, or incubated in SCRafGal for 2 h before a 2-h treatment with 0.2 μg/ml rapamycin (Rapa). Atg1 complex formation and Atg1 kinase activity were examined as for Fig. 3A. α, anti. (B) Pho8Δ60-expressing cells harboring the indicated p306GAL1[ATG13] plasmids were treated as for panel A and subjected to the ALP assay. Error bars represent the SD from three independent experiments.
We next monitored autophagy by observing autophagic body formation by phase-contrast and electron microscopy. Accumulation of autophagic bodies in the vacuole was observed in ATG13-8SA cells but not in ATG13WT or ATG13-8SA atg2Δ cells (Fig. 4C and D). It should also be noted that cell growth requires Tor function, and expression of Atg13-8SA with resulting Atg1 activation did not inhibit Tor activity or affect cell growth.
We next wished to identify individual serine residues whose phosphorylation prevented autophagy from proceeding. As shown in Fig. 6A, expression of Atg13-4SAa failed to induce autophagy to an extent comparable to that for wild-type Atg13, indicating that the presence of Ser residues 348, 496, and 535/541 is needed for phosphorylation. In particular, reversion of residue 496 to serine, a group 3 residue located in the Atg1-binding domain, completely abrogated autophagy induction (comparison of Atg13-7SAb and Atg13-8SA). However, other residues are clearly involved in autophagy induction because, even in the presence of S496A, revertant mutants still abrogated autophagy induction (i.e., Atg13-5SA). Thus, there may be some functional redundancy present in Atg13. Notably, the results of the ALP assay correlated well with those of the TORC1 phosphorylation assay (Fig. 6C). Considering the results together, Atg13-8SA induced autophagy to the greatest extent, confirming that the eight Ser residues identified above are involved in autophagy regulation.
FIG. 6.
Autophagy induction by Atg13 constructs. (A) Pho8Δ60-expressing cells harboring the indicated p416GAL1[ATG13] plasmids were subjected to the ALP assay before or after a 4-h incubation with SCRafGal. Error bars represent the SD from at least three independent experiments. (B) Atg13 proteins were detected by immunoblotting. (C) Summary of in vitro TORC1 assay and ALP assay using various Atg13 constructs. The shaded box represents the Atg1-binding site.
We also generated an Atg13 Ser-to-Asp mutant (Atg13-8SD), and this mutant induced autophagy to an intermediate extent compared to Atg13-8SA and Atg13WT. Thus, the Asp residues may mimic a phosphorylated form of Atg13 (Fig. 6A), and when overexpressed in starved cells, it was still able to induce autophagy (data not shown).
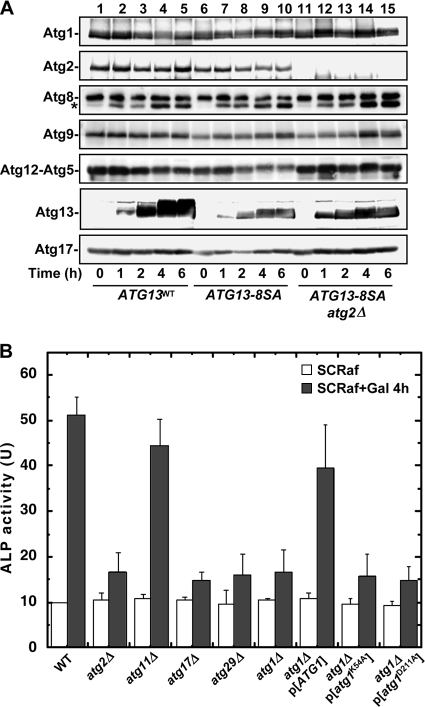
Atg8 expression is increased following Tor inactivation (23), and we wished to examine Atg8 in greater detail. Using an antibody with 12-fold-greater affinity for conjugated Atg8-PE (11), we estimated the amounts of unconjugated and total Atg8, and overall Atg8 expression appeared to be unchanged by Atg13 expression (Fig. 7A). However, Atg8-PE formation was stimulated by Atg13 expression, but there were no differences between ATG13WT and ATG13-8SA cells. These similarities may partially explain why autophagy is induced by Atg13-8SA expression to a lesser extent than nitrogen starvation, because Atg8 levels directly affect autophagosome size (48). We also examined the levels of other Atg proteins by immunoblotting, and there were no substantial differences between ATG13WT and ATG13-8SA cells (Fig. 7A).
FIG. 7.
Effect of Atg13-8SA on other Atg proteins. (A) Atg13-8SA expression does not significantly affect Atg protein expression. Atg protein levels in cells used for panel A were assessed by immunoblotting. The asterisk denotes the phosphatidylethanolamine-conjugated form of Atg8. (B) Requirement for ATG genes for autophagy induced by Atg13-8SA expression. Cells harboring the indicated mutations carrying the p416GAL1[ATG13-8SA] plasmid were subjected to the ALP assay before or after a 4-h incubation with SCRafGal to induce Atg13-8SA expression. Error bars represent the SD form three independent experiments.
As expected, the induction of autophagy by Atg13-8SA expression required other ATG genes essential for starvation-inducing autophagy, including ATG1, ATG2, ATG17, and ATG29 (Fig. 7B). Additionally, Atg13-8SA-induced autophagy was not inhibited by deletion of ATG11/CVT9; it is dispensable for starvation-induced autophagy (22). Finally, Atg1 catalytic activity is required for Atg13-8SA-induced autophagy as shown the abrogation of autophagy in the Atg1 kinase-deficient atg1K54A and atg1D211A mutants (19, 28). When all of our results are considered together, our data indicate that expression of Atg13-8SA is sufficient for autophagy induction by mimicking autophagy-inducing conditions, bypassing TORC1 signaling.
DISCUSSION
In this study we investigated the mechanism(s) regulating autophagy, and we found that TORC1 prevents autophagy by directly phosphorylating Atg13 at multiple Ser residues. Atg13 is dephosphorylated upon TORC1 inactivation, allowing it to associate with Atg1. Atg1-Atg13 complex formation leads to Atg1 activation, and the recruitment of other Atg proteins promotes PAS assembly. We further confirmed that Atg1-Atg13 does not induce Atg8 expression, despite the increased Atg8 levels seen with TORC1 inactivation (23). Therefore, TORC1 likely regulates autophagy through at least two independent pathways: the induction of autophagy by phosphorylation of Atg13 and enlargement of the autophagosomal membrane by expression of Atg8.
Protein kinase A (PKA) (a cyclic AMP [cAMP]-dependent protein kinase) also directly phosphorylates Atg13 (41, 49). Both Tor and PKA are required for cell growth, but the addition of cAMP prevents rapamycin-induced autophagy (34). However, TORC1 plays a larger role in Atg13 phosphorylation than PKA, and inactivation of PKA does not affect the mobility of Atg13 in SDS-PAGE (41, 49). There is evidence that PKA is under the control of the TORC1 pathway (27), and further studies are needed to clarify the interaction of PKA and TORC1 in cell growth and autophagy. To better understand the factors regulating Atg13 activity, we sought to identify a phosphatase that acted upon Atg13. Toward that end, we screened 30 nonessential phosphatase disruptants, but the phosphorylation state of Atg13 was not affected in any of the mutants examined (T. Funakoshi and Y. Ohsumi, unpublished results). Thus, it is possible that Atg13 might not be dephosphorylated by a specific phosphatase activated by cell starvation. Consequently, we favor a model wherein the phosphorylation status of Atg13 is predominantly regulated by TORC1 activity.
Two TORC1 substrates have been identified in Saccharomyces cerevisiae, Tap42 (13) and Sch9 (46). Inactivation of neither Tap42 nor Sch9 induces autophagy (19, 49), and our findings indicate that the TORC1-Atg13-Atg1 axis comprises a novel branch of TORC1 signaling specific for autophagy induction. Mammalian and fly homologs of the Atg1 complex were recently reported (3, 8, 10), and the TORC1-Atg13-Atg1 complex may be conserved among eukaryotes. Unlike the case for yeast, formation of the mammalian Atg1 (ULK1) complex does not appear to be regulated by nutrient conditions; the Atg13-Atg1 association is constitutively detected (3, 4, 10, 14). This difference likely arises from the dual functions of ULK1 in autophagy (as Atg1) and axonal elongation (as UNC-51) (36). Thus, constitutive binding of Atg13 to ULK1 may restrict its function in autophagy (as Atg1) and neural development (as UNC-51).
Expression of Atg13-8SA, an unphosphorylatable form of Atg13, is sufficient for induction of autophagy irrespective of TORC1 activity and cellular nutrient conditions. This is the first report demonstrating an experimental system leading to autophagy induction in normally growing cells under nutrient-rich conditions. Overexpression of dAtg1 in Drosophila induces autophagy (39). However, Atg1 homologs in multicellular organisms also function in axonal elongation (like UNC-51 of Caenorhabditis elegans) (36), and overexpression of dAtg1 itself may have other effects than autophagy. Therefore, these studies did not exclude the possibilities that other cellular effects or stress could promote autophagy induction. Moreover, Drosophila cells overexpressing dAtg1 are not able to maintain normal growth and ultimately die.
A recurring theme in the autophagy literature is the identification of roles for autophagy in unexpected physiologic phenomena (29). However, most of these studies have examined nutrient-starved or rapamycin-treated cells in which Tor function and its diverse downstream targets are affected. Thus, it is important to distinguish the role of autophagy per se from the many cellular responses to nutrient starvation. Based on this study, Atg13-8SA may be a useful tool for these purposes.
Acknowledgments
We thank A. Nakashima, M. Ohneda, H. Yamamoto, and M. Nishimura for providing yeast strains and materials; H. Nakatogawa, K. Okamoto, and members of the Ohsumi laboratory for discussions; and the National Institute for Basic Biology Center for Analytical Instruments for technical assistance.
This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
We declare no conflict of interest.
Footnotes
Published ahead of print on 7 December 2009.
REFERENCES
- 1.Baba, M., K. Takeshige, N. Baba, and Y. Ohsumi. 1994. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124:903-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 3.Chan, E. Y., A. Longatti, N. C. McKnight, and S. A. Tooze. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29:157-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, Y. Y., and T. P. Neufeld. 2009. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 20:2004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funakoshi, T., A. Matsuura, T. Noda, and Y. Ohsumi. 1997. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192:207-213. [DOI] [PubMed] [Google Scholar]
- 6.Hanada, T., N. N. Noda, Y. Satomi, Y. Ichimura, Y. Fujioka, T. Takao, F. Inagaki, and Y. Ohsumi. 2007. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282:37298-37302. [DOI] [PubMed] [Google Scholar]
- 7.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 8.Hara, T., A. Takamura, C. Kishi, S. Iemura, T. Natsume, J. L. Guan, and N. Mizushima. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, C., M. Baba, Y. Cao, and D. J. Klionsky. 2008. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol. Biol. Cell 19:5506-5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosokawa, N., T. Hara, T. Kaizuka, C. Kishi, A. Takamura, Y. Miura, S. I. Iemura, T. Natsume, K. Takehana, N. Yamada, J. L. Guan, N. Oshiro, and N. Mizushima. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20:1981-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichimura, Y., Y. Imamura, K. Emoto, M. Umeda, T. Noda, and Y. Ohsumi. 2004. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 279:40584-40592. [DOI] [PubMed] [Google Scholar]
- 12.Jacinto, E., and M. N. Hall. 2003. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell Biol. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung, C. H., C. B. Jun, S. H. Ro, Y. M. Kim, N. M. Otto, J. Cao, M. Kundu, and D. H. Kim. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabeya, Y., Y. Kamada, M. Baba, H. Takikawa, M. Sasaki, and Y. Ohsumi. 2005. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16:2544-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeya, Y., T. Kawamata, K. Suzuki, and Y. Ohsumi. 2007. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 356:405-410. [DOI] [PubMed] [Google Scholar]
- 17.Kabeya, Y., N. N. Noda, Y. Fujioka, K. Suzuki, F. Inagaki, and Y. Ohsumi. 2009. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 389:612-615. [DOI] [PubMed] [Google Scholar]
- 18.Kamada, Y., Y. Fujioka, N. N. Suzuki, F. Inagaki, S. Wullschleger, R. Loewith, M. N. Hall, and Y. Ohsumi. 2005. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25:7239-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawamata, T., Y. Kamada, Y. Kabeya, T. Sekito, and Y. Ohsumi. 2008. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 19:2039-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamata, T., Y. Kamada, K. Suzuki, N. Kuboshima, H. Akimatsu, S. Ota, M. Ohsumi, and Y. Ohsumi. 2005. Characterization of a novel autophagy-specific gene, ATG29. Biochem. Biophys. Res. Commun. 338:1884-1889. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J., Y. Kamada, P. E. Stromhaug, J. Guan, A. Hefner-Gravink, M. Baba, S. V. Scott, Y. Ohsumi, W. A. Dunn, Jr., and D. J. Klionsky. 2001. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153:381-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirisako, T., M. Baba, N. Ishihara, K. Miyazawa, M. Ohsumi, T. Yoshimori, T. Noda, and Y. Ohsumi. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klionsky, D. J. 2005. The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky, D. J., and Y. Ohsumi. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1-32. [DOI] [PubMed] [Google Scholar]
- 26.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 27.Martin, D. E., A. Soulard, and M. N. Hall. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969-979. [DOI] [PubMed] [Google Scholar]
- 28.Matsuura, A., M. Tsukada, Y. Wada, and Y. Ohsumi. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192:245-250. [DOI] [PubMed] [Google Scholar]
- 29.Mizushima, N., B. Levine, A. M. Cuervo, and D. J. Klionsky. 2008. Autophagy fights disease through cellular self-digestion. Nature 451:1069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakashima, A., Y. Maruki, Y. Imamura, C. Kondo, T. Kawamata, I. Kawanishi, H. Takata, A. Matsuura, K. S. Lee, U. Kikkawa, Y. Ohsumi, K. Yonezawa, and Y. Kamada. 2008. The yeast Tor signaling pathway is involved in G2/M transition via polo-kinase. PLoS One 3:e2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakatogawa, H., Y. Ichimura, and Y. Ohsumi. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130:165-178. [DOI] [PubMed] [Google Scholar]
- 32.Nakatogawa, H., K. Suzuki, Y. Kamada, and Y. Ohsumi. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458-467. [DOI] [PubMed] [Google Scholar]
- 33.Noda, T., A. Matsuura, Y. Wada, and Y. Ohsumi. 1995. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210:126-132. [DOI] [PubMed] [Google Scholar]
- 34.Noda, T., and Y. Ohsumi. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963-3966. [DOI] [PubMed] [Google Scholar]
- 35.Obara, K., T. Noda, K. Niimi, and Y. Ohsumi. 2008. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells 13:537-547. [DOI] [PubMed] [Google Scholar]
- 36.Ogura, K., C. Wicky, L. Magnenat, H. Tobler, I. Mori, F. Muller, and Y. Ohshima. 1994. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 8:2389-2400. [DOI] [PubMed] [Google Scholar]
- 37.Oshiro, N., R. Takahashi, K. Yoshino, K. Tanimura, A. Nakashima, S. Eguchi, T. Miyamoto, K. Hara, K. Takehana, J. Avruch, U. Kikkawa, and K. Yonezawa. 2007. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J. Biol. Chem. 282:20329-20339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 39.Scott, R. C., G. Juhasz, and T. P. Neufeld. 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shintani, T., and D. J. Klionsky. 2004. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279:29889-29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephan, J. S., Y.-Y. Yeh, V. Ramachandran, S. J. Deminoff, and P. K. Herman. 2009. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. U. S. A. 106:17049-17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki, K., T. Kirisako, Y. Kamada, N. Mizushima, T. Noda, and Y. Ohsumi. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki, K., Y. Kubota, T. Sekito, and Y. Ohsumi. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12:209-218. [DOI] [PubMed] [Google Scholar]
- 44.Takeshige, K., M. Baba, S. Tsuboi, T. Noda, and Y. Ohsumi. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169-174. [DOI] [PubMed] [Google Scholar]
- 46.Urban, J., A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, J. R. Broach, C. De Virgilio, M. N. Hall, and R. Loewith. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26:663-674. [DOI] [PubMed] [Google Scholar]
- 47.Xie, Z., and D. J. Klionsky. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102-1109. [DOI] [PubMed] [Google Scholar]
- 48.Xie, Z., U. Nair, and D. J. Klionsky. 2008. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19:3290-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yorimitsu, T., S. Zaman, J. R. Broach, and D. J. Klionsky. 2007. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 18:4180-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]