Abstract
All cells rely on DNA polymerases to duplicate their genetic material and to repair or bypass DNA lesions. In humans, 16 polymerases have been identified, and each bears specific functions in genome maintenance. We identified here the recently discovered polymerase POLN to be involved in repair of DNA cross-links. Such DNA lesions are highly toxic and are believed to be repaired by the sequential activity of nucleotide excision repair, translesion synthesis, and homologous recombination mechanisms. By functionally assaying its role in these processes, we unraveled an unexpected involvement of POLN in homologous recombination. Moreover, we obtained evidence for physical and functional interaction of POLN with factors belonging to the Fanconi anemia pathway, a master regulator of cross-link repair. Finally, we show that POLN interacts and cooperates in DNA repair with the helicase HEL308, which shares a common origin with POLN in the Drosophila mus308 gene. Our data indicate that this novel polymerase-helicase complex participates in homologous recombination repair and is essential for cellular protection against DNA cross-links.
Cells are continuously threatened by DNA damage that can potentially alter their genetic information. Numerous overlapping mechanisms exist to deal with DNA lesions. During S phase, unrepaired DNA lesions can block the progression of the replication machinery, causing cell cycle arrest and cell death. Cells have evolved mechanisms that signal the stalling of replication forks and recruit enzymes that can quickly restart these structures. Thus, replication forks can bypass the lesion without removing it. Homologous recombination (HR) and translesion synthesis (TLS) represent the most common mechanisms for dealing with stalled replication forks. In HR, the information on the newly replicated sister chromatid is used to overcome the lesion (32). In contrast, TLS uses special DNA polymerases that are able to replicate damaged DNA. Often, however, TLS polymerases insert wrong nucleotides across lesions, leading to mutations. Most polymerases involved in TLS belong to the Y family of DNA polymerases (2, 7, 29), which in addition to their characteristic catalytic domains and PCNA interaction protein (PIP) motifs, also have ubiquitin-binding domains toward their C termini. These domains are required for interaction with ubiquitinated PCNA and are essential for their recruitment to stalled replication forks. PCNA is an essential cofactor of DNA polymerases during replication, and it becomes monoubiquitinated when the replication fork is stalled at a damaged site (22).
POLN (DNA polymerase ν) is a recently discovered enzyme, belonging to the A family of DNA polymerases (16). In vitro, POLN is capable of DNA-templated synthesis, but it shows high mutagenicity, with a preference for inserting T across G and G across T (37). However, its lesion bypass activity is restricted to a subclass of DNA damage, namely, thymine glycol lesions (37). This suggests that POLN might not act as a TLS polymerase in vivo. Surprisingly, however, POLN was shown to have a high strand displacement activity (37). POLN has homology with the C-terminal part of the Drosophila gene mus308, which is specifically required for repair of interstrand DNA cross-links (3, 9, 16). Mus308 contains an N-terminal helicase domain and a C-terminal polymerase domain. The vertebrate homologues of Mus308 also include POLQ (DNA polymerase θ), containing both domains, and HEL308, only encompassing the helicase domain. Experiments in avian DT40 cells showed that concomitant loss of POLN and POLQ, as well as of HEL308 and POLQ, does not result in hypersensitivity to DNA cross-link-inducing agents; instead, a function in base excision repair was identified for POLQ (40).
DNA interstrand cross-links are highly toxic lesions, since they covalently couple the two DNA strands, and block unwinding by the replication and transcription machineries (33). Repair of such lesions occurs predominantly during replication, when it is initiated by the stalling of the replication machinery (30). Cross-link repair appears to require the successive participation of three classic DNA repair mechanisms: nucleotide excision repair (NER), TLS, and HR (25). According to the current model, dual incision on each side of the cross-link by the NER endonuclease complexes MUS81-EME1 and XPF-ERCC1 unhook the lesion, allowing bypass by the TLS polymerases REV1 (inserts a nucleotide across the cross-linked base) and Polζ (extends a few bases from that original insertion); finally, HR is used to copy the processed strand on the sister chromatid (8, 26, 27, 30). A second, recombination-independent, minor pathway has also been proposed, involving two consecutive steps of NER and TLS (33). Several polymerases, including REV1, Polζ, POLH, and POLK, have been shown to participate in cross-link bypass, which also involves PCNA ubiquitination (19, 21, 34). It is possible that the exact nature of the cross-link dictates the type of repair and the enzymes involved.
How the factors participating in cross-link repair are coordinated is not yet understood. However, a group of genes with regulatory activities required for interstrand cross-link (ICL) repair has been identified by studying the human syndrome Fanconi anemia (FA) (5, 13). FA is characterized by bone marrow failure, developmental abnormalities, and cancer proneness. Cells from FA patients are hypersensitive to cross-link-inducing agents, and it is believed that the pathological characteristics of the disease are caused by accumulation of DNA damage in progenitor stem cells. Thirteen FA genes have been identified, and most of the encoded proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and the associated FAAP100) form a large nuclear complex with ubiquitin ligase activity. During S phase, this complex is targeted to sites of DNA damage by the FANCM-FAAP24 heterodimer and monoubiquitinates the substrates FANCI and FANCD2 in a highly specific reaction (4, 6, 36). This leads to relocalization of the ubiquitinated substrates to chromatin foci, together with other FA gene products (FANCD1/BRCA2, FANCN, and FANCJ) and several DNA repair factors (RAD51, PCNA, NBS1, and H2AX) (13, 31). How these events control removal of DNA cross-links is unknown.
Experiments measuring different repair pathways in FA patient cells revealed deficiencies in TLS and HR, suggesting that the FA pathway normally regulates these activities (20, 24, 27, 28). Although HR was shown to be dependent on ubiquitination of FANCD2-I by the core complex (24), TLS seems to be controlled by the core complex independent of this event and independent of PCNA ubiquitination (20). Thus, the FA pathway appears to be forked, with the core complex controlling two branches: one that activates HR via FANCD2-I and one that promotes mutagenic repair via an unknown mechanism of recruiting TLS polymerases.
Here, we show that POLN is required for repair of DNA cross-links in mammalian cells. By dissecting the different steps of ICL repair, we found that, surprisingly, POLN is involved in HR, and it interacts physically and genetically with FA proteins.
MATERIALS AND METHODS
Cell culture and protein techniques.
HeLa, 293T, and U2OS cells were grown in Dulbecco modified Eagle medium (Invitrogen) supplemented with 15% fetal calf serum. For cell cycle synchronization, cells were incubated with 2.5 mM thymidine for 18 h, washed twice with fresh medium, incubated again with thymidine for 18 h, washed, and released in medium containing 100 ng of nocodazole/ml. Preparation of whole-cell extracts, chromatin fractionation, immunoprecipitation assays, and immunofluorescence experiments were performed as previously described (14). The immunoprecipitation experiments were performed in the presence of the DNA-digesting enzyme benzonase. For immunoprecipitation from chromatin, protein complexes were eluted from chromatin by 300 mM NaCl for 5 min on ice. Eluates were diluted threefold to reduce salt concentration and subjected to immunoprecipitation.
Antibodies.
Rabbit polyclonal anti-POLN (R) antibodies were obtained by immunizing with a recombinant POLN fragment spanning the N-terminal 100 amino acids. Mouse polyclonal anti-POLN (M) was purchased from Abnova. Other antibodies used included γH2AX and PCNA (both from Abcam), FANCA and FANCG (39), Rad51 (Novus), green fluorescent protein (GFP; Clontech), FANCD2, vinculin, cyclin B1, HEL308, hemagglutinin (HA), and FLAG M5 (all from Santa Cruz Biotechnology) and FANCI (a rabbit polyclonal antibody generated in our lab). For Flag immunoprecipitation, anti-Flag beads were used (M2; Santa Cruz).
Plasmids and small interfering RNA (siRNA).
POLN cDNA was kindly provided by Richard Wood (MD Anderson, Houston, TX). The cDNA for HEL308 was purchased from PlasmID. For transient transfection, the open reading frames were cloned into pFLAG-080, obtained from Jurg Tschopp (University of Lausanne, Lausanne, Switzerland). Plasmid transfections were performed with Lipofectamine 2000 (Invitrogen) or GeneJuice (Novagen). For stable expression of POLN, the pWZL retroviral system (Millennium Pharmaceuticals) was used.
All siRNA oligonucleotides were purchased from Qiagen, with the exception of siHEL308, which was obtained from Invitrogen (for sequences, see the supplemental material). As controls, AllStars Negative Control (Qiagen) and Stealth RNAi negative controls (Invitrogen) were used. Lipofectamine RNAiMAX (Invitrogen) and Hiperfect (Qiagen) were used for siRNA transfection, For creating stable shRNA-mediated POLN knockdown cells, the lentiviral Plko system (Open Biosystems) was used.
Drug sensitivity assays.
For survival assays, cells were transfected with siRNA in 96-well plates. After 2 days, the respective drugs were added to the wells. Three days later, cellular viability was assayed by using the CellTiterGlo reagent (Promega). Unless otherwise indicated in the figure legends, the results shown are averages of at least five experiments, and error bars represent the standard deviations. For clonogenic assays, two rounds of siRNA were performed, 24 h apart. The day after the second siRNA round, 250 to 2,000 cells were plated in each dish, and 8 h later the drug was added. Colony formation was scored after 2 weeks. The results shown are averages of at least two experiments, and error bars represent the standard deviations.
Functional assays.
SupF assays for measuring mutagenesis frequency (20) and HR assays using the DR-GFP reporter (24) were performed as previously described. At least two or three assays were averaged.
RESULTS
POLN is required for cross-link repair.
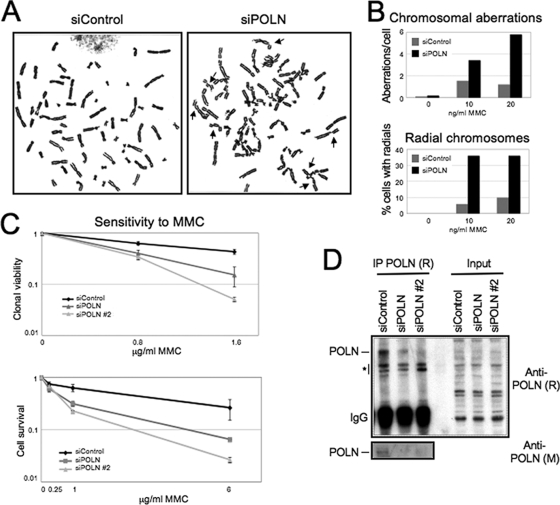
In an effort to identify DNA polymerases involved in cross-link repair, we performed a small-scale RNA interference screen using a siRNA library targeting human DNA polymerases, in HeLa cells (see Fig. S1 in the supplemental material). Increased sensitivity to the cross-link-inducing drug cisplatin was scored. Two hits were identified: REV1, previously implicated in cross-link repair in human and avian cells (20, 27), and POLN. To confirm that POLN is required for cross-link repair, we analyzed metaphase spreads of 293T cells depleted of POLN by siRNA (Fig. 1A). We observed that the loss of POLN resulted in a dramatic increase in chromosomal aberrations after treatment with mitomycin C (MMC) (Fig. 1B). Among these aberrations, there was a marked increase in radial chromosomes, which are considered specific cytological hallmarks of cells deficient in the FA pathway. Expression of a siRNA resistant POLN variant could suppress MMC-induced chromosomal aberrations, as well as radial formation (see Fig. S2 in the supplemental material). In contrast, the POLN D624A mutant, which was shown to lack enzymatic activity in vitro (16), did not reduce MMC-induced radial chromosome formation, the way that wild-type POLN did (see Fig. S3 in the supplemental material). Next, we investigated whether the increase in chromosome aberrations after POLN depletion leads to cellular sensitivity to cross-linking damage. We knocked down POLN, using two different siRNA oligonucleotides or used lentivirus-mediated shRNA stable depletion in different cell lines. Cells depleted of POLN were consistently sensitive to the DNA ICL-inducing agent MMC (Fig. 2C; also see Fig. S4 in the supplemental material).
FIG. 1.
POLN is involved in cross-link repair. (A) Metaphase spreads of 293T cells treated with POLN siRNA or control siRNA and exposed to 15 ng of MMC/ml. Arrows indicate examples of chromatid breaks and radial chromosomes in POLN-depleted cells. (B) Quantification of total chromosomal aberrations and of radial chromosomes after siRNA-mediated POLN or control depletion from 293T cells. For each condition, 50 cells were analyzed. (C) Depletion of POLN by siRNA confers MMC sensitivity to HeLa cells in both clonogenic assays and short-term cell survival assays. (D) Western blot showing POLN knockdown by siRNA. Endogenous POLN could only be detected after enrichment by immunoprecipitation, but not in whole-cell extracts (input samples). The two anti-POLN antibodies used are identified by the species in which they were raised: rabbit (R) or mouse (M). Both of them could detect overexpressed POLN in extracts (see Fig. S5 in the supplemental material). The asterisk marks cross-reactive bands.
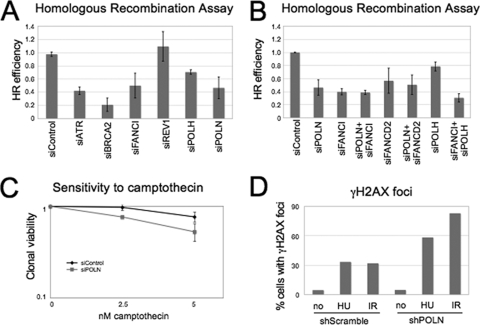
FIG. 2.
Efficient HR requires POLN. (A) POLN knockdown decreases HR. U2OS cells bearing the DR-GFP reporter, transfected with siRNA as indicated, were analyzed by flow cytometry. A different siRNA targeting POLN gave similar results (results not shown). (B) POLN is epistatic to FANCD2-I. HR proficiency was analyzed as in panel A. The efficiency of siRNA treatment is shown in Fig. S7 in the supplemental material. (A and B) Error bars represent the standard deviations. (C) Depletion of POLN by siRNA in HeLa cells confers sensitivity to DSB-inducing agent camptothecin, as assayed by clonal viability experiments. (D) HeLa cells bearing stable shRNA-mediated knockdown of POLN show increased γH2AX staining after treatment with 20 mM HU for 24 or 12 h after 5 Gy of IR irradiation. At least 50 cells were analyzed for each condition.
To detect endogenous POLN, we raised rabbit polyclonal antibodies against the amino terminus of the protein. Due to the very low levels of expression of this protein, the antibodies could not directly detect endogenous POLN in Western blots of whole-cell extracts. However, after enriching POLN by immunoprecipitation, the antibodies could specifically detect the endogenous protein by immunoblot, with the predicted molecular mass of 100 kDa. Moreover, a commercially available mouse anti-POLN antibody could detect the immunoprecipitated protein. Thus, we were able to verify that the siRNA oligonucleotides used diminish POLN protein levels (Fig. 1D).
We concluded that POLN is involved in the repair of DNA cross-links, and cells depleted of POLN have cytological phenotypes similar to FA cells.
POLN is required for efficient HR.
HR is considered an essential step in cross-link repair (25). The Fanconi anemia pathway has been shown to be required for efficient HR, as depletion of FANCD2 or FANCI leads to a 50% decrease in HR efficiency, when measured by a GFP-based HR reporter assay (24, 36). We investigated whether also POLN is involved in HR. In cells depleted of POLN, we observed an identical phenotype with FA-deficient cells, namely, reduction by 50% of HR efficiency (Fig. 2A). In contrast, we could not detect a HR defect after REV1 knockdown, confirming that the main function of REV1 is in TLS rather than in HR. The reduction in HR efficiency following POLN knockdown is not due to a deficiency in FA pathway activation, as FANCD2 is normally ubiquitinated in the absence of POLN (see Fig. S6 in the supplemental material). POLH (DNA polymerase η), another polymerase involved in HR (11, 18), had a mild effect on recombination, namely, reduction to 75% (although this might also reflect insufficient depletion). Codepletion of FANCD2 (or FANCI) and POLN did not further decrease HR frequency (Fig. 2B), suggesting that POLN is epistatic with the FANCD2-I complex in this context. In contrast, knockdown of POLH and FANCI showed a possibly additive effect (Fig. 2B).
We next examined whether POLN is involved in repair of double-strand breaks (DSBs). DSB repair during the S and G2 phases mainly uses HR-based mechanisms (35). POLN knockdown conferred mild sensitivity to camptothecin (Fig. 2C) and bleomycin (see Fig. S8A in the supplemental material). Camptothecin is a topoisomerase I inhibitor that stabilizes this enzyme on DNA creating structures transformed into DSBs by the replication machinery, whereas bleomycin is a chemotherapeutic agent that causes DNA breaks. Notably, in cells bearing stable knockdown of POLN by shRNA we detected an increase in γH2AX staining after treatment with hydroxyurea or exposure to ionizing irradiation (IR), agents that also cause double strand breaks (Fig. 2D; also see Fig. S8B in the supplemental material). This result suggests the accumulation of unrepaired DSBs in the absence of POLN. Moreover, immunofluorescence studies concluded that POLN forms chromatin foci in response to exposure to several agents that can cause DSBs, including MMC and IR (see Fig. S9 in the supplemental material), again suggesting a participation of POLN in the repair of these structures. In conclusion, we identified a novel function of POLN in promoting HR repair and DSB resistance, which may be regulated by the FA proteins FANCD2 and FANCI.
Since POLN was previously characterized as a high-error-rate polymerase in vitro (1, 37), we also analyzed the mutation frequency of 293T cells depleted of POLN, by using a plasmid-based assay that allows accurate identification of point mutations and small insertions and/or deletions (15, 20). Surprisingly, the total number of mutations was not decreased by knockdown of POLN. Instead, mutation frequency was slightly increased (see Fig. S10 in the supplemental material). This increase was consistently found when undamaged plasmid was used, or when the plasmid was damaged by UVC or cross-link damage (obtained by UVA and psoralen treatment), before transfection into mammalian cells. These results suggest that POLN suppresses accumulation of mutations in vivo.
POLN interacts physically with FANCD2-I and RAD51.
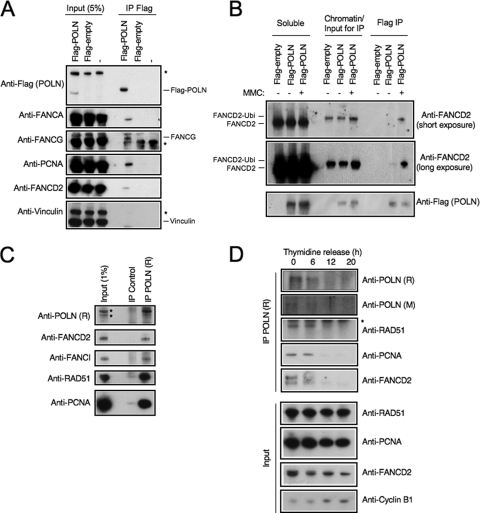
Prompted by the genetic connection between FANCD2-I and POLN for controlling HR, we tested for a possible physical interaction between these proteins. A transiently overexpressed Flag-tagged version of POLN coimmunoprecipitated with endogenous FANCD2 from 293T cells, as well as the core complex members, FANCA and FANCG (Fig. 3A). In addition, overexpressed HA-tagged POLN partly colocalized with FANCD2 in DNA damage-induced foci in HeLa cells (data not shown). Also interacting with Flag-POLN was PCNA, a known cofactor of DNA polymerases (Fig. 3A).
FIG. 3.
POLN interacts with proteins in the FA pathway. (A) 293T cells overexpressing Flag-tagged POLN or empty vector were subjected to anti-Flag immunoprecipitation. Samples were analyzed by Western blotting. Vinculin was used as negative control. Asterisks denote cross-reactive bands. (B) POLN interacts with ubiquitinated FANCD2 on chromatin. 293T cells expressing Flag-tagged POLN or empty vector, treated or not with MMC, were subjected to lysis and separation into soluble and chromatin-bound fractions. The chromatin faction was then used for Flag immunoprecipitation. (C) Interactions of endogenous POLN. Shown are anti-POLN immunoprecipitations from HeLa cell extracts. The bands in the Input lane, anti-POLN panel (marked with asterisks), represent cross-reactive proteins and not POLN. (D) Endogenous POLN is only detectable during S phase. HeLa cells were synchronized by double thymidine arrest, followed by release in nocodazole-containing media. At the indicated time points, corresponding to early S phase, late S phase, G2 phase, and mitosis, cells were harvested, and anti-POLN immunoprecipitation was performed. Samples were analyzed by Western blotting. The asterisk denotes a cross-reactive band.
Upon activation of the FA pathway by DNA damage, the core complex monoubiquitinates FANCD2 and FANCI, leading to their localization to chromatin foci. In order to investigate whether POLN interacts with the activated form of FANCD2, we treated cells expressing Flag-POLN with MMC and, after lysis, separated soluble proteins from the chromatin-bound fraction (Fig. 3B). As expected, unmodified FANCD2 was found in the soluble fraction, whereas ubiquitinated FANCD2, induced by MMC treatment, was on chromatin. When we used the chromatin fraction for Flag immunoprecipitations, we observed that ubiquitinated FANCD2 interacts specifically with POLN after MMC treatment, on chromatin, suggesting that POLN participates in FANCD2-mediated repair.
We next investigated these interactions at the endogenous level, by performing immunoprecipitations from HeLa cellular extracts with anti-POLN antibodies. FANCD2 and FANCI, as well as PCNA, interacted with POLN (Fig. 3C). Interestingly, the HR factor RAD51 was pulled down with POLN, further strengthening the hypothesis that POLN is involved in recombination-mediated repair.
The FA pathway is specifically activated during S-phase (38). In light of the physical and genetic connections between FA and POLN, we examined whether these interactions are regulated differentially during the cell cycle. We synchronized HeLa cells in S phase by double thymidine arrest and released them in nocodazole-containing medium, thus trapping cells in mitosis. Cells harvested at different time points after release were analyzed for POLN. The levels of POLN decreased as cells exited S phase (Fig. 3D). Concordantly, FANCD2, FANCI, RAD51, and PCNA coprecipitated with anti-POLN antibodies only in S phase. This result confirms the specificity of the POLN interactions we uncovered and rules out that these proteins are precipitated nonspecifically by the anti-POLN antibodies. Indeed, in immunoprecipitates of mitotic cells (in which POLN is absent), none of the tested interactors coimmunoprecipitated.
Preliminary experiments suggest that POLN depletion after S phase is caused mainly by increased protein degradation, rather than by transcriptional shutoff. A Flag-tagged variant of POLN, stably expressed in HeLa cells under the cytomegalovirus promoter, also showed decreased levels after S phase (see Fig. S11 in the supplemental material). The mechanism of this degradation is currently under investigation. We conclude that POLN interacts with FANCD2-I during S phase, when the FA pathway is active. Degradation of POLN following S-phase completion ensures that these interactions occur specifically during this phase of the cell cycle.
HEL308 cooperates with POLN in HR.
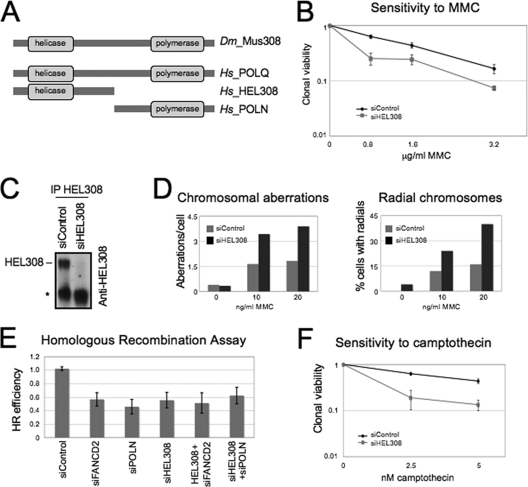
Mammalian homologues of the Drosophila gene mus308 include POLN, POLQ and HEL308 (Fig. 4A) (3, 9, 16, 17). POLQ is likely involved in base excision repair (40). HEL308, however, was of particular interest, since disruption of the HEL308 gene in C. elegans causes cross-link sensitivity (23). Moreover, in this system, HEL308 was epistatic with FANCD2 in cross-link repair (23). In light of the parallels between HEL308 phenotypes in nematodes and our own findings of POLN function in mammals, we investigated whether human HEL308 is involved in cross-link repair. Indeed, knockdown of HEL308 by siRNA in HeLa cells resulted in increased sensitivity to MMC (Fig. 4B). Since commercially available anti-HEL308 antibodies did not detect endogenous protein from cell lysates, we used immunoprecipitation to confirm the knockdown efficiency of the siRNA (Fig. 4C). Cytological analyses showed an increased in MMC-induced chromosomal aberrations, including radial chromosomes, in 293T cells depleted of HEL308 (Fig. 4D), confirming an important function for HEL308 in cross-link repair.
FIG. 4.
The HEL308 helicase cooperates with POLN in cross-link repair and HR. (A) Graphic representation (not to scale) of Drosophila mus308 and its human homologs. (B) Depletion of HEL308 by siRNA sensitizes HeLa cells to the cross-link-inducing drug MMC. (C) The siRNA targeting HEL308 confers efficient knockdown of the endogenous protein in HeLa cells, as evidenced by immunoprecipitation experiments. The endogenous protein (125 kDa) was depleted after treatment with siRNA. The asterisk denotes a cross-reactive band. (D) HEL308 protects cells against cross-link-induced genetic instability. Shown are quantifications of chromosomal aberrations observed after control or POLN depletion from 293T cells. For each condition, 50 cells were analyzed. (E) Examination of HR frequency using the DR-GFP reporter in HeLa cells reveals a function for HEL308 in promoting HR in cooperation with POLN and FANCD2. Error bars represent the standard deviations. (F) Depletion of HEL308 by siRNA leads to camptothecin sensitivity in clonogenic assays.
Next, we analyzed whether HEL308 is involved in HR. Knockdown of HEL308 resulted in a significant decrease in HR efficiency (Fig. 4E) and in cellular sensitivity to camptothecin (Fig. 4F) after HEL308 depletion, suggesting that, like POLN, HEL308 is involved in HR. Codepletion of HEL308 and POLN did not further reduce HR, suggesting that these factors are epistatic for HR (Fig. 4E). Interestingly, in this system, HEL308, like POLN, was epistatic with FANCD2. Moreover, overexpression of inactive HEL308 K365M mutant (17) resulted in reduced HR frequency (see Fig. S12 in the supplemental material), suggesting that the helicase activity is important for HR. Similar to POLN, HEL308 knockdown did not decrease mutagenesis efficiency, as measured by the SupF assay (data not shown). Knockdown of either POLN or HEL308 did not reduce RPA or RAD51 focus formation after IR irradiation (see Fig. S13 in the supplemental material), showing that they act downstream of these factors in DSB repair.
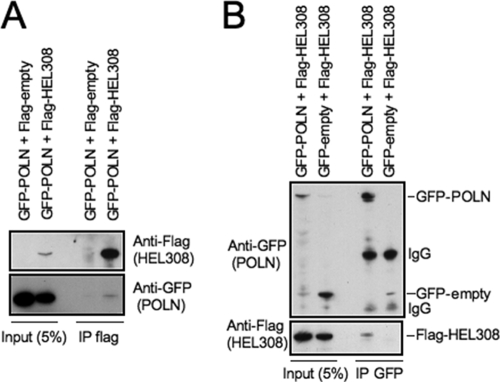
Overall, HEL308 or POLN knockdowns yield similar phenotypes, suggesting a common function. Thus, we investigated a possible physical interaction. Flag-tagged HEL308 and GFP-tagged POLN overexpressed in 293T cells, interacted in coimmunoprecipitation assays (Fig. 5). Taken together, our results suggest that POLN and HEL308 form a complex involved in HR-mediated repair.
FIG. 5.
HEL308 interacts with POLN. (A and B) Reciprocal coimmunoprecipitation assays, from 293T cells overexpressing Flag-HEL308 and GFP-POLN, revealed an interaction between the two proteins.
DISCUSSION
POLN and HEL308 cooperate in HR.
Here, we demonstrate that the recently described A family DNA polymerase POLN is required for efficient HR in mammalian cells. It is likely that POLN is directly involved in some step of HR requiring DNA polymerization. Recently, the Y family polymerase POLH was shown to perform D loop extension in vitro (11, 18). POLH therefore may function in one of the latest steps of HR, after strand invasion. Accordingly, loss of POLH in avian cells reduced HR efficiency (11). Indeed, we also observed a small decrease in HR frequency in human cells depleted of POLH (Fig. 2A and B). It is possible that POLN, like POLH, is involved in D-loop extension. POLN has marked strand displacement activity in vitro (37), a finding consistent with a role in D-loop extension. As do other DNA repair proteins, POLN shows high expression in testis (16). HR activity is known to be required for proper meiosis, and high POLN expression in testis might be associated with a possible involvement in meiotic recombination.
We also identified HEL308 as a POLN interacting protein (Fig. 5), and showed that these proteins may work together to promote HR (Fig. 4E). HEL308 is an active helicase in vitro (17), and its activity may further stimulate POLN-dependent D-loop extension. The two activities, helicase and polymerase, are present in a single polypeptide in Drosophila (3, 9), and it remains unclear why higher eukaryotes have evolved two separate proteins.
Under certain conditions, cells may need to use mutagenic rather than error free polymerases in the DNA synthesis step of HR repair. Since only short patches of DNA are synthesized during HR, a low processivity polymerase may be better suited to perform this function. Such an enzyme might dissociate from DNA after synthesizing a few tens of bases. In contrast, a high processivity replicative polymerase might perform synthesis over a long stretch of DNA and require special regulation to terminate the reaction. Alternatively, POLN may be deployed in a subset of HR reactions, namely, those requiring increased strand displacement activity and, possibly, those involving a cross-linked template.
Previously, double-knockout POLN/POLQ and HEL308/POLQ avian cells were competent for cross-link repair (40). In our hands, knockdown of POLN or HEL308 by RNA interference resulted in increased sensitivity to cross-link- and DSB-inducing agents. The reason for this discrepancy between avian and human systems is unclear. We observed that after several passages, HeLa cells bearing a stable shRNA-mediated knockdown of POLN become more resistant to damage, without restoring POLN levels (data not shown). Possibly, another polymerase is upregulated and takes over its function.
Novel POLN interacting proteins.
Endogenous POLN interacts with several DNA repair proteins, including FANCD2, FANCI, PCNA, and RAD51 (Fig. 3). PCNA may function as a cofactor for POLN, as it is the case for other polymerases (22). We could not identify a PIP box in POLN. It is possible that this interaction is mediated by some other type of PCNA interaction domain or that the PIP box is highly degenerate. Interestingly, we identified a putative PIP box in HEL308, raising the intriguing possibility that this helicase might mediate the PCNA-POLN interaction (G. L. Moldovan, unpublished observations). We could not exclude the possibility that the interaction between POLN and these repair proteins is indirect, reflecting a colocalization of these proteins to DNA repair structures.
Previous studies have shown that POLH interacts with RAD51, and this molecule can enhance the D-loop extension activity of POLH (18). RAD51 is an essential HR factor, which coats the single-stranded DNA formed after resection at the DSB and catalyzes the strand invasion step leading to the creation of the D loop (32). Thus, the interaction between POLN and RAD51 (Fig. 3), suggests that POLN might also be stimulated by RAD51 to perform DNA synthesis after strand invasion.
Finally, we reported an interaction between POLN and FANCD2 and FANCI (Fig. 3). Coupled with the genetic epistasis between POLN and FANCD2-I for HR efficiency (Fig. 2B), these interactions suggest an intimate connection among POLN and FA proteins. Perhaps, in a subset of HR reactions (namely, those initiated by the FA pathway), POLN, rather than POLH, is involved in DNA synthesis (see the model in Fig. S14 in the supplemental material). How this regulation is achieved remains unclear. An HR reaction initiated at a DNA cross-link might represent a specific structure, for which POLN is the most suitable polymerase. Still, POLN may also function in DNA repair outside the FA pathway. Only a fraction of POLN foci overlap with FANCD2 in immunofluorescence experiments (data not shown).
Surprisingly, depletion of POLN slightly increased the overall mutation frequency, as measured using the SupF assay (see Fig. S10 in the supplemental material). Although FA mutants are specifically hypersensitive to DNA cross-links, the FA pathway is activated by a variety of agents, including UV, IR, or HU. Moreover, FANCD2 depletion can also affect the outcome of the repair of other lesions than cross-links: UV-induced mutagenesis is increased (20), and DSB-induced HR repair is reduced (36). Thus, it is possible that the FA pathway also coordinates cellular responses to other types of DNA damages than DNA cross-links. During cross-link repair, a stepwise involvement of NER, TLS, and HR factors is required, and the FA pathway may control their orderly participation. However, in response to damages other than cross-links, which can be bypassed by either one repair mechanism or the other, the FA proteins might be involved in channeling the reaction into either REV1- or FANCD2-FANCI-POLN-dependent repair (see Fig. S14 in the supplemental material). In the absence of the latter branch, upregulation of REV1 may increase mutagenesis.
POLN interactions are regulated by cell cycle-dependent degradation.
POLN is specifically regulated during the cell cycle, with protein levels peaking in S phase (Fig. 3D). The mechanism of this regulation is unclear, but preliminary experiments suggest that it involves protein degradation rather than transcriptional shut off (see Fig. S11 in the supplemental material). The differential control of DNA repair pathways during the cell cycle is currently emerging as a major theme in the field. The unscheduled activation of certain repair mechanisms can be inefficient or detrimental for the global repair response. The preferential use of HR in S and G2 and NHEJ in G1 is well documented, involving CDK activity (10, 35). Recently, we showed that the FA protein FANCM, which recruits the FA core complex to sites of DNA damage, is degraded in mitosis, leading to inactivation of the FA pathway (12). Degradation of POLN after S-phase termination might ensure that this mutagenic polymerase does not negatively affect genomic integrity.
Different polymerases have evolved in higher eukaryotes to perform DNA synthesis in specialized conditions (2). Our identification of POLN (ν) function in HR adds a new entry to the long list of specialized DNA polymerase functions: POLL (λ), POLM (μ) and terminal deoxytransferase in nonhomologous end joining, POLB (β) and POLQ (θ) in base excision repair, POLA (α) in Okazaki fragment maturation, POLE (ɛ) and POLD1 (δ) on leading and, respectively, lagging strand synthesis during unperturbed replication, Y family polymerases in TLS, and POLH (η) in HR. Our results demonstrate the usage of noncanonical polymerases in recombinational repair and substantiate the idea that particular DNA synthesis reactions require specific DNA polymerases. How the cell controls these polymerases and what makes them best fitted to act in their specific function is currently of fundamental interest.
Supplementary Material
Acknowledgments
We thank Richard Wood for the gift of GFP-POLN construct; Michael Seidman for the SupF plasmid; Maria Jasin for the DR-GFP reporter; Jurg Tschopp for the pFLAG-080 vector; Hans Widlund and Jay Morgenstern for the pWZL retroviral system; Lisa Moreau for chromosome aberration analyses; Patricia Stuckert for the clonogenic sensitivity assays; and Jungmin Kim, Younghoon Kee, and Richard Kennedy for materials and advice.
G.-L.M. is supported by a Human Frontiers Science Program Organization postdoctoral fellowship. P.V. is recipient of a postdoctoral fellowship from the Swiss Foundation for Grants in Medicine and Biology. This study was supported by NIH grants RO1DK43889, RO1HL52725, PO1DK50654, and U19A1067751 to A.D.D.
Footnotes
Published ahead of print on 7 December 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arana, M. E., K. Takata, M. Garcia-Diaz, R. D. Wood, and T. A. Kunkel. 2007. A unique error signature for human DNA polymerase nu. DNA Repair 6:213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebenek, K., and T. A. Kunkel. 2004. Functions of DNA polymerases. Adv. Protein Chem. 69:137-165. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, J. B., K. Sakaguchi, and P. V. Harris. 1990. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics 125:813-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciccia, A., C. Ling, R. Coulthard, Z. Yan, Y. Xue, A. R. Meetei, H. Laghmani el, H. Joenje, N. McDonald, J. P. de Winter, W. Wang, and S. C. West. 2007. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell 25:331-343. [DOI] [PubMed] [Google Scholar]
- 5.de Winter, J. P., and H. Joenje. 2009. The genetic and molecular basis of Fanconi anemia. Mutat. Res. 668:11-19. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 7.Guo, C., J. N. Kosarek-Stancel, T. S. Tang, and E. C. Friedberg. 2009. Y-family DNA polymerases in mammalian cells. Cell Mol. Life Sci. 66:2363-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanada, K., M. Budzowska, M. Modesti, A. Maas, C. Wyman, J. Essers, and R. Kanaar. 2006. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 25:4921-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris, P. V., O. M. Mazina, E. A. Leonhardt, R. B. Case, J. B. Boyd, and K. C. Burtis. 1996. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell. Biol. 16:5764-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huertas, P., F. Cortes-Ledesma, A. A. Sartori, A. Aguilera, and S. P. Jackson. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455:689-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamoto, T., K. Araki, E. Sonoda, Y. M. Yamashita, K. Harada, K. Kikuchi, C. Masutani, F. Hanaoka, K. Nozaki, N. Hashimoto, and S. Takeda. 2005. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell 20:793-799. [DOI] [PubMed] [Google Scholar]
- 12.Kee, Y., J. M. Kim, and A. D. D'Andrea. 2009. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev. 23:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy, R. D., and A. D. D'Andrea. 2005. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19:2925-2940. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J. M., Y. Kee, A. Gurtan, and A. D. D'Andrea. 2008. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood 111:5215-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraemer, K. H., and M. M. Seidman. 1989. Use of supF, an Escherichia coli tyrosine suppressor tRNA gene, as a mutagenic target in shuttle-vector plasmids. Mutat. Res. 220:61-72. [DOI] [PubMed] [Google Scholar]
- 16.Marini, F., N. Kim, A. Schuffert, and R. D. Wood. 2003. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 278:32014-32019. [DOI] [PubMed] [Google Scholar]
- 17.Marini, F., and R. D. Wood. 2002. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 277:8716-8723. [DOI] [PubMed] [Google Scholar]
- 18.McIlwraith, M. J., A. Vaisman, Y. Liu, E. Fanning, R. Woodgate, and S. C. West. 2005. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell 20:783-792. [DOI] [PubMed] [Google Scholar]
- 19.Minko, I. G., M. B. Harbut, I. D. Kozekov, A. Kozekova, P. M. Jakobs, S. B. Olson, R. E. Moses, T. M. Harris, C. J. Rizzo, and R. S. Lloyd. 2008. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J. Biol. Chem. 283:17075-17082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirchandani, K. D., R. M. McCaffrey, and A. D. D'Andrea. 2008. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair 7:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mogi, S., C. E. Butcher, and D. H. Oh. 2008. DNA polymerase eta reduces the gamma-H2AX response to psoralen interstrand crosslinks in human cells. Exp. Cell Res. 314:887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moldovan, G. L., B. Pfander, and S. Jentsch. 2007. PCNA, the maestro of the replication fork. Cell 129:665-679. [DOI] [PubMed] [Google Scholar]
- 23.Muzzini, D. M., P. Plevani, S. J. Boulton, G. Cassata, and F. Marini. 2008. Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair 7:941-950. [DOI] [PubMed] [Google Scholar]
- 24.Nakanishi, K., Y. G. Yang, A. J. Pierce, T. Taniguchi, M. Digweed, A. D. D'Andrea, Z. Q. Wang, and M. Jasin. 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. U. S. A. 102:1110-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niedernhofer, L. J., A. S. Lalai, and J. H. Hoeijmakers. 2005. Fanconi anemia (cross)linked to DNA repair. Cell 123:1191-1198. [DOI] [PubMed] [Google Scholar]
- 26.Niedernhofer, L. J., H. Odijk, M. Budzowska, E. van Drunen, A. Maas, A. F. Theil, J. de Wit, N. G. Jaspers, H. B. Beverloo, J. H. Hoeijmakers, and R. Kanaar. 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niedzwiedz, W., G. Mosedale, M. Johnson, C. Y. Ong, P. Pace, and K. J. Patel. 2004. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell 15:607-620. [DOI] [PubMed] [Google Scholar]
- 28.Papadopoulo, D., C. Guillouf, H. Mohrenweiser, and E. Moustacchi. 1990. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc. Natl. Acad. Sci. U. S. A. 87:8383-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prakash, S., R. E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317-353. [DOI] [PubMed] [Google Scholar]
- 30.Raschle, M., P. Knipsheer, M. Enoiu, T. Angelov, J. Sun, J. D. Griffith, T. E. Ellenberger, O. D. Scharer, and J. C. Walter. 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134:969-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rego, M. A., F. W. t. Kolling, and N. G. Howlett. 2009. The Fanconi anemia protein interaction network: casting a wide net. Mutat. Res. 668:27-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.San Filippo, J., P. Sung, and H. Klein. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77:229-257. [DOI] [PubMed] [Google Scholar]
- 33.Scharer, O. D. 2005. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem 6:27-32. [DOI] [PubMed] [Google Scholar]
- 34.Shen, X., S. Jun, L. E. O'Neal, E. Sonoda, M. Bemark, J. E. Sale, and L. Li. 2006. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA). J. Biol. Chem. 281:13869-13872. [DOI] [PubMed] [Google Scholar]
- 35.Shrivastav, M., L. P. De Haro, and J. A. Nickoloff. 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18:134-147. [DOI] [PubMed] [Google Scholar]
- 36.Smogorzewska, A., S. Matsuoka, P. Vinciguerra, E. R. McDonald III, K. E. Hurov, J. Luo, B. A. Ballif, S. P. Gygi, K. Hofmann, A. D. D'Andrea, and S. J. Elledge. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129:289-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takata, K., T. Shimizu, S. Iwai, and R. D. Wood. 2006. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 281:23445-23455. [DOI] [PubMed] [Google Scholar]
- 38.Taniguchi, T., I. Garcia-Higuera, P. R. Andreassen, R. C. Gregory, M. Grompe, and A. D. D'Andrea. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100:2414-2420. [DOI] [PubMed] [Google Scholar]
- 39.Taniguchi, T., I. Garcia-Higuera, B. Xu, P. R. Andreassen, R. C. Gregory, S. T. Kim, W. S. Lane, M. B. Kastan, and A. D. D'Andrea. 2002. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109:459-472. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimura, M., M. Kohzaki, J. Nakamura, K. Asagoshi, E. Sonoda, E. Hou, R. Prasad, S. H. Wilson, K. Tano, A. Yasui, L. Lan, M. Seki, R. D. Wood, H. Arakawa, J. M. Buerstedde, H. Hochegger, T. Okada, M. Hiraoka, and S. Takeda. 2006. Vertebrate POLQ and POLβ cooperate in base excision repair of oxidative DNA damage. Mol. Cell 24:115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.