Abstract
Caries and gingivitis are the most prevalent oral infectious diseases of humans and are due to the accumulation of dental plaque (a microbial biofilm) on the tooth surface and at the gingival margin, respectively. Several in vitro and in vivo studies have shown that many natural components of foods and beverages inhibit the adhesion of and/or exert activity against oral bacteria. These biological activities have mainly been attributed to the polyphenol fraction. In order to explore the possibility that diet can alter the dental plaque community, in this study we evaluated the composition of the microbiota of supra- and subgingival plaque samples collected from 75 adult subjects with different drinking habits (drinkers of coffee, red wine, or water for at least 2 years) by analyzing the microbial population through the separation of PCR-amplified fragments using the denaturing gradient gel electrophoresis (DGGE) technique. The mean numbers of bands of the DGGE profiles from all three categories were evaluated. There were no significant differences between the two kinds of plaque collected from the control group (water drinkers), and this group showed the highest number of bands (supragingival plaque, 18.98 ± 3.16 bands; subgingival plaque, 18.7 ± 3.23 bands). The coffee and wine drinker groups generated the lowest numbers of bands for both supragingival plaque (coffee drinkers, 8.25 ± 3.53 bands; wine drinkers, 7.93 ± 2.55 bands) and subgingival plaque (coffee drinkers, 8.3 ± 3.03 bands; wine drinkers, 7.65 ± 1.68 bands). The differences between coffee drinkers or wine drinkers and the control group (water drinkers) were statistically significant. A total of 34 microorganisms were identified, and the frequency of their distribution in the three subject categories was analyzed. A greater percentage of subjects were positive for facultative aerobes when supragingival plaque was analyzed, while anaerobes were more frequent in subgingival plaque samples. It is noteworthy that the frequency of identification of anaerobes was significantly reduced when the frequencies for coffee and wine drinkers were compared with the frequencies for subjects in the control group. The DGGE profiles of the organisms in both plaque samples from all groups were generated and were used to construct dendrograms. A number of distinct clusters of organisms from water, coffee, and wine drinkers were formed. The clustering of some of the DGGE results into cohort-specific clusters implies similarities in the microbiotas within these groups and relevant differences in the microbiotas between cohorts. This supports the notion that the drinking habits of the subjects may influence the microbiota at both the supragingival and the subgingival levels.
The oral cavity hosts a complex ecosystem composed of hundreds of different microbial species, the vast majority of which are bacteria (14, 18, 35). A fundamental component of this complex ecosystem is dental plaque (a microbial biofilm) (17, 21), which, in addition to comprising a heterogeneous microbial population, is characterized by the presence of environments that differ in their physicochemical parameters and that vary over time (14, 18). These features make the dental plaque a very rich and diverse habitat from which we can culture only about 50% of the species present because nonculturable fastidious facultative anaerobes and strict anaerobes are commonly included in dental plaque (24, 35). Numerous methods applying molecular biology techniques which can overcome this culture bias have been developed and used for bacterial detection. A number of these allow the qualitative or quantitative detection of single bacterial species (13), while others can detect and profile whole heterogeneous populations (25). In order to more fully study the complex oral microbiota, techniques such as denaturing gradient gel electrophoresis (DGGE) can be used (8, 9, 15, 16, 26). This technique separates PCR-amplified DNA fragments from a heterogeneous population according to the sequence information. By use of this technique with global primers for the 16S rRNA gene, fragments of the same size but with different base pair sequences can be individually separated and identified (19, 20).
Pioneer oral bacteria (e.g., the Streptococcus mitis group) colonize biotic and abiotic structures, and over time and following numerous specific interactions with other species, a climax community is reached (14, 18, 35). This bacterial proliferation is usually a consequence of a diet rich in sugars such as sucrose and/or of poor oral hygiene and may lead to the commonest oral pathologies, such as caries, gingivitis, and periodontitis (14, 18, 35). Many strategies for reducing the accumulation of plaque have been proposed, ranging from the use of sugar restriction and sugar substitutes to the use of vaccination against specific odontopathogenic bacteria (e.g., Streptococcus mutans) and antimicrobial agents such as antibiotics and antiplaque agents in mouth rinses and toothpastes (6, 30). In addition to these classical approaches, it is noteworthy that several in vitro and in vivo studies have shown that many natural components of foods and beverages inhibit the adhesion of and/or exert activity against oral bacteria (for an update and review, see reference 28). These biological activities have mainly been attributed to the polyphenol fraction. Polyphenols are naturally present in a number of foods and drinks, such as tea (12), coffee (5), wine and grape juice (3, 33), beer and hops (32), cranberry juice (11, 34, 36, 37), cocoa (23), apple juice (38), and many others (28). In a previous study, we have also shown that people who regularly consume foods and drinks containing polyphenols have lower levels of culturable bacteria in their saliva and dental plaque (27).
In this study we evaluated the compositions of the microbiotas of supra- and subgingival plaque samples collected from subjects with different drinking habits by analyzing the microbial population through the separation of PCR-amplified fragments using the DGGE technique.
MATERIALS AND METHODS
Patient selection.
Seventy-five adult volunteers ranging in age from 22 to 65 years were selected from among the persons attending the Dental Clinic of the University of Verona. The inclusion and exclusion criteria were as follows: (i) only dentate subjects were examined; (ii) none of the subjects recruited had received antibiotic treatment for at least 6 months, women could not be pregnant, and all the subjects were free of systemic diseases; and (iii) the subjects were expected to have been drinking any one of the beverages evaluated in this study (coffee, red wine, or water) for at least 2 years. Only subjects who declared that they drank at least five cups of espresso coffee (30 to 40 ml each) or a couple of glasses (200 ml each) of red wine a day were recruited. Subjects who declared that they did not consume or only occasionally consumed coffee or wine and that they mainly drank water constituted the control population. The subjects were informed of the study procedures and were included on the basis of their ability to comply with specific guidelines. Informed consent was obtained from each subject. The numbers of subjects recruited, divided by drinking habits, age range, and mean age, are indicated in Table 1.
TABLE 1.
Subjects enrolled in the study, by group, gender, and age
| Subject group | No. of subjects | % of subjects |
Age (yr) |
||
|---|---|---|---|---|---|
| Male | Female | Range | Mean | ||
| Water drinkers (control) | 27 | 30 | 70 | 28-60 | 44 ± 8 |
| Coffee drinkers | 31 | 36 | 64 | 22-64 | 43 ± 11 |
| Wine drinkers | 17 | 70 | 30 | 38-65 | 52 ± 7 |
Collection of dental plaque.
Samples of supragingival plaque (approximately 1 mg) were collected with a sterile toothpick from a buccal and lingual lower molar surface of either the left or the right side of the mouth. Subgingival plaque was collected from the dental pocket of the same tooth used for supragingival plaque sampling. The material was dispersed in a sterile tube containing 500 μl of TE buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA), and the tube was stored at −80°C until further processing.
DNA extraction.
The 500-μl samples were utilized for the extraction of genomic DNA. Briefly, the sample was treated with lysis buffer (10% SDS and 0.2 mg/ml proteinase K in 25 mM Tris-HCl, pH 8) for 1 h at 58°C. The proteinase K was inactivated by incubation at 80°C for 10 min (40). The DNA was purified from the lysate by repeated extraction with phenol-chloroform-isoamyl alcohol (according to standard protocols), precipitated with sodium acetate and ethanol, and then dissolved in sterile Milli-Q water (Millipore) and stored at −20°C in aliquots.
PCR-DGGE of DNA from plaque.
The V2-V3 region of the 16S rRNA gene was amplified with universal bacterial primers (22, 39, 40) F968 GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC-3′), which contains a GC clamp that makes it suitable for DGGE, and R-1401 (5′-CGG TGT GTA CAA GAC GG-3′). The PCR mixture contained 200 μM each deoxynucleoside triphosphate, 0.5 U of AB Super Taq polymerase (AB Analitica, Padua, Italy), 1× Taq buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 2 mM MgCl2), 1 μM each primer, and 5 μl of DNA template in a final volume of 50 μl. The cycling parameters were 1 cycle of 94°C for 4 min; 5 cycles of 94°C for 90 s, 52°C for 30 s, and 72°C for 45 s; 5 cycles of 94°C for 90 s, 60°C for 30 s, and 72°C for 45 s; 25 cycles of 94°C for 90 s, 55°C for 30 s, and 72°C for 45 s; and a final cycle of 72°C for 5 min; the temperature was held at 4°C following the final cycle. The PCR products were separated and visualized by electrophoresis on a 1.5% agarose gel containing 0.5 μg/ml of ethidium bromide.
DGGE.
DGGE of the PCR-generated products was performed with a D-Code universal mutation detection system (Bio-Rad Laboratories), as described by Muyzer et al. (19). The denaturing gradient gel was formed with 8% acrylamide stock solutions containing either low (35%) or high (55%) concentrations of urea and formamide that increased in the direction of electrophoresis (100% denaturant equals 7 M urea and 40% formamide). The gradient was obtained with a model 475 gradient delivery system (Bio-Rad). Samples (ca. 2 μg of DNA, as determined spectrophotometrically) were mixed with a loading buffer containing bromophenol blue, xylene cyanol, and 70% glycerol in TAE buffer (0.02 M Tris base, 0.01 M acetic acid, 1 mM EDTA, pH 7.5) before they were loaded into the gels. About 2 μg of the reference species mixture (Fusobacterium nucleatum, Lactobacillus rhamnosus, and Porphyromonas gingivalis) was combined with loading buffer, and the mixture was loaded so that it flanked the plaque samples. Electrophoresis was performed for 16 h at 120 V and 60°C. The gels were stained with ethidium bromide (0.5 μg/ml) for 30 min. DGGE images were digitally captured and recorded with an Image Master VDS apparatus (Pharmacia Biotech).
Gel analysis of all plaque samples was carried out in duplicate, to check the reproducibility. A representative selection of bands was excised from the gel, and the DNA was eluted with sterile Milli-Q water (Millipore) overnight at room temperature. A volume of 10 μl of the eluted DNA was used for further amplification with the same primers and parameters described above, and the amplified material was sent for sequencing commercially (Eurofins MWG Operon/M-Medical srl, Milan, Italy). The identification of the sequence was made by submission to the NCBI BLAST database.
Analysis of microbial profiles by DGGE.
The DGGE gel images were converted and transferred into a microbial profile database with FPQuest software, version 4.5 (Bio-Rad). Each gel was normalized according to a DGGE standard marker (F. nucleatum, L. rhamnosus, P. gingivalis). The similarities between the band patterns generated were analyzed by using the Pearson correlation coefficient (29). The clustering algorithm used to calculate the dendrograms was a nonweighted pair group method with arithmetic averages (UPGMA) (7). The results were displayed graphically as a dendrogram.
Cloning of excised DGGE bands.
The DGGE bands were excised, and the DNA fragments were eluted and purified by using a QuiaexII agarose gel extraction system (Qiagen Ltd., Crawley, United Kingdom), according to the manufacturer's protocol. The purified fragment was then amplified with primers F968 (without the CG clamp) and R-1401. The amplified material was cloned into the pPrime cloning vector (5Prime), according to the manufacturer's instruction. The vector was then cloned into Escherichia coli DH5α. The extracted plasmid was purified with a Qiagen plasmid kit (Promega) and sequenced as described above.
Statistical analysis.
Statistical and demographic analyses were performed by Fisher's exact test and one-way and three-way analyses of variance (ANOVAs) with Statgraphics Centurion XV software.
RESULTS
DGGE profiles of supra- and subgingival plaque samples from subjects with different drinking habits.
A total of 75 subjects were enrolled in this study. The grouping of the subjects by drinking habit, gender, and age is shown in Table 1. Thirty-one of the subjects were coffee drinkers, 17 were wine drinkers, and 27 were water drinkers. The mean ages of the volunteer coffee and water drinkers were similar (43 and 44 years, respectively), but the mean age of the wine drinkers was slightly higher (52 years). Females were predominant in the groups that drank coffee and water, and vice versa, males predominated among the wine drinkers.
DGGE profiles were obtained for both the supragingival and the subgingival plaque samples collected from all the subjects enrolled in this study. Both intense and faint bands were generated in all the DGGE profiles. Figure 1 shows a representative DGGE profile of supra- and subgingival plaque from the three groups of subjects enrolled. Some profiles of plaque samples collected from the control group showed up to 30 distinct bands, while plaque samples collected from coffee and wine drinkers showed no more than 20 bands. The mean numbers of bands in the profiles from all three categories are shown in Table 2. There were no significant differences in the number of bands between the two kinds of plaque collected from the control group, and plague from this group showed the highest number of bands (supragingival plaque, 18.98 ± 3.16 bands; subgingival plaque, 18.70 ± 3.23 bands). The coffee and wine drinker groups generated the lowest numbers of bands for both supragingival plaque (coffee drinkers, 8.25 ± 3.53 bands; wine drinkers, 7.93 ± 2.55 bands) and subgingival plaque (coffee drinkers, 8.30 ± 3.03; wine drinkers, 7.65 ± 1.68 bands). The differences between coffee drinkers or wine drinkers and the control group were highly statistically significant for wine drinkers (P < 0.01) and were significant for coffee drinkers (P < 0.05). Furthermore, the type of beverage consumed was significantly associated with gender (Fisher's exact text, P = 0.023) and age (one-way ANOVA, P = 0.005). Hence, we evaluated the effect of the beverage consumed on the number of DGGE bands, controlling for gender and age by a three-way ANOVA. Only beverage was a significant predictor of the number of DGGE bands, while gender and age did not significantly affect this. The interaction between beverage and either gender or age was also not significant, suggesting that gender and age do not modify the effect of beverage on the number of DGGE bands.
FIG. 1.
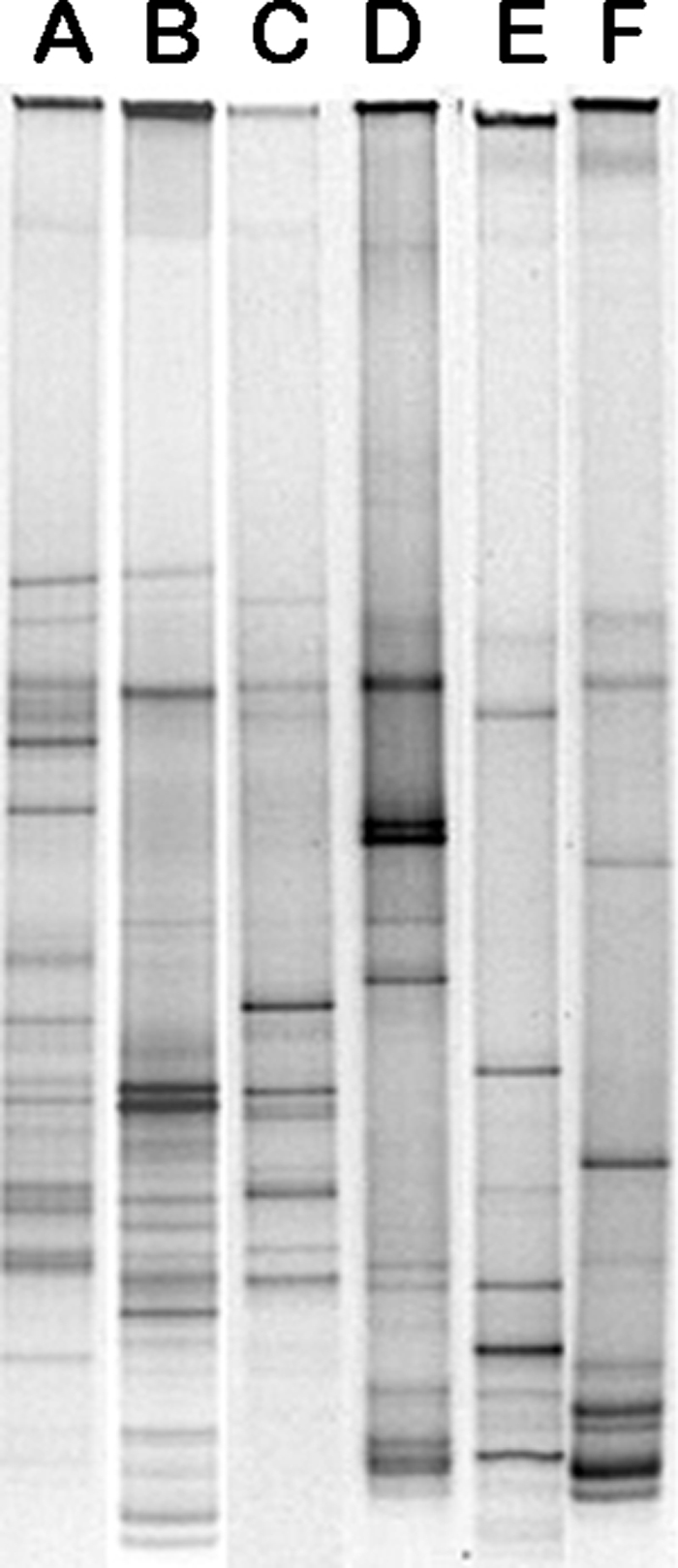
Examples of DGGE profiles for supragingival plaque (lane A) and subgingival plaque (lane B) from a subject belonging to the control group, supragingival plaque (lane C) and subgingival plaque (lane D) from a coffee drinker, and supragingival plaque (lane E) and subgingival plaque (lane F) from a wine drinker.
TABLE 2.
Number of bands in DGGE profiles of both supra- and subgingival plaque samples of subjects included in the three categories under investigation
| Subject group | Mean no. of bands in plaque sample ±SD (P valuea) |
|
|---|---|---|
| Supragingival | Subgingival | |
| Water drinkers (control) | 18.98 ± 3.16 | 18.70 ± 3.23 |
| Coffee drinkers | 8.25 ± 3.53 (0.0195) | 8.30 ± 3.03 (0.0453) |
| Wine drinkers | 7.93 ± 2.55 (0.000594) | 7.65 ± 1.68 (0.00441) |
P values comparing the coffee or wine drinkers with the control group (water drinkers).
It has previously been shown that DNA fragments from different taxa can comigrate to the same position within DGGE gels (10). The 10 most intense bands with different relative mobilities in the gels were eluted, and the fragments were subcloned into pPrime cloning vector (5Prime). Sequencing of these clones (from the 10 bands) was performed, and an unequivocal identification of a single taxon was obtained for the 10 different amplification bands, suggesting the purity of the DNA band (data not shown). In addition and to confirm the band identification, bands from different lanes with the same relative mobility were excised and sequenced. These data confirm that the sequencing of bands with the same relative mobility identified the same microorganism (data not shown).
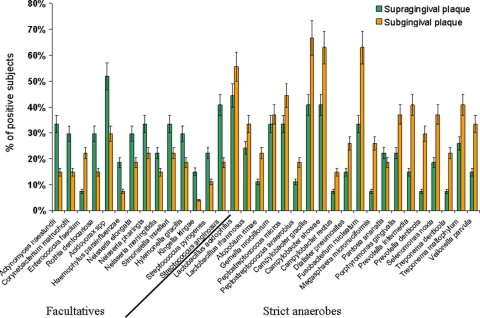
A total of 34 microorganisms were identified, and the frequencies of their distribution into the three subject categories were analyzed. Figure 2 shows the percentage of water drinkers who hosted the identified microorganisms in supra- and subgingival plaque samples. Differences in the prevalence of facultative aerobes and anaerobes at the two sampling sites (supragingival and subgingival plaque) were observed. A greater percentage of subjects were positive for facultative aerobes when supragingival plaque was analyzed, while anaerobes were more frequent in subgingival plaque samples. Similar results were observed when the microbial populations in the two plaque samples from both coffee drinkers and wine drinkers were evaluated (data not shown). Since anaerobic microorganisms are those mainly involved with gingivitis and periodontitis (14, 18, 35), we compared the microorganisms present in both supra- and subgingival plaque samples in the three categories of subjects examined in this study. Table 3 shows that the majority of the microorganisms evaluated, all of which were strict anaerobes, were found more frequently in the subgingival plaque than in the supragingival plaque. It is noteworthy that the frequency of their isolation was reduced when the results for the coffee and wine drinkers were compared with those for the subjects in the control group. This is even more evident for subgingival plaque. Statistical analysis indicated that the differences between coffee drinkers or wine drinkers and water drinkers (the control group) were significant or highly significant for all the microorganisms in the subgingival plaque and were significant for several microorganisms in the supragingival plaque.
FIG. 2.
Percentage of positive subjects belonging to the control group (water drinkers) whose supragingival and subgingival plaque samples are colonized by the indicated microorganisms. Bars represent standard deviations.
TABLE 3.
Percentage of subjects belonging to the three categories (water, coffee, and wine drinkers) who host specific periodontopathogenic bacteria in their supra- and subgingival plaque samples
| Microorganism | % of subjects |
|||||
|---|---|---|---|---|---|---|
| Supragingival plaquea |
Subgingival plaquea |
|||||
| Water drinkers | Coffee drinkers | Wine drinkers | Water drinkers | Coffee drinkers | Wine drinkers | |
| Actinomyces naeslundiib | 33 | 19g | 18g | 15 | 10g | 13f |
| Rothia dentocariosac | 30 | 26e | 24f | 15 | 13f | 6g |
| Peptostreptococcus microsd | 33 | 26f | 29e | 44 | 32g | 34g |
| Peptostreptococcus anaerobiusd | 11 | 4g | 9f | 19 | 13g | 12g |
| Dialister pneumositesd | 15 | 10g | 12f | 26 | 23f | 19g |
| Magasphera micromuciformisd | 7 | 6e | 6e | 27 | 19g | 18g |
| Porphyromonas gingivalisd | 22 | 23e | 18f | 37 | 28g | 31f |
| Prevotella intermediad | 7 | 6e | 6e | 30 | 24g | 26f |
| Fusobacterium nucleatumd | 32 | 28e | 27f | 64 | 51f | 53f |
| Treponema denticolad | 7 | 5f | 5f | 22 | 17f | 16g |
The standard deviations ranged from 8 to 13%. Statistical evaluations were performed by comparing the results for the coffee or wine drinkers with those for the water drinkers (control).
Anaerobe/microaerophilic.
Facultative aerobe.
Strict anaerobe.
Value not statistically significant.
Value statistically significant (P < 0.05).
Value highly statistically significant (P < 0.01).
Evaluation of DGGE profiles.
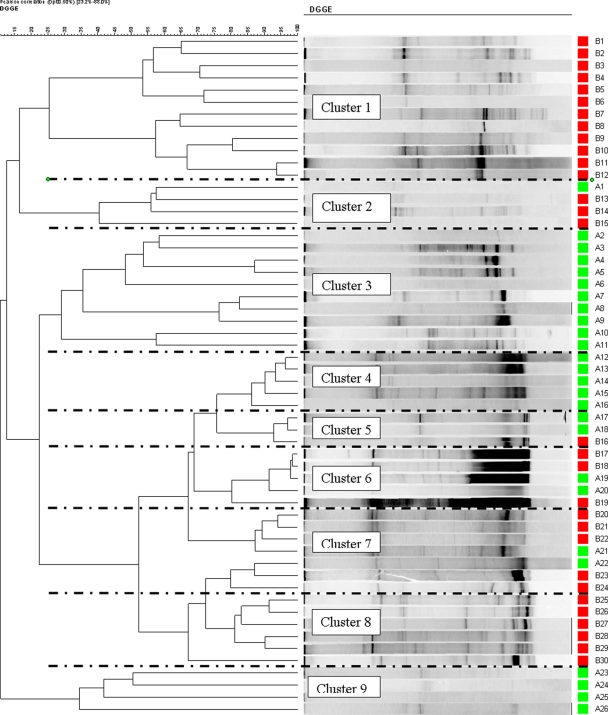
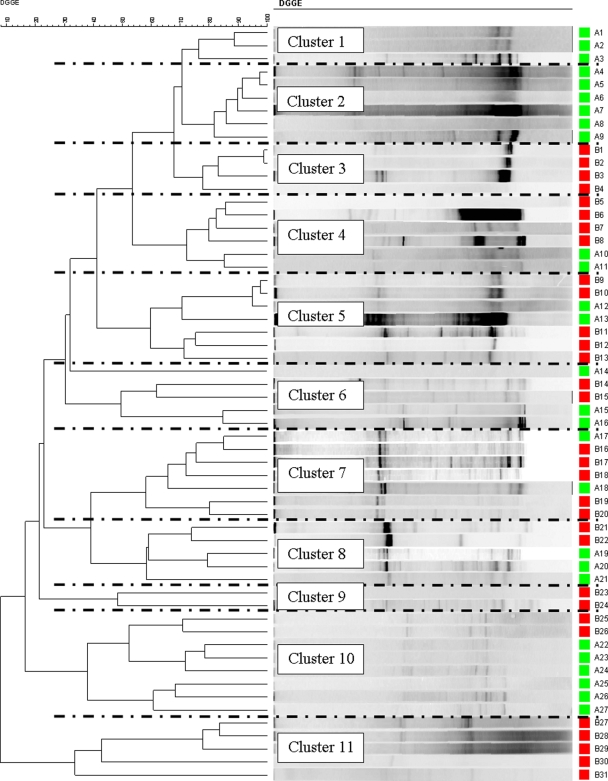
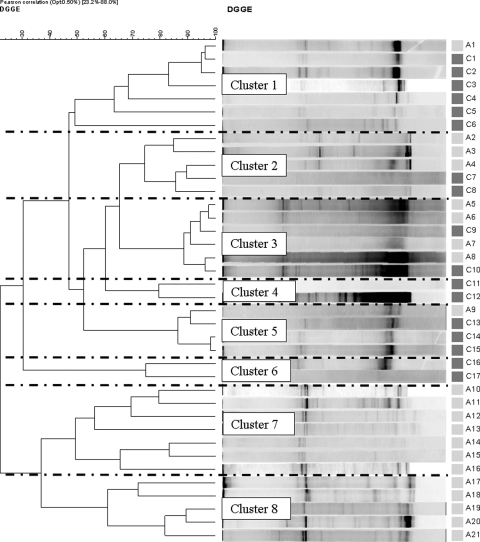
The DGGE profiles of both types of plaque samples from all groups were generated and used to construct dendrograms. Figures 3 and 4 show the results of hierarchical cluster analyses of the DGGE profiles of the supragingival and subgingival plaque samples, respectively, from the control subjects and coffee drinkers. A number of distinct clusters were formed and are shown in Fig. 3 and 4. In Fig. 3 (supragingival plaque), clusters 1 and 8 comprised samples only from coffee drinkers, while clusters 3, 4, and 9 comprised samples only from the controls. Additionally, in Fig. 4 (subgingival plaque), clusters 3, 9, and 11 solely comprised samples from the coffee drinkers and clusters 1 and 2 completely comprised samples from the controls.
FIG. 3.
Hierarchical cluster analysis results of all the DGGE profiles for supragingival plaque from both control subjects (green) and coffee drinkers (red) demonstrated graphically as a UPGMA dendrogram.
FIG. 4.
Hierarchical cluster analysis results of all the DDGE profiles for subgingival plaque from both control subjects (green) and coffee drinkers (red) demonstrated graphically as a UPGMA dendrogram.
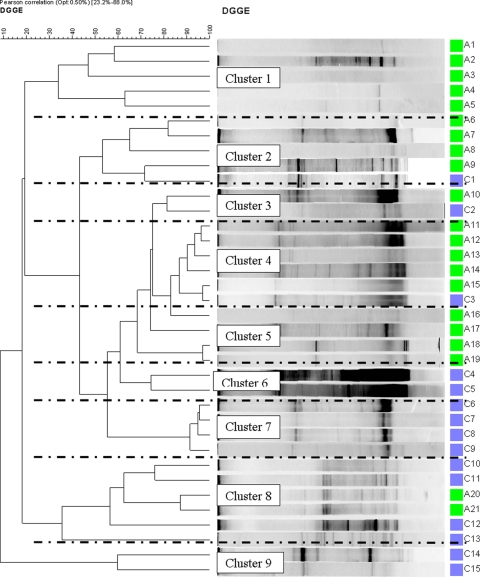
Similarly, Fig. 5 and 6 show the results of the hierarchical cluster analyses of supragingival and subgingival plaque samples, respectively, from wine drinkers, and again, a number of clusters which consisted only of samples from the wine drinkers or, indeed, only of samples from the controls were formed. In Fig. 5, clusters 1 and 5 are composed entirely of samples from controls and clusters 6, 7, and 9 are composed entirely of samples from wine drinkers, and in Fig. 6, clusters 7 and 8 are composed of samples only from the controls. The clustering of the some of the DGGE results into cohort-specific clusters implies similarities in the microbiota within these groups and relevant differences in the microbiota between cohorts (e.g., Fig. 3, cluster 1 versus cluster 3). It is noteworthy that a mixed population (by age and gender) was always included within the clusters identified in Fig. 3 to 6.
FIG. 5.
Hierarchical cluster analysis results of all the DDGE profiles for supragingival plaque from both control subjects (green) and wine drinkers (blue) demonstrated graphically as a UPGMA dendrogram.
FIG. 6.
Hierarchical cluster analysis results of all the DDGE profiles for subgingival plaque from both control subjects (green) and wine drinkers (blue) demonstrated graphically as a UPGMA dendrogram.
DISCUSSION
A major challenge in microbial ecology is the assessment of the community structure present in a defined environment. It is also important to be able to analyze such structures in a time- and resource-efficient manner. Recent technical advances in molecular biology have allowed some of these issues to be realized and have allowed us to better characterize complex microbial communities. Given that we believe that at least 50% of the human oral microbiota is unculturable (24, 31, 35), we chose to use a culture-independent technique (DGGE) to characterize the differences in microbial populations between the three cohorts in an effort to include the uncultivable portion of the oral microbiota in the analysis. The advantages of using DGGE as a means of studying microbial ecology are well established, in that it provides a more accurate means of visualizing whole microbial communities (since there should be a reduced bias in the detection of unculturable species) and it is less labor-intensive than the more conventional culture-independent techniques of PCR cloning and sequencing.
Three cohorts were recruited for the study: coffee drinkers, wine drinkers, and water drinkers (the control group). The nonhomogeneity of the three groups by gender may be explained in terms of the Italian drinking tradition: the control group, which included water drinkers, had more woman than men; the coffee drinker group had features similar to those of the water-drinking group, but they were less evident; and the wine drinker group was, on the contrary, composed overall of men, who made up two-thirds of the wine-drinking group. Similarly, the mean age of the wine drinkers was slightly older than the mean ages of the coffee and water drinkers.
A wide range of appropriate taxa were identified from supragingival and subgingival plaque samples collected from the control group (Fig. 2). These are broadly distributed according to atmospheric growth requirements; i.e., the strict anaerobes were predominantly found in the subgingival plaque, where the oxygen tension is far lower than that in the supragingival plaque. Conversely, the facultatively anaerobic taxa were more frequently detected in the supragingival plaque.
The DGGE profiles generated were analyzed. In the first instance, the numbers of visible bands were counted for each profile. This showed striking and significant differences between the cohorts. The samples from the coffee and wine drinkers had fewer bands than the samples from the control groups (7 or 8 and 18, respectively), which suggests that the samples were less species rich. This also implies that a form of microbial community control is elicited by the coffee and wine. Interestingly, the frequency of periodontopathic taxa (Table 3) detected was reduced significantly in the subgingival plaque samples from the coffee drinkers (all 10 taxa) and the red wine drinkers (all 10 taxa) and to a lesser extent in the samples taken from supragingival plaque (red wine drinkers, 7 taxa; coffee drinkers, 5 taxa). Again, the implication is that the drinking habits of the subjects are associated with changes in their subgingival microbial communities.
The issues of comigration highlighted by Gafan and Spratt (10) were not apparent in this study, as cloning and sequencing of single DGGE bands (in a manner similar to that done in the previous work) resulted in the identification of single taxa from each band.
Cluster analysis of the profiles and the generation of dendrograms showed some very interesting relationships. Distinct clusters were formed, and a number of these comprised samples from a single cohort (Fig. 3, supragingival plaque, clusters 1 and 8). This also supports the notion that the drinking habits of the subjects influence the microbiota at both the supraginigival plaque and the subgingival plaque levels. There were clearly a number of different monocohort clusters, indicating that while there are similarities between the microbiota of some subjects, e.g., Fig. 3, cluster 1 (12 subjects), there are also differences within the cohort; i.e., Fig. 3, cluster 8, comprised samples from 6 subjects with similar microbiotas that were quite different from the microbiotas of the 12 subjects of cluster 1. Furthermore, the observation that each cluster includes subjects diverse by gender and age clearly indicates that these peculiar DGGE profiles and, therefore, the oral microbiotas are not linked to the gender or the age of the volunteers but most likely are linked to their drinking habits. It would be interesting to expand this work and try to determine the microbiological differences between the cohorts and, indeed, between the clusters.
Red wine and coffee have a number of compounds which are known to possess antimicrobial, antiadhesive, and antiplaque activities. Coffee brew, widely consumed, is obtained from a hot infusion of grounded roasted coffee beans. Natural green coffee beans include dozens of bioactive compounds, such as caffeine, trigonelline, and nicotinic and chlorogenic acids. During the roasting process their concentrations are partially reduced, but new compounds (not polyphenols) are formed as a result of the Maillard reaction: carbohydrate caramelization and the pyrolysis of organic compounds (2). In particular, high-molecular-weight melanoidins, which confer the brown color, are endowed with potent antioxidant activities. The activity of roasted coffee against S. mutans and Staphylococcus aureus (4) and against enterobacteria (1) has been detected only as a result of the formation of α-dicarbonyl compounds during the roasting process (4). Green and roasted coffees also have the ability to interfere in vitro with S. mutans sucrose-independent adsorption to hydroxyapatite (HA) beads (5). Coffee components such as trigonelline and nicotinic and chlorogenic acids possess strong antiadhesive properties, as well as a fraction with a molecular mass ranging from 1,000 to 3,500 Da, commonly considered low-molecular-mass melanoidins. As far as wine is concerned, both dealcoholized red and white wines have been shown to have activity against oral streptococci. Organic acids, such as succinic, malic, lactic, tartaric, citric, and acetic acids, are responsible for the antimicrobial activity, while wine polyphenols are completely devoid of this activity (3). More recently, both potent antiadhesive and antibiofilm properties and bacterial detachment from HA beads have been shown in S. mutans, and these properties correlated to the fraction of red wine that included proanthocyanidin components (4a). Grapes and pomace (a residual product of winemaking containing grape skin, seeds, and stalks) are rich sources of natural polyphenols that have been demonstrated to have activity against S. mutans glucosyltransferases (GTFs) and to produce acids (33).
In conclusion, this study has demonstrated that it is possible to successfully profile the oral microbiota by culture-independent techniques, and indeed, the method used detected and characterized shifts in the microbial community which may be associated with the consumption of different beverages.
Acknowledgments
We acknowledge Laura Giansanti and Giulia Zanconato, students in the degree course in oral hygiene at the University of Verona, for their invaluable collaboration with the management of the subjects recruited for this research and Giuseppe Verlato for his help with the statistical calculations. The technical assistance of Anna Bertoncelli is greatly appreciated.
This work was supported by grant PRIN 2005 from the Ministero dell'Università e della Ricerca (MIUR), Rome, Italy.
Footnotes
Published ahead of print on 2 December 2009.
REFERENCES
- 1.Almeida, A. P., A. Farah, D. A. M. Silva, E. A. Nunan, and M. B. A. Gloria. 2006. Antibacterial activity of coffee extracts and selected coffee chemical components against enterobacteria. J. Agric. Food Chem. 54:8738-8743. [DOI] [PubMed] [Google Scholar]
- 2.Daglia, M. A., A. Papetti, C. Gregotti, F. Bertè, and G. Gazzani. 2000. In vitro antioxidant and ex vivo protective activities of green and roasted coffee. J. Agric. Food Chem. 48:1549-1554. [DOI] [PubMed] [Google Scholar]
- 3.Daglia, M., A. Papetti, P. Grisoli, C. Aceti, C. Dacarro, and G. Gazzani. 2007. Antibacterial activity of red and white wine against oral streptococci. J. Agric. Food Chem. 55:5038-5042. [DOI] [PubMed] [Google Scholar]
- 4.Daglia, M., A. Papetti, P. Grisoli, C. Aceti, V. Spini, C. Dacarro, and G. Gazzani. 2007. Isolation, identification and quantification of roasted coffee antibacterial compounds. J. Agric. Food Chem. 55:10208-10213. [DOI] [PubMed] [Google Scholar]
- 4a.Daglia, M., M. Stauder, A. Papetti, C. Signoretto, G. Giusto, P. Canepari, C. Pruzzo, and G. Gazzani. 2010. Isolation of red wine components with anti-adhesion and anti-biofilm activity against Streptococcus mutans. Food Chem. 119:1182-1188. [Google Scholar]
- 5.Daglia, M., R. Tarsi, A. Papetti, P. Grisoli, C. Da Carro, C. Pruzzo, and G. Gazzani. 2002. Antiadhesive effect of green and roasted coffee on Streptococcus mutans adhesive properties on saliva-coated hydroxyapatite beads. J. Agric. Food Chem. 50:1225-1229. [DOI] [PubMed] [Google Scholar]
- 6.Featherstone, J. 2006. Delivery challenges for fluoride, chlorhexidine and xylitol. BMC Oral Health 6(Suppl. 1):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 8.Fujimoto, C., H. Maeda, S. Kokeguchi, S. Tkashiba, F. Nishimura, H. Arai, K. Fukui, and Y. Murayama. 2003. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of microbial communities of subgingival plaque. J. Periodontal Res. 38:440-445. [DOI] [PubMed] [Google Scholar]
- 9.Gafan, G. P., V. S. Lucas, G. J. Roberts, A. Petrie, M. Wilson, and D. A. Spratt. 2005. Statistical analyses of complex denaturing gradient gel electrophoresis profiles. J. Clin. Microbiol. 43:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gafan, G. P., and D. A. Spratt. 2005. Denaturing gradient gel electrophoresis expansion (DGGEGE)—an attempt to resolve the limitations of co-migration in the DGGE of complex polymicrobial communities. FEMS Microbiol. Lett. 253:303-307. [DOI] [PubMed] [Google Scholar]
- 11.Gregoire, S., A. P. Singh, N. Vorsa, and H. Koo. 2007. Influence of cranberry phenolics on glucan synthesis by glucosyltransferases and Streptococcus mutans acidogenicity, J. Appl. Microbiol. 103:1960-1968. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton-Miller, J. M. T. 2001. Anti-cariogenic properties of tea (Camellia sinensis). J. Med. Microbiol. 50:299-302. [DOI] [PubMed] [Google Scholar]
- 13.Kuboniwa, M., A. Amano, R. K. Kimura, S. Sekine, S. Kato, Y. Yamamoto, N. Okahashi, T. Iida, and S. Shizukuishi. 2004. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol. Immunol. 19:168-176. [DOI] [PubMed] [Google Scholar]
- 14.Lamont, R. J., R. A. Burne, M. S. Lantz, and D. J. Leblanc. 2006. Oral microbiology and immunology. ASM Press, Washington, DC.
- 15.Ledder, R. G., P. Gilbert, S. A. Huws, L. Aarons, M. P. Ashley, I. P. S. Hul, and A. J. McBain. 2007. Molecular analysis of the subgingival microbiota in health and disease. Appl. Environ. Microbiol. 73:516-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y., D. Saxena, V. M. Barnes, H. M. Trivedi, Y. Ge, and T. Xu. 2006. Polymerase chain reaction-based denaturing gradient gel electrophoresis in the evaluation of oral microbiota. Oral Microbiol. Immunol. 21:333-339. [DOI] [PubMed] [Google Scholar]
- 17.Marsh, P. D. 2005. Dental plaque: biological significance of a biofilm and community life-style. J. Clin. Periodontol. 32(Suppl. 6):7-15. [DOI] [PubMed] [Google Scholar]
- 18.Marsh, P., and M. V. Martin. 1999. Oral microbiology. Wright, Oxford, United Kingdom.
- 19.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 21.Newman, H. N., and M. Wilson. 1999. Dental plaque revisited: oral biofilms in health and diseases. Bioline, Cardiff, United Kingdom.
- 22.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ooshima, T., Y. Osaka, H. Sasaki, K. Osawa, H. Yasuda, M. Matsumura, S. Sobue, and M. Matsumoto. 2000. Caries inhibitory activity of cocoa bean husk extract in in-vitro and animal experiments. Arch. Oral Biol. 45:639-645. [DOI] [PubMed] [Google Scholar]
- 24.Paster, B. J., S. K. Boches, J. L. Galvin, L. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paster, B. J., I. Olsen, J. A. Aas, and F. E. Dewhirst. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000. 42:80-87. [DOI] [PubMed] [Google Scholar]
- 26.Sakamato, M., M. Umada, and Y. Benno. 2005. Molecular analysis of human oral microbiota. J. Periodontal Res. 40:277-285. [DOI] [PubMed] [Google Scholar]
- 27.Signoretto, C., G. Burlacchini, F. Bianchi, G. Cavalleri, and P. Canepari. 2006. Differences in microbiological composition of saliva and dental plaque in subjects with different drinking habits. New Microbiol. 29:293-302. [PubMed] [Google Scholar]
- 28.Signoretto, C., P. Canepari, C. Pruzzo, and G. Gazzani. 2009. Anticaries and antiadhesive properties of food constituents and plant extract and implication for oral health, p. 240-262. In M. Wilson (ed.), Food constituents and oral health: current status and future prospects. Woodhead Publishing Limited, Cambridge, United Kingdom.
- 29.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, D. J. 2002. Dental caries vaccines: prospects and concerns. Crit. Rev. Oral Biol. Med. 13:335-349. [DOI] [PubMed] [Google Scholar]
- 31.Socransky, S. S., R. J. Gibbons, A. C. Dale, L. Bortnick, E. Rosenthal, and J. B. McDonald. 1963. The microbiota of the gingival crevice area of man. I. Total microscopic and viable counts and counts of specific organisms. Arch. Oral Biol. 8:275-280. [DOI] [PubMed] [Google Scholar]
- 32.Tagashira, A., K. Uchiyama, T. Yoshimura, M. Shirota, and N. Uemitsu. 1997. Inhibition by hop bract polyphenols of cellular adherence and water-insoluble glucan synthesis of mutans streptococci. Biosci. Biotech. Biochem. 61:332-335. [DOI] [PubMed] [Google Scholar]
- 33.Thimothe, J., I. A. Bonsi, I. P. Padilla-Zakour, and H. Koo. 2007. Chemical characterization of red wine grape (Vitis vinifera and Vitis interspecific hybrids) and pomace phenolic extracts and their biological activity against Streptococcus mutans. J. Agric. Food Chem. 55:10200-10207. [DOI] [PubMed] [Google Scholar]
- 34.Weiss, E. I., A. Kozlovshy, D. Steinberg, R. Lev-Dor, R. B. N. Greenstein, M. Feldman, N. Sharon, and I. Ofek. 2004. A high molecular mass cranberry constituent reduces mutans streptococci level in saliva and inhibits in vitro adhesion to hydroxyapatite. FEMS Microbiol. Lett. 232:89-92. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, M. 2008. Bacteriology of humans: an ecological perspective. Blackwell Publishing, Oxford, United Kingdom.
- 36.Yamanaka, A., R. Kimizuka, T. Kato, and K. Okuda. 2004. Inhibitory effect of cranberry juice on attachment of oral streptococci and biofilm formation. Oral Microbiol. Immunol. 19:150-154. [DOI] [PubMed] [Google Scholar]
- 37.Yamanaka, A., T. Kouchi, K. Kasai, T. Kato, K. Hishihara, and K. Okuda. 2007. Inhibitory effect of cranberry polyphenols on biofilm formation and cysteine proteases of Porphyromonas gingivalis. J. Periodontal Res. 42:589-592. [DOI] [PubMed] [Google Scholar]
- 38.Yanagida, A., T. Kanda, M. Tanabe, F. Matsudaira, and J. G. Oliveira Cordeiro. 2000. Inhibitory effects of apple polyphenols and related compounds on cariogenic factors of mutans streptococci. J. Agric. Food Chem. 48:5666-5671. [DOI] [PubMed] [Google Scholar]
- 39.Zijnge, V., H. J. M. Harmsen, J. W. Kleinfelder, M. E. van der Rest, J. E. Degener, and G. W. Welling. 2003. Denaturing gradient gel electrophoresis analysis to study bacterial community structure in pockets of periodontitis patients. Oral Microbiol. Immunol. 18:59-65. [DOI] [PubMed] [Google Scholar]
- 40.Zijnge, V., G. W. Welling, J. E. Degener, A. J. van Winkelhoff, F. Abbas, and H. J. M. Harmsen. 2006. Denaturing gradient gel electrophoresis as a diagnostic tool in periodontal microbiology. J. Clin. Microbiol. 44:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]