Abstract
Six patients with Acinetobacter genomic species 10 bacteremia were identified. The clinical features of the patients, phenotypic and genotypic identifications, antimicrobial susceptibilities, and genes flanking ISAba1 of the bacteria were described. The results revealed that this bacterium is a potentially lethal pathogen that can cause health care-associated infections in debilitated patients.
Acinetobacter species that do not belong to the Acinetobacter calcoaceticus-Acinetobacter baumannii complex are rarely encountered in clinical practice. Moreover, they cannot be reliably identified by commercially available systems (1, 6, 7). As a result, their clinical significance remains elusive. With the advancement of molecular methods, infections caused by uncommon Acinetobacter species, such as Acinetobacter ursingii, Acinetobacter schindleri (6), and Acinetobacter septicus sp. nov. (9), have been described recently.
Although isolates of Acinetobacter genomic species 10 (AGS 10), renamed Acinetobacter bereziniae sp. nov. (13), have previously been identified in clinical samples, they were subjected to studies of identification methods without mention of clinical relevance (10, 15). In this study, we identified AGS 10 isolates from six blood samples. The clinical features of the patients, phenotypic and genotypic identifications, and antimicrobial susceptibilities of the bacteria were described. To our knowledge, this is the largest case series report of AGS 10 infection.
Six of the 47 non-A. calcoaceticus-A. baumannii complex strains from a previous study (4) were identified as AGS 10 on the basis of sequence analyses of the 16S-23S rRNA intergenic spacer (ITS) and recA gene (2, 10). The bacteria were initially isolated from blood culture by using the BACTEC NR-660 system (Becton Dickinson Diagnostic Instrument Systems, Sparks, MD) and phenotypically identified as Acinetobacter junii by the 32GN system (bioMérieux, Marcy l'Etoile, France). The infected patients were men with multiple comorbidities and/or invasive devices (Table 1). Five of the six episodes of bacteremia occurred nosocomially. Patient 2 had just been discharged from our hospital and thus had acquired a health care-associated infection. The matters of infection origin and other concurrent infection remained elusive. Fevers (range, 38.5 to 39.5°C; median, 39.0°C) during bacteremia occurred in five of the six subjects, although patient 3, with a 37.5°C temperature, had presented with chills, dyspnea, and oxygen desaturation during bacteremia. All patients had sepsis, while patients 2, 3, and 5 developed septic shock. Blood cultures of all patients yielded only AGS 10. Five patients survived to discharge. One critically ill patient (patient 5) who received inadequate antibiotic therapy succumbed. No other cause of death was identified.
TABLE 1.
Clinical characteristics of patients with Acinetobacter genomic species 10 bacteremiaa
| Case | No. of isolates (yr of isolation) | Age (yr)/sexb | Underlying condition or diseases | Procedure or invasive device(s) | Ward/dayc | Treatment | Appropriate antimicrobial therapy | Outcome | WBC count (1010)/liter | CRP concn (mg/liter) | Apache II score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 568 (2002) | 79/M | Squamous cell carcinoma of lung postlobectomy with recurrence, postradiation therapy, hypertension, DM, old CVA, complete AV block postpacemaker | Tracheostomy, ventilator, Foley catheter, CVC, NG | ICU/45 | Ciprofloxacin for 14 days | Yes | Survived | 1.07 | 21 | 27 |
| 2 | 634 (2002) | 78/M | Parkinson's disease; patient bedridden | Endotracheal tube, ventilator, Foley catheter, CVC, NG | ICU/1 | Ampicillin-sulbactam for 2 days followed by piperacillin and amikacin for 14 days | Yes | Survived | 0.54 | 44 | 28 |
| 3 | 649 (2002) | 81/M | Chronic renal insufficiency, COPD, hypertension, DM, congestive heart failure | Endotracheal tube, ventilator, NG | ICU/26 | Ceftazidime for 14 days | Yes | Survived | 1.18 | 55 | 31 |
| 4 | 1091 (2005) | 38/M | Chronic myeloid leukemia with accelerated blast crisis post nonrelative bone marrow transplant, GVHD under steroid treatment, chronic hepatitis B | Port-A | Ordinary ward/31 | Ciprofloxacin for 12 days | Yes | Survived | 0.12 | 165 | 16 |
| 5 | 1279 (2000) | 89/M | CVA; patient bedridden | Tracheostomy, ventilator, Foley catheter, arterial line, CVC, NG | RCU/65 | Aztreonam for 13 days | No | Died of disease | 1.05 | ND | 31 |
| 6 | 1312 (2000) | 82/M | Prior pulmonary tuberculosis, duodenal ulcer postsubtotal gastrectomy, hypertension, AVNRT postablation | Nil | Ordinary ward/4 | Cefazolin for 15 days plus gentamicin for 9 days | Yes | Survived | 1.63 | 39 | 21 |
AV, atrioventricular; AVNRT, atrioventricular nodal reentry tachycardia; COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; CVA, cerebrovascular accident; CVC, central venous catheter; DM, diabetes mellitus; GVHD, graft-versus-host disease; ICU, intensive care unit; ND, no data; NG, nasogastric tube; RCU, respiratory intensive care unit; WBC, white blood cell.
M, male; F, female.
Day of bacteremia after hospitalization.
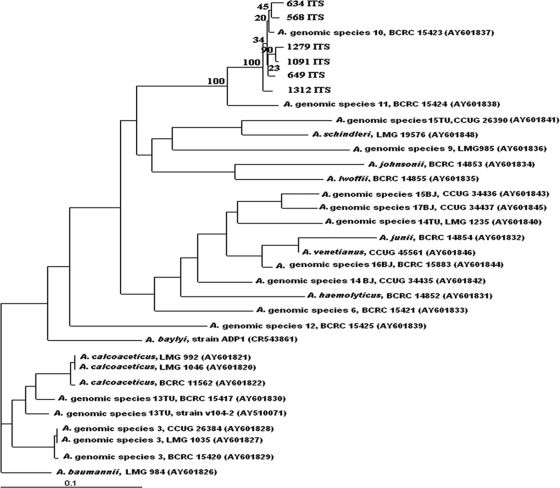
The ITS sequence (793 bp) showed 98 to 99% similarity to the reference strain in GenBank (BCRC 15423, equal to ATCC 17924), while the intraspecies similarity was 97.85 to 99.33%. Neighbor-joining phylogenetic analysis (Fig. 1) utilized sequences of ITS genes from other different Acinetobacter species and grouped the clinical isolates and BCRC 15423 into the same cluster. The three most related species were Acinetobacter genomic species 11 (BCRC 15424, equal to ATCC 11171), Acinetobacter genomic species 14 BJ (CCUG 34435), and Acinetobacter haemolyticus (BCRC 14852, equal to ATCC 17906), with similarities of 92.18 to 93.52%, 79.75 to 80.53%, and 79.27 to 80.13%, respectively. Our data demonstrated the specificity of ITS sequencing in identification of AGS 10 in addition to species belonging to the A. calcoaceticus-A. baumannii complex (2). The recA gene of our strains also showed high sequence similarity (98 to 99%) to the GenBank reference strain (GenBank accession number AF191139) but only 88 to 89% similarity to genomic species 11 (GenBank accession number AF191140). The results revealed that recA may be an acceptable and specific tool for identifying AGS 10. Pulsed-field gel electrophoresis (PFGE) (3) revealed no clonal relationship among them (Fig. 2).
FIG. 1.
Neighbor-joining phylogenetic tree based on the 16S-23S rRNA gene ITS sequences of 27 Acinetobacter reference strains. Selected bootstrap analysis (100×) values are shown at the branching points. The scale bar represents a 0.1-base change per nucleotide position. The GenBank accession numbers of the sequences are in parentheses.
FIG. 2.
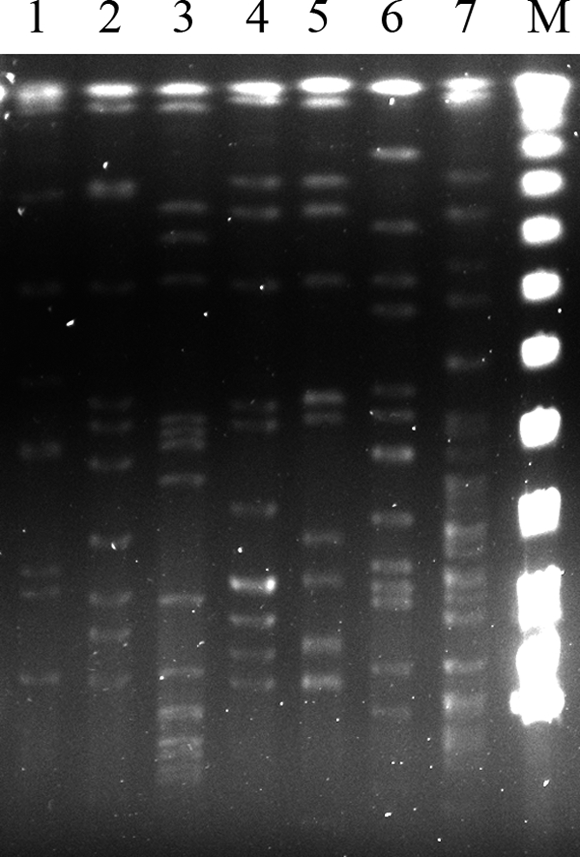
PFGE of six clinical isolates (lanes 1 to 6) and a reference strain (lane 7) of Acinetobacter genomic species 10. Lanes 1 to 6 correspond to isolates 568, 634, 649, 1091, 1279, and 1312, respectively. Lane M is a marker.
It has been assumed that identification of Acinetobacter species to the genus level by use of colorimetric Vitek 2 cards (bioMérieux, Marcy l'Etoile, France) is reliable (17). In our study, the colorimetric Vitek 2 cards had identified all clinical strains as Acinetobacter lwoffii (95 to 99% probability for five isolates and 50.27% for one). The reference strain was identified as A. lwoffii with lower probability (33.75%). The 32GN system identified the six clinical isolates as Acinetobacter junii. Neither the Vitek 2 nor the 32GN system misidentified clinical isolates as members of the A. calcoaceticus-A. baumannii complex or another genus.
The in vitro susceptibility testing of these clinical isolates and the reference strain was determined by the AST-GN09 card of the Vitek 2 system or the agar dilution method (colistin). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains. The results were interpreted according to CLSI recommendations (Table 2) (5). The six AGS 10 clinical strains had lower levels of susceptibility to ceftazidime (50%) and ceftriaxone (16.7%) than A. junii strains from the same hospital, which exhibited 97.1% and 84.2% susceptibility, respectively, to these two antibiotics (8). Unexpectedly, the reference strain and three clinical strains were resistant to colistin.
TABLE 2.
Antimicrobial susceptibility of a reference strain and six clinical isolates of Acinetobacter genomic species 10
| Antimicrobial agent(s) | MIC (μg/ml) for indicated strain |
||||||
|---|---|---|---|---|---|---|---|
| 568 | 634 | 649 | 1091 | 1279 | 1312 | ATCC 17924 | |
| Amikacin | <2 | <2 | <2 | <2 | 16 | <2 | <2 |
| Gentamicin | <1 | <1 | <1 | <1 | >16 | <1 | <1 |
| Tobramycin | <1 | <1 | <1 | <1 | 8 | <1 | <1 |
| Ampicillin | >32 | >32 | 16 | >32 | >32 | >32 | 16 |
| Ampicillin-sulbactam | <2 | <2 | <2 | <2 | <2 | <2 | <2 |
| Piperacillin | 8 | 8 | 8 | 8 | 8 | 8 | <4 |
| Piperacillin-tazobactam | <4 | <4 | <4 | <4 | <4 | <4 | <4 |
| Cefazolin | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| Cefuroxime | 16 | 32 | 16 | 32 | 32 | 16 | 16 |
| Cefotetan | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| Ceftazidime | 8 | 16 | 4 | 16 | 16 | 8 | 4 |
| Ceftriaxone | 16 | 16 | 8 | 16 | 16 | 16 | 4 |
| Cefepime | 2 | 2 | <1 | 2 | 2 | 2 | 2 |
| Aztreonam | 32 | >64 | 16 | >64 | >64 | >64 | 16 |
| Imipenem | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| Meropenem | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Ciprofloxacin | <0.25 | >4 | 2 | <0.25 | >4 | <0.25 | <0.25 |
| Levofloxacin | <0.25 | 2 | 1 | <0.25 | 1 | <0.25 | <0.25 |
| TMP-SMXa | <20 | <20 | >320 | <20 | >320 | <20 | 160 |
| Colistin | 2 | 2 | 4 | 16 | 8 | 1 | 4 |
TMP-SMX, trimethoprim-sulfamethoxazole.
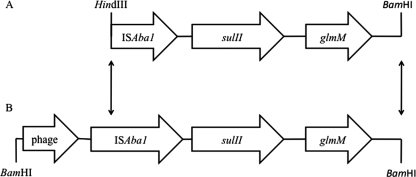
Production of beta-lactamases in our clinical isolates and the reference strain was confirmed by a positive colorimetric nitrocefin disk assay (Remel, Lenexa, KS). In spite of resistance to broad-spectrum cephalosporins, five clinical strains and the reference strain lacked detectable extended-spectrum beta-lactamases (ESBLs) and AmpC-type beta-lactamases on the basis of a phenotypic assay (3). Strain 634 expressed AmpC beta-lactamases phenotypically. However, PCR with specific primers failed to detect genes encoding ESBLs and AmpC-type beta-lactamases, including blaPER, blaSHV, blaTEM, blaVEB, blaCTX-M-1, blaCTX-M-2 (TOHO-1), blaCTX-M-9 (TOHO-2), blaADC-1 to blaADC-7, blaADC-8, blaMOX-1, blaMOX-2, blaCMY-1, blaCMY-8 to blaCMY-11, blaLAT-1 to blaLAT-4, blaCMY-2 to blaCMY-7, blaBIL-1, blaDHA-1, blaDHA-2, blaACC, blaMIR-1T, blaACT-1, and blaFOX-1 to blaFOX-5b (3), as well as carbapenemase genes, including blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, blaIMP, blaVIM, blaSIM, blaSPM, blaGIM-1, blaGES, and blaKPC (11, 12, 16). ISAba1 (3) was found in strains 634 and 649 without downstream carbapenemase genes. Genomic DNA was digested by BamHI, self-ligated, and subjected to inverse PCR with primers ISABAINVF (5′-GCCATTTTTGGTAAAGACCGT-3′) and ISABAINVR (5′-GATGAGCGCAAAGCACTTTAA-3′). These PCR primers targeted ISAba1 to identify its flanking regions. The PCR yielded a 3,184-bp product in strain 649 (Fig. 3) (GenBank accession number GQ421466). A part of the region of the DNA sequence was similar to a previously reported DNA sequence of A. baumannii (14), indicating a horizontal gene transfer. Using the BPROM program, a putative promoter composed of −35 (TTGAGA) and −10 (CGTTAGAAA) elements was located upstream of sulII and within ISAba1. Its proximity suggested that ISAba1 may provide putative promoters for sulII and may have contributed to the sulfonamide resistance of strain 649.
FIG. 3.
Comparison of a 2.29-kb HindIII-BamHI fragment from an A. baumannii strain (GenBank accession number AY823412) (A) with a 3.18-kb BamHI fragment from strain 649 (GenBank accession number GQ421466) (B). The region between nucleotides 17 and 637 contained a protein domain resembling the phage-related portal vertex protein of A. baumannii AYE (GenBank accession number Y_P001712545), with 60% identity. ISAba1 was located in the region between nucleotides 1861 and 2676. A total of 18 nucleotides downstream of ISAba1 initiated an open reading frame matched to sulfonamide resistance gene sulII (GenBank accession number EU360945; 100% similarity), followed by a glmM gene (GenBank accession number EU855788; 99% similarity), which encoded phosphoglucosamine mutase. Arrows indicate that the sequences in between two fragments are homologous.
In conclusion, our study revealed that AGS 10 is a potentially lethal pathogen causing health care-associated infections in humans. The identification was achieved by the analysis of ITS and recA gene sequences. Sepsis or even septic shock occurred in vulnerable patients with multiple comorbidities and invasive devices. Death may ensue in severely debilitated patients.
Acknowledgments
This study was supported by grants V98B2-001 from Taipei Veterans General Hospital.
Footnotes
Published ahead of print on 2 December 2009.
REFERENCES
- 1.Bernards, A. T., J. van der Toorn, C. P. van Boven, and L. Dijkshoorn. 1996. Evaluation of the ability of a commercial system to identify Acinetobacter genomic species. Eur. J. Clin. Microbiol. Infect. Dis. 15:303-308. [DOI] [PubMed] [Google Scholar]
- 2.Chang, H. C., Y. F. Wei, L. Dijkshoorn, M. Vaneechoutte, C. T. Tang, and T. C. Chang. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, T. L., L. K. Siu, Y. T. Lee, C. P. Chen, L. Y. Huang, R. C. C. Wu, W. L. Cho, and C. P. Fung. 2008. Acinetobacter baylyi as a pathogen for opportunistic infection. J. Clin. Microbiol. 46:2938-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, T. L., L. K. Siu, R. C. C. Wu, M. F. Shaio, L. Y. Huang, C. P. Fung, C. M. Lee, and W. L. Cho. 2007. Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:801-806. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing, 19th informational supplement. M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Dortet, L., P. Legrand, C.-J. Soussy, and V. Cattoir. 2006. Bacterial identification, clinical significance, and antimicrobial susceptibilities of Acinetobacter ursingii and Acinetobacter schindleri, two frequently misidentified opportunistic pathogens. J. Clin. Microbiol. 44:4471-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerner-Smidt, P., I. Tjernberg, and J. Ursing. 1991. Reliability of phenotypic tests for identification of Acinetobacter species. J. Clin. Microbiol. 29:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung, Y. T., Y. T. Lee, L. J. Huang, T. L. Chen, K. W. Yu, C. P. Fung, W. L. Cho, and C. Y. Liu. 2009. Clinical characteristics of patients with Acinetobacter junii infection. J. Microbiol. Immunol. Infect. 42:47-53. [PubMed] [Google Scholar]
- 9.Kilic, A., H. Li, A. Mellmann, A. C. Basustaoglu, M. Kul, Z. Senses, H. Aydogan, C. W. Stratton, D. Harmsen, and Y.-W. Tang. 2008. Acinetobacter septicus sp. nov. association with a nosocomial outbreak of bacteremia in a neonatal intensive care unit. J. Clin. Microbiol. 46:902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krawczyk, B., K. Lewandowski, and J. Kur. 2002. Comparative studies of the Acinetobacter genus and the species identification method based on the recA sequences. Mol. Cell. Probes. 16:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Lee, M. F., C. F. Peng, H. J. Hsu, and Y. H. Chen. 2008. Molecular characterisation of the metallo-beta-lactamase genes in imipenem-resistant Gram-negative bacteria from a university hospital in southern Taiwan. Int. J. Antimicrob. Agents 32:475-480. [DOI] [PubMed] [Google Scholar]
- 12.Moubareck, C., S. Bremont, M. C. Conroy, P. Courvalin, and T. Lambert. 2009. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3579-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemec, A., M. Musilek, O. Sedo, T. De Baere, M. Maixnerova, T. J. van der Reijden, Z. Zdrahal, M. Vaneechoutte, and L. Dijkshoorn. 6 August 2009, posting date. Acinetobacter berezinae sp. nov. and Acinetobacter guillouiae sp. nov., to accommodate, respectively, Acinetobacter genomic species 10 and Acinetobacter genomic species 11. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijs.0.013656-0. [DOI] [PubMed]
- 14.Segal, H., S. Garny, and B. G. Elisha. 2005. Is IS(ABA-1) customized for Acinetobacter? FEMS Microbiol. Lett. 243:425-429. [DOI] [PubMed] [Google Scholar]
- 15.Tjernberg, I., and J. Ursing. 1989. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS 97:595-605. [DOI] [PubMed] [Google Scholar]
- 16.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 17.Zbinden, A., E. C. Bottger, P. P. Bosshard, and R. Zbinden. 2007. Evaluation of the colorimetric VITEK 2 card for identification of gram-negative nonfermentative rods: comparison to 16S rRNA gene sequencing. J. Clin. Microbiol. 45:2270-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]