Abstract
We evaluated a single membrane device assay for simultaneously detecting both Clostridium difficile glutamate dehydrogenase (GDH) and toxin A/B antigens against a standard that combines two PCR assays and cytotoxigenic culture. Results showing dual GDH and toxin A/B antigen positives and negatives can be reported immediately as true positives and negatives, respectively. Specimens with discrepant results for GDH and toxins A/B, which comprised 13.2% of the specimens, need to be retested.
Rapid and accurate diagnosis of Clostridium difficile infection (CDI) is crucial for patient care, infection control, and efficient surveillance. The well-accepted standard is cytotoxigenic culture, which is done by culturing C. difficile from the stool and then performing a cytotoxin assay on the isolate (9). The cytotoxigenic culture is labor-intensive, subjective, and time-consuming, which has limited its wide use in the clinical setting. Enzyme immunoassays (EIAs) are the most common diagnostic laboratory methods used for rapid detection of C. difficile-specific glutamate dehydrogenase (GDH) and/or toxin A/B antigens in stool specimens. However, traditional EIAs lack sensitivity and specificity (4, 5, 11, 12, 18).
Recently, a C. Diff Quik Chek Complete dual-antigen EIA (D-EIA; TechLab, Blacksburg VA) became commercially available; this assay comprises rapid detection of both GDH antigen and toxin A/B with one easy-to-use cartridge (Fig. 1). Previous studies based on two membrane-bound enzyme immunoassays for GDH and toxins A/B indicated that the single GDH testing was more sensitive than that for detection of C. difficile toxins A/B; however, false-positive results were recognized upon comparison with results for culture (10, 15). It has been recommended that GDH be used as the first-line screening test, followed by cell culture for toxin testing (2, 15).
FIG. 1.
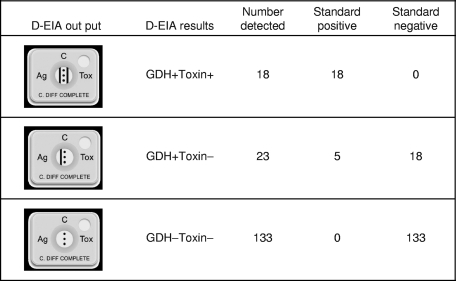
Performance of D-EIA for detection of C. difficile in stool specimens. Results for GDH− Toxin+ specimens were not observed. D-EIA, C. Diff Quik Chek Complete dual-antigen EIA; GDH, glutamate dehydrogenase.
More than 700,000 patient visits occur each year, with approximately 35,000 patients being admitted at the Vanderbilt University Medical Center (VUMC). Approximately 9,000 stool specimens were submitted for C. difficile testing for the year 2008. Currently, the Premier toxin A and B EIA (A/B EIA; Meridian Bioscience, Inc., Cincinnati, OH) is used in the clinical microbiology laboratory for detection of C. difficile toxin in stool samples: this test uses a 96-well microtiter format to detect both toxins A and B (7, 8, 11, 17). In this study, we validated the D-EIA in comparison to a standard that combines two PCR assays and a cytotoxigenic culture.
Patients and specimens.
Consecutive stool specimens submitted for C. difficile testing were collected between 2 and 14 May 2009. Liquid or soft stool specimens with sufficient leftover volumes were included. Multiple specimens, up to three, from the same patient may be included during the study period. This study was approved by the VUMC Institutional Review Board. Specimens were stored refrigerated and tested by enzyme immunoassays and molecular assays within 48 h after collection. Specimens were stored at −80°C and sent out for anaerobic C. difficile culture.
C. difficile antigen assays.
The C. Diff Quik Chek Complete dual-antigen EIA (D-EIA) (lot numbers 0309067 and 0809131) was performed according to the manufacturer's instructions. In brief, 25 μl or an equivalent volume of stool specimens was added in a tube containing the diluent and conjugate (TechLab), and the mixture was transferred to the device sample well. After incubation for 15 min at room temperature, the wash buffer and then the substrate (TechLab) were added to the reaction window. Results were read 10 min later. GDH antigen and/or toxins were reported positive if a visible band was seen on the antigen and/or the toxin side of the device display window, respectively. The Premier A/B EIA was performed according to manufacturer's instructions (7, 8, 11, 17).
Cytotoxigenic cell culture.
Frozen stool specimens were thawed, and 0.5 to 1.0 ml of stool was heated to 80°C for 10 min for spore enrichment. After cooling, 2 to 3 drops were inoculated onto cycloserine-cetoxitin-fructose agar (CCFA) with horse blood (Remel, Inc., Lenexa, KS) and also into prereduced chopped-meat broth. Plates were incubated anaerobically for as long as 5 days at 35°C and were examined for suspicious colonies at 24-h intervals. Suspicious colonies were subcultured and further identified as previously described (9). The broths were subcultured if the plates were negative at 5 days. For positive cultures, single isolated C. difficile colonies were tested for toxin production as follows. Colonies were inoculated into chopped-meat broth, incubated for 48 h, diluted 1:2 and 1:10, and incubated with human foreskin fibroblasts at 37°C under 5% CO2 in duplicate with and without toxin-specific antibody; the plates were examined after 24 and 48 h of incubation with a microscope at ×100 magnification; and ≥50% cell rounding was interpreted as a positive result (9, 13, 15).
BD-PCR.
The BD GeneOhm Cdiff assay uses a real-time PCR format to detect toxin B gene in stool samples and was performed as previously described (13). Every PCR run included a PCR-positive control and a negative control. The reaction tubes were placed in the SmartCycler I-CORE module (Cepheid, Sunnyvale, CA) and run using Cepheid SmartCycler software with the BD GeneOhm Cdiff real-time PCR (BD-PCR) assay amplification protocol. Results were automatically interpreted by the software as follows: “negative” indicated that no tcdB gene was detected, “positive” indicated that the tcdB gene was detected, “unresolved” indicated that either the internal control was inhibited or there was reagent failure, “invalid assay run” indicated that the PCR control (positive or negative) failed, and “not determined” indicated that there was an I-CORE module malfunction (13).
Stool processing and nucleic acid extraction.
To extract nucleic acid for the laboratory-developed PCR (LD-PCR), a Dacron swab was inserted into the stool specimen at 3 to 5 locations and swirled into a tube containing 1 ml of sterile water, making an approximate mixture of 10%. The mixture was vortexed and allowed to settle (11). For each 0.2-ml supernatant, 0.9 ml of lysis buffer (bioMérieux, Inc., Durham, NC) was added, and the mixture was directly placed in the NucliSens easyMAG system (bioMérieux, Inc.), using the default extraction protocol (14). Total nucleic acids were eluted in 55 μl of elution buffer (bioMérieux, Inc.), and 5 μl of each extract was used for nucleic acid amplification (see below). The human β-actin gene was detected as an internal amplification control (3).
LD-PCR.
The laboratory-developed PCR, followed by automated fluorescence capillary electrophoresis, was developed to amplify and detect the tcdC gene of the C. difficile pathogenicity locus. A primer set (forward primer, 5′-CAA ATT GTC TGA TGC TGA ACC-3′; reverse primer, 5′-TCA GAT GTT CTA GCT AAT TGG TCA-3′) was designed to amplify a 178- or 196-bp fragment as described previously (1). The 5′ end of the forward primer was labeled with 6-carboxyfluorescein for fragment analysis by capillary electrophoresis on an ABI 3730xl automated sequencer, and data were analyzed using GeneMapper 4.0 software. Positive specimens demonstrated fragment lengths of 178 or 196 bp, dependent on the C. difficile strain (1).
Data analysis.
A combination standard was defined as concordant results for two or more of the following assays: BD-PCR, LD-PCR, and toxigenic culture for diagnosis of CDI. Sensitivity, specificity, and positive and negative predictive values (PPV and NPV, respectively) were determined. Test hands-on time and turnaround time were calculated. Total costs, including those of reagents and labor as well as reimbursements from Medcare/Medcaid, were determined. P values were calculated, and values of ≤0.05 were considered statistically significant.
During the study period, a total of 180 stool specimens were submitted for C. difficile antigen testing. Six specimens were excluded due to insufficient volumes, and 174 specimens were included in the study. All specimens were first tested by the D-EIA (lot number 0309067), and sensitivities, specificities, and predictive values in comparison to the levels for the combined standard were determined. GDH antigen was detected in 41 specimens, with a sensitivity of 100.0% and a specificity of 88.1%. Simultaneously, in the same cartridge, toxins A/B were detected in 18 specimens, with a sensitivity of 78.3% and a specificity of 100.0% (Fig. 1). Five specimens which were GDH positive but toxin A/B negative (GDH+ Toxin−) were falsely negative in comparison to the reference standard (Fig. 1). Among the total 174 stool specimens tested, 18 (10.3%) tested positive for both GDH and toxins A/B, 23 (13.2%) tested positive for GDH but negative for toxins A/B, and 133 (76.4%) tested negative for both GDH and toxins A/B. No D-EIA results showing GDH-negative but toxin-A/B positive specimens were revealed (Fig. 1). Of the total 174 specimens, 50 (18 GDH+ Toxin+, 23 GDH+ Toxin−, and 9 GDH− Toxin−) were retested by another D-EIA device (lot number 0809131), and a perfect reproducibility was achieved.
All five assays presented satisfactory specificities, ranging from 96.7 to 100.0% (Table 1). When the combined reference standard was used, the sensitivity of the Premier A/B EIA was 69.6% while the D-EIA possessed a better sensitivity (78.3%). Both A/B EIA and D-EIA had statistically lower sensitivity than the combined standard, which had an average sensitivity of 92.8% (P = 0.023; Fisher exact test). These data indicate that a more sensitive assay, preferably in molecular platforms, is needed to retest those specimens testing positive for GDH but negative for A/B toxin by the D-EIA.
TABLE 1.
Performance, test hands-on time, turnaround time, costs, and reimbursements for five C. difficile assaysa
| Assayb | Sensitivity (%) | Specificity (%) | Hands-on time (min) | Test turnaround time (min) | Cost per test | Medcare/Medcaid reimbursement |
|---|---|---|---|---|---|---|
| A/B-EIA | 69.6 | 100.0 | 20 | 55 | $5.05 | $29.27 |
| D-EIA | 78.3 | 100.0 | 35 | 50 | $15.50 | $29.27 |
| BD-PCR | 95.7 | 100.0 | 60 | 130 | $26.70 | $51.25 |
| LD-PCR | 91.3 | 96.7 | 60 | 1,640 | $14.08 | $51.25 |
| Toxigenic culture | 91.3 | 98.3 | 45 | 7,200 | $22.00 | $42.64 |
Hands-on time and test turnaround time were calculated on the basis of a full run of 16 specimens.
A/B-EIA, Premier toxin A and B EIA; D-EIA, C. Diff Quik Chek Complete dual-antigen EIA; BD-PCR, BD GeneOhm Cdiff real-time PCR; LD-PCR, laboratory-developed PCR.
Values representing hands-on time, test turnaround time, cost, and reimbursement for the four assays are contrasted in Table 1. Both rapid antigen EIAs had test turnaround times within 1 h. The D-EIA costs three times more than the A/B EIA, with a benefit margin of $13.77. BD-PCR and LD-PCR resulted in greater reimbursements, ranging from $24.55 to $37.17. The long test turnaround time of LD-PCR will limit the wide application of this assay in clinical services (Table 1). In comparison to the combined standard, the 10.3% GDH and toxin A/B dual positives were true positives and the 76.4% GDH and toxin A/B dual negatives were true negatives. These results can be finalized within hours. The 13.2% GDH-positive but toxin A/B-negative specimens need to be retested by another assay, such as PCR, which has higher sensitivity, longer test turnaround time, and higher costs.
In summary, the C. Diff Quik Chek Complete D-EIA provides a rapid and reproducible first-line screening assay for laboratory diagnosis of C. difficile infection. The majority of test results can be performed and reported within 1 h with satisfactory sensitivity and specificity. As costs associated with devices decrease and FDA-cleared user-friendly products become available, GDH-positive, toxin-negative results can be confirmed by a molecular assay on-site even in smaller hospitals.
Footnotes
Published ahead of print on 2 December 2009.
REFERENCES
- 1.Chow, W. H., C. McCloskey, Y. Tong, L. Hu, Q. You, C. P. Kelly, H. Kong, Y. W. Tang, and W. Tang. 2008. Application of isothermal helicase-dependent amplification with a disposable detection device in a simple sensitive stool test for toxigenic Clostridium difficile. J. Mol. Diagn. 10:452-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilligan, P. H. 2008. Is a two-step glutamate dehyrogenase antigen-cytotoxicity neutralization assay algorithm superior to the premier toxin A and B enzyme immunoassay for laboratory detection of Clostridium difficile? J. Clin. Microbiol. 46:1523-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, H., J. S. Dummer, W. R. Estes, S. Meng, P. F. Wright, and Y. W. Tang. 2003. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J. Clin. Microbiol. 41:187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyerly, D. M., L. M. Neville, D. T. Evans, J. Fill, S. Allen, W. Greene, R. Sautter, P. Hnatuck, D. J. Torpey, and R. Schwalbe. 1998. Multicenter evaluation of the Clostridium difficile TOX A/B TEST. J. Clin. Microbiol. 36:184-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musher, D. M., A. Manhas, P. Jain, F. Nuila, A. Waqar, N. Logan, B. Marino, and E. A. Graviss. 2007. Detection of Clostridium difficile toxin: comparison of enzyme immunoassay results with results obtained by cytotoxicity assay. J. Clin. Microbiol. 45:2737-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reference deleted.
- 7.Novak-Weekley, S. M., and M. H. Hollingsworth. 2008. Comparison of the premier toxin A and B assay and the TOX A/B II assay for diagnosis of Clostridium difficile infection. Clin. Vaccine Immunol. 15:575-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Connor, D., P. Hynes, M. Cormican, E. Collins, G. Corbett-Feeney, and M. Cassidy. 2001. Evaluation of methods for detection of toxins in specimens of feces submitted for diagnosis of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 39:2846-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reller, M. E., C. A. Lema, T. M. Perl, M. Cai, T. L. Ross, K. A. Speck, and K. C. Carroll. 2007. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J. Clin. Microbiol. 45:3601-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes, R. C., M. A. John, D. L. Ayotte, A. Covacich, S. Milburn, and Z. Hussain. 2007. Performance of TechLab C. DIFF QUIK CHEK and TechLab C. DIFFICILE TOX A/B II for the detection of Clostridium difficile in stool samples. Diagn. Microbiol. Infect. Dis. 59:33-37. [DOI] [PubMed] [Google Scholar]
- 11.Sloan, L. M., B. J. Duresko, D. R. Gustafson, and J. E. Rosenblatt. 2008. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 46:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snell, H., M. Ramos, S. Longo, M. John, and Z. Hussain. 2004. Performance of the TechLab C. DIFF CHEK-60 enzyme immunoassay (EIA) in combination with the C. difficile Tox A/B II EIA kit, the Triage C. difficile panel immunoassay, and a cytotoxin assay for diagnosis of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 42:4863-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamper, P. D., R. Alcabasa, D. Aird, W. Babiker, J. Wehrlin, I. Ikpeama, and K. C. Carroll. 2009. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing Clostridium difficile in clinical samples. J. Clin. Microbiol. 47:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang, Y. W., S. E. Sefers, H. Li, D. J. Kohn, and G. W. Procop. 2005. Comparative evaluation of three commercial systems for nucleic acid extraction from urine specimens. J. Clin. Microbiol. 43:4830-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ticehurst, J. R., D. Z. Aird, L. M. Dam, A. P. Borek, J. T. Hargrove, and K. C. Carroll. 2006. Effective detection of toxigenic Clostridium difficile by a two-step algorithm including tests for antigen and cytotoxin. J. Clin. Microbiol. 44:1145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Whittier, S., D. S. Shapiro, W. F. Kelly, T. P. Walden, K. J. Wait, L. T. McMillon, and P. H. Gilligan. 1993. Evaluation of four commercially available enzyme immunoassays for laboratory diagnosis of Clostridium difficile-associated diseases. J. Clin. Microbiol. 31:2861-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng, L., S. F. Keller, D. M. Lyerly, R. J. Carman, C. W. Genheimer, C. A. Gleaves, S. J. Kohlhepp, S. Young, S. Perez, and K. Ye. 2004. Multicenter evaluation of a new screening test that detects Clostridium difficile in fecal specimens. J. Clin. Microbiol. 42:3837-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]