Abstract
Among nonhemolytic Streptococcus pyogenes (group A streptococcus) strains (n = 9) isolated from patients with pharyngitis or acute otitis media, we identified three deletions in the region from the epf gene, encoding the extracellular matrix binding protein, to the sag operon, mediating streptolysin S production.
In clinical laboratories, the property of beta-hemolysis on a blood agar plate is a characteristic used to preliminarily detect pyogenic streptococci such as Streptococcus pyogenes (group A streptococcus [GAS]), Streptococcus agalactiae (group B streptococcus), and Streptococcus dysgalactiae subsp. equisimilis (12). GAS produces two hemolysins: oxygen-dependent, labile streptolysin O, encoded by the slo gene (8), and oxygen-stable streptolysin S (SLS), encoded by the sag operon extending from the sagA gene to sagI (5). SLS, a potent cytolytic toxin produced by nearly all strains of GAS, is responsible for the zone of hemolysis surrounding GAS colonies grown under routine CO2 culture conditions.
The sagA gene, which is positioned upstream in the sag operon, encodes a prepropeptide consisting of 53 amino acid (aa) residues, including a Gly-Gly proteolytic cleavage site that has been predicted to release a propeptide of 30 aa from a 23-aa leader sequence. The propeptide is considered to be the structural element of SLS. The remaining genes in the operon have features consistent with export functions, posttranslational modification of the SLS peptide, and a possible immunity protein (3).
Rarely, nonhemolytic variants of GAS have been isolated from patients with pharyngitis (6, 10), pneumonia (13), sepsis (2, 14), and cellulitis (11). These isolates were probably not producers of SLS, but the molecular cause had previously not been explained. Recently, based on mutational analysis, it was reported that all genetic components of the sag operon are required for the expression of functional SLS as an important virulence factor in the pathogenesis of invasive infection (3). In this study, we aimed to determine the reason for nonhemolysis by GAS clinical isolates at the molecular level.
A total of 1,690 samples, including throat swabs (n = 1,513) from patients with pharyngitis/tonsillitis and middle ear fluid (n = 177) from patients with acute otitis media (AOM), were sent to our laboratory by clinical physicians. Real-time PCR was immediately carried out routinely, in parallel with culturing, for all clinical samples on the day they were received. The real-time PCR used in this study was an application of the methods using molecular beacon probes and primers that we had constructed to detect six pathogens, including GAS, in samples from patients with respiratory tract infection (9). The set of primers and the probe for 16S rRNA genes used for the identification of GAS are as follows: sense primer, 5′-GAGAGACTAACGCATGTTAGTA-3′; reverse primer, 5′-TAGTTACCGTCACTTGGTGG-3′; and probe, 6-carboxyfluorescein-CGCGATCGCGACGATACATAGCCGACCTGGATCGCG-Black Hole Quencher 1. DNA extraction with the Extragen II kit (TOSOH, Tokyo, Japan) and subsequent DNA amplification with the Mx3000P system (Stratagene, La Jolla, CA) were performed by our protocol (9). On the following day, when no colonies with hemolysis were observed on the blood agar plate, Gram staining and reexamination by real-time PCR were carried out for some nonhemolytic colonies having different shapes, regardless of the positive PCR results for GAS on the preceding day. Next, colonies were confirmed to have characteristics of GAS by (i) an agglutination test for Lancefield group A antigen (Streptex; Mitsubishi Chemical Medience, Tokyo, Japan), (ii) use of the API Strep system (bioMérieux, Tokyo, Japan), and (iii) evaluation for the pyrrolidonyl arylamidase reaction (Oxoid, Hampshire, United Kingdom) in accordance with the Manual of Clinical Microbiology (12).
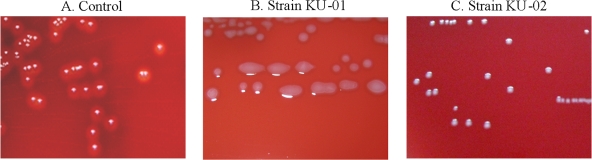
We finally identified nine nonhemolytic GAS strains from among 818 clinical isolates (1.1%) obtained from patients with pharyngitis/tonsillitis or AOM between November 2006 and March 2009. Colonies of these GAS isolates remained non-beta-hemolytic under aerobic, 5% CO2, and anaerobic conditions. Representative examples are shown in Fig. 1. The clinical and epidemiologic features of these isolates, including the emm type, the sequence type determined by multilocus sequence typing (MLST), and the type of deletion in the sag region, are listed in Table 1.
FIG. 1.
Nonhemolytic S. pyogenes colonies grown on 5% sheep blood agar plates for 18 h at 37°C. (A) Control strain; (B and C) nonhemolytic strains.
TABLE 1.
Clinical and epidemiologic features of nonhemolytic S. pyogenes isolates
| Strain no. | Date of isolation | District | Patient |
emm type | Sequence type | Deletion type involving sag operon | ||
|---|---|---|---|---|---|---|---|---|
| Age (yr) | Gender | Disease | ||||||
| KU-01 | Nov. 2006 | Gunma | 30 | M | AOM | emm1.0 | 28 | 1 |
| KU-02 | Feb. 2008 | Chiba | 2 | M | Pharyngitis | emm12.0 | 36 | 2 |
| KU-03 | Apr. 2008 | Chiba | 4 | F | Pharyngitis | emm12.0 | 36 | 2 |
| KU-04 | May 2008 | Chiba | 5 | F | Pharyngitis | emm12.0 | 36 | 2 |
| KU-05 | May 2008 | Chiba | 6 | M | Pharyngitis | emm12.0 | 36 | 2 |
| KU-06 | May 2008 | Chiba | 2 | F | Pharyngitis | emm12.0 | 36 | 2 |
| KU-07 | Nov. 2008 | Niigata | 4 | M | Pharyngitis | emm12.0 | 36 | 2 |
| KU-08 | Jan. 2009 | Sendai | 5 | F | Pharyngitis | emm1.0 | 28 | 3 |
| KU-09 | Jan. 2009 | Sendai | 5 | M | Pharyngitis | emm1.0 | 28 | 3 |
The emm types of these strains were determined based on DNA sequence homology by comparison of sequences with entries in the CDC database using the Streptococci Group A Subtyping Request Form Blast 2.0 Server (http://www.cdc. gov/ncidod/biotech/strep/strepblast.htm). DNA sequences approximately 14,000 bp in length, extending from the epf gene, encoding an extracellular matrix binding protein, to the sag operon, encoding SLS, were determined for all nonhemolytic GAS strains. Primers used initially for long-DNA-fragment amplification included a sense primer, 5′-TGTGGATGCCGTTTAGAACA-3′, and a reverse primer, 5′-GAATAGCGACACGCCTTAGC-3′.
For MLST of the nine GAS strains, DNA sequences from seven housekeeping loci were determined by the methods described by Enright et al. (4), and the sequence results were compared with the data in the S. pyogenes database (http://spyogenes.mlst.net/misc/info.asp).
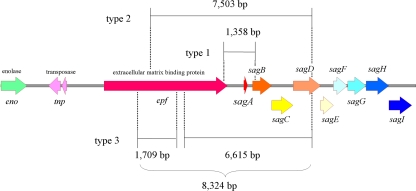
Figure 2 depicts three types of DNA deletions in the region from the epf gene to the sag operon that were identified among the strains. Deletion type 1 was exhibited by one strain (strain KU-01; emm1.0) that lacked a 1,358-bp segment extending from the 3′-terminal region of the epf gene to the 5′-terminal region of the sagB gene. Six other strains (strains KU-02 to strain KU-07; emm12.0) with deletion type 2 lacked a 7,503-bp segment extending from the middle region of the epf gene to the sagD gene. The remaining two strains (KU-08 and KU-09; emm1.0) represented deletion type 3 and had discontinuous deletions in two regions: a 1,709-bp segment in the epf gene and another segment of 6,615 bp extending from the middle region of the epf gene to the sagD gene. All these deletions encompassed the region of the promoter and the sagA gene encoding the precursor of SLS.
FIG. 2.
Three deletion types identified near and/or in the sag operon encoding SLS. Large deletions encompassed the regions of the promoter and sagA, encoding the precursor of SLS. Deletion type 1, accession number AB518308; deletion type 2, accession number AB518309; deletion type 3, accession number AB518310.
Two types of sag regions have been identified using a DNA database for GAS genomes. In one, the ordinary type, the genes are aligned beginning with the eno gene, encoding enolase, and extending through sagA to sagI. The other type possesses both a tnp gene, encoding transposase, and an epf gene between the eno and the sagA genes. GAS strains identified as having emm2, emm3, and emm5 represented the former type, while the emm1, emm4, emm12, and emm28 strains carried the latter type. All nine strains analyzed in this study contained the latter type. Although these unique deletions suggest some associations with a transposon or insertion sequence, such details remain to be clarified.
Transcripts of the nga and the slo genes are known to be produced by read-through from the nga promoter (7). Although the data are not shown here, nucleotide sequences of 4,754 bp in length from the nga gene, including the promoter region, to the end of open reading frame of the slo gene from the nine strains were identified. No mutations or nucleotide deletions were detected in this region; therefore, the slo gene was intact in all strains.
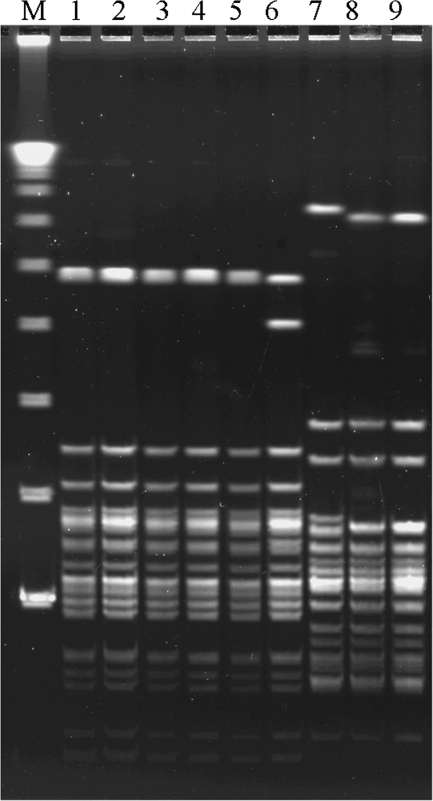
DNA profiles of the nine nonhemolytic strains after pulsed-field gel electrophoresis (PFGE) are shown in Fig. 3. PFGE was performed with the ApaI restriction enzyme (Takara Bio, Kyoto, Japan). The DNA fragments were separated on a 1% agarose gel by using a contour-clamped homogeneous electric field mapper system (Bio-Rad, Tokyo, Japan) for 18 h at 14°C in 0.5× TBE buffer (0.05 M Tris, 0.05 M boric acid, and 1 mM EDTA [pH 8.0]) at 5.7 V/cm with pulse times of 3 to 20 s at an angle of 120° (1). Six strains with emm12.0 isolated from patients in different regions, i.e., the Chiba and Niigata prefectures, Japan, showed similar DNA restriction patterns. Furthermore, three strains from Gunma prefecture and the Sendai City area identified as emm1.0 strains showed very similar DNA restriction patterns.
FIG. 3.
PFGE patterns for nine nonhemolytic strains. Lanes: M, lambda ladder; 1, KU-02 (emm12.0); 2, KU-03 (emm12.0); 3, KU-04 (emm12.0); 4, KU-05 (emm12.0); 5, KU-06 (emm12.0); 6, KU-07 (emm12.0); 7, KU-01 (emm1.0); 8, KU-08 (emm1.0); and 9, KU-09 (emm1.0).
Previously described nonhemolytic GAS strains have included various T antigen types and emm types (2, 6, 10, 11, 13, 14). In this study, we analyzed nonhemolytic phenotypes of the emm1 and emm12 strains. Evidence suggests that nonhemolytic emm12 variants spread horizontally among children, considering that several cases occurred in the same area (Chiba prefecture). We also isolated three emm1 GAS strains, one mucoid type and two nonmucoid types, from samples obtained from different areas. These strains displayed different deletion types in the sag operon but showed highly similar PFGE profiles, suggestive of a common origin.
Emergence of the GAS strains described herein suggests that the routine bioassay poses a risk of missing nonhemolytic GAS colonies on blood agar plates, although nonhemolytic GAS variants are considered to be rare.
Nucleotide sequence accession numbers.
The deletion type sequences determined in this study have been deposited in GenBank under the following accession numbers: deletion type 1, AB518308; deletion type 2, AB518309; and deletion type 3, AB518310.
Acknowledgments
This work was supported by a grant for a Research Project for Emerging and Re-emerging Infectious Diseases (no. H-20-002) from the Japanese Ministry of Health, Labor and Welfare.
Footnotes
Published ahead of print on 16 December 2009.
REFERENCES
- 1.Chiba, N., M. Morozumi, K. Sunaoshi, S. Takahashi, M. Takano, T. Komori, K. Sunakawa, and K. Ubukata. 2009. Serotype and antibiotic resistance of isolates from patients with invasive pneumococcal disease in Japan. Epidemiol. Infect. 138:61-68. [DOI] [PubMed] [Google Scholar]
- 2.Cimolai, N., C. Trombley, and N. M. Bhanju. 2002. Nonhemolytic Streptococcus pyogenes causing invasive infection. Clin. Pediatr. (Philadelphia) 41:453. [DOI] [PubMed] [Google Scholar]
- 3.Datta, V., S. M. Myskowski, L. A. Kwinn, D. N. Chiem, N. Varki, R. G. Kansal, M. Kotb, and V. Nizet. 2005. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol. Microbiol. 56:681-695. [DOI] [PubMed] [Google Scholar]
- 4.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James, L., and R. B. McFarland. 1971. An epidemic of pharyngitis due to a nonhemolytic group A streptococcus at Lowry air force base. N. Engl. J. Med. 284:750-752. [DOI] [PubMed] [Google Scholar]
- 7.Kimoto, H., Y. Fujii, Y. Yokota, and A. Taketo. 2005. Molecular characterization of NADase-streptolysin O operon of hemolytic streptococci. Biochim. Biophys. Acta 1681:134-149. [DOI] [PubMed] [Google Scholar]
- 8.McCormick, J. K., M. L. Peterson, and P. M. Schlievert. 2006. Toxins and superantigens of group A streptococci, p. 47-58. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens, 2nd ed. ASM Press, Washington, DC.
- 9.Morozumi, M., E. Nakayama, S. Iwata, Y. Aoki, K. Hasegawa, R. Kobayashi, N. Chiba, T. Tajima, and K. Ubukata. 2006. Simultaneous detection of pathogens in clinical samples from patients with community-acquired pneumonia by real-time PCR with pathogen-specific molecular beacon probes. J. Clin. Microbiol. 44:1440-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin, L. G., and G. S. Mirkin. 2000. Apparent false positive detection of group A Streptococcus antigen resulting from pharyngeal infection with a nonhemolytic Streptococcus pyogenes. Pediatr. Infect. Dis. J. 19:672-674. [DOI] [PubMed] [Google Scholar]
- 11.Sönksen, U. W., K. Ekelund, and B. G. Bruun. 2007. Case of bacteraemic cellulitis by a non-haemolytic strain of Streptococcus pyogenes. Scand. J. Infect. Dis. 39:262-264. [DOI] [PubMed] [Google Scholar]
- 12.Spellerberg, B., and C. Brandt. 2007. Streptococcus, p. 412-429. In P. A. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 13.Taylor, M. B., and T. Barkham. 2002. Fatal case of pneumonia caused by a nonhemolytic strain of Streptococcus pyogenes. J. Clin. Microbiol. 40:2311-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner, D. P., and S. L. Gunn. 2007. Fatal case of sepsis caused by a non-haemolytic strain of Streptococcus pyogenes. J. Clin. Pathol. 60:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]