Abstract
We report a skin and soft-tissue infection outbreak among football team members due to a USA300 methicillin-susceptible Staphylococcus aureus (MRSA) strain with genes coding for Panton-Valentine leukocidin and the arginine catabolic mobile element. We postulate that the strain is a community-associated USA300 MRSA strain that lost methicillin resistance but retained important virulence factors.
In the United States, most community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections have been caused by the USA300 clone (12). Outbreaks caused by USA300 have been reported in at least 38 states, and it accounts for >50% of all S. aureus infections (12). USA300 belongs to multilocus sequence type clonal complex 8, sequence type 8, and spa type 008. It is characterized by the presence of SCCmec IV, which confers methicillin resistance, the arginine catabolic mobile element (ACME), and genes coding for Panton-Valentine leukocidin (PVL) (2).
During the fall of 2007, we detected an outbreak of skin infections among football players at a New England college. The outbreak was characterized by rapid spread among players and abscess formation, features characteristic of previously described CA-MRSA outbreaks among members of athletic teams. The causative organism was methicillin-susceptible S. aureus (MSSA), however, and it showed an unusual resistance profile (resistance to erythromycin and ciprofloxacin). The epidemiologic and clinical pattern of the outbreak and the resistance profile prompted us to investigate the cases and look for molecular similarities between the outbreak strain and USA300.
A case was defined as a college football player with a ciprofloxacin-resistant MSSA skin infection with onset between preseason training and the end of the season. Cases were detected by active surveillance for cellulitis and/or skin abscess among players and athletic trainers. Incision and drainage were performed on all abscesses, and specimens were sent for routine cultures. On day 16 of the outbreak, all 110 football players and eight athletic trainers were screened for S. aureus colonization. The anterior nares and pharynx were sampled using rayon-tipped swabs (Fisherfinest*; Fisher Scientific, Ontario, Canada), and specimens from each person were combined for microbiological evaluation.
Isolates from cases were typed by pulsed-field gel electrophoresis (PFGE) using restriction endonuclease SmaI (11). Bionumerics software (version 5.0; Applied Maths, Kortrijk, Belgium) was used to compare isolates to each other and to the USA300 MRSA control strain. spa typing (6) and PCR to determine the presence or absence of PVL genes (9) and the ACME-specific arcA gene (2) were also performed.
Eight of the 110 football players met the outbreak case definition (attack rate, 7%; Table 1). All of the isolates had the same antibiotic susceptibility profile, with susceptibility to oxacillin, cefazolin, ceftriaxone, piperacillin-tazobactam, clindamycin, gentamicin, rifampin, tetracycline, trimethoprim-sulfamethoxazole, and vancomycin and resistance to penicillin, erythromycin, ciprofloxacin, and bacitracin. The isolates were indistinguishable by PFGE, with a macrorestriction fragment pattern typical of USA300 (Fig. 1). The outbreak isolate was spa type 008 and contained PVL-encoding and ACME-specific arcA genes, consistent with a USA300 designation. Compared with the USA300 control, the outbreak isolates were missing an approximately 30-kb portion of the SmaI fragment of mecA.
TABLE 1.
Demographic and clinical description of the eight cases of MSSA infection associated with a New England college football team outbreak in 2007
| Case no. | Age (yr) | Outbreak day | Infection date | Infection site | I and Da plus antibiotics | Skin infection in past year |
|---|---|---|---|---|---|---|
| 1 | 19 | 1 | 8/27/2007 | Leg | Yes | Yesb |
| 2 | 20 | 2 | 8/28/2007 | Knee | Yes | No |
| 3 | 22 | 3 | 8/29/2007 | Forearm | Yes | No |
| 4 | 18 | 4 | 8/30/2007 | Forearm | Yes | No |
| 5 | 19 | 12 | 9/7/2007 | Thigh | Yes | No |
| 6 | 19 | 12 | 9/7/2007 | Chest | Yes | No |
| 7 | 18 | 22 | 9/17/2007 | Knee | Yes | No |
| 8 | 21 | 55 | 10/20/2007 | Elbow | Yes | No |
I and D, incision and drainage.
Arm lesion 3 weeks before outbreak.
FIG. 1.
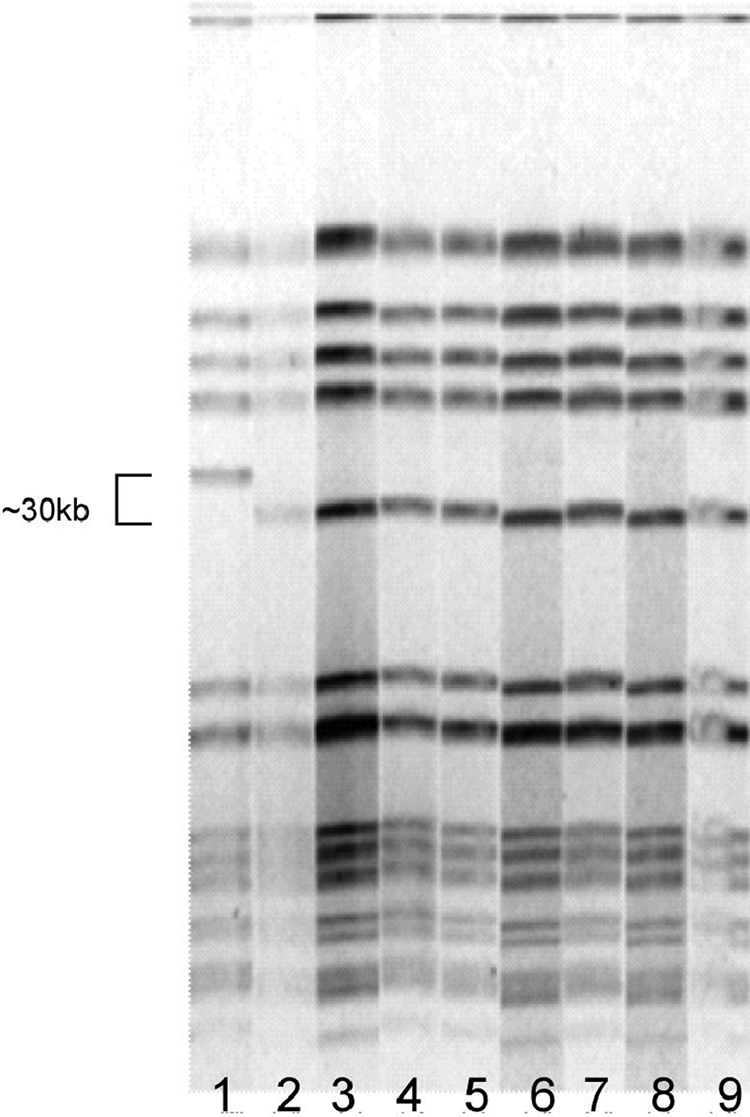
PFGE results of the eight cases of MSSA infection associated with a New England college football team outbreak in 2007, suggesting a single source of infection (lanes 2 to 9). Lane 1 is a reference strain of USA300 MRSA which differs from the outbreak strain by a single band.
Of the 118 players and athletic trainers, 62 (53%) were colonized with MSSA but none was colonized with the outbreak strain. Five (4%) were colonized with MRSA. One of the eight infected players was colonized with a nonoutbreak MSSA strain.
Several features of our outbreak are similar to those previously described in reported USA300 MRSA outbreaks, such as the rapidity of spread and the severity of tissue inflammation and abscess formation. The finding that none of the players or trainers was colonized with the outbreak strain, despite a high MSSA colonization rate, is also consistent with USA300 MRSA outbreaks in football teams (1, 8). It is important to note, however, that it is possible that some carriers were not detected because broth enrichment was not used with our selective culture media.
The outbreak strain is a USA300 strain by PFGE and contains genes coding for PVL and ACME. The outbreak strain differs from USA300 MRSA only by an approximately 30-kb deletion consistent with loss of SCCmec IV. Our isolate is similar to USA300 MSSA strains previously described in non-outbreak-associated settings (3, 10, 13).
We believe that our outbreak isolate represents a strain of MSSA that evolved from USA300 by losing SCCmec IV. USA300 MRSA is thought to have evolved from MSSA through horizontal transfer of mobile elements (2). SCCmec IV and ACME were likely acquired from S. epidermidis, given that they were highly prevalent in S. epidermidis years before they were detected in S. aureus (4). Since SCCmec IV and ACME integrate at the same attachment site, orfX, it has been hypothesized that they may have been acquired in association (2, 7). Diep et al. (2) proposed that ACME from S. epidermidis is excised by ccrAB recombinase from an adjacent SCC element and then integrated into the USA300 MRSA genome by ccrAB from an existing SCCmec IV element (2). The dependence of ACME on ccrAB from an existing SCCmec element may explain the common attachment site.
Our outbreak isolate is similar to the ACME-positive, SCCmec IV-negative USA300 strain described by Goering et al. (5). Their analysis of the junction between ACME and the attachment site, orfX, did not reveal any SCCmec IV elements. Since the insertion of ACME is most likely dependent on SCCmec recombinase (ccr) genes, the absence of SCCmec IV in this isolate suggests that it has been deleted. Donnio et al. (3) showed that an MSSA strain with an unusual resistance pattern (resistant to erythromycin and fluoroquinolones) could be derived from a dominant epidemic MRSA strain with loss of a 40-kb DNA fragment corresponding to the SCCmec region (3). Further analysis of their USA300 MSSA isolates revealed small fragments of SCCmec, further supporting this idea (3).
This outbreak of severe skin and soft-tissue infections among healthy athletes provides evidence that USA300 MRSA is capable of losing its methicillin resistance while maintaining its virulence. If loss of resistance represents the latest evolutionary development for S. aureus, it has important implications for infection surveillance and prevention and control programs, which are increasingly based on detection of methicillin resistance as a trigger for interventions.
Footnotes
Published ahead of print on 9 December 2009.
REFERENCES
- 1.Begier, E. M., K. Frenette, N. L. Barrett, P. Mshar, S. Petit, D. J. Boxrud, K. Watkins-Colwell, S. Wheeler, E. A. Cebelinski, A. Glennen, D. Nguyen, and J. L. Hadler. 2004. A high-morbidity outbreak of methicillin-resistant Staphylococcus aureus among players on a college football team, facilitated by cosmetic body shaving and turf burns. Clin. Infect. Dis. 39:1446-1453. [DOI] [PubMed] [Google Scholar]
- 2.Diep, B. A., S. R. Gill, R. F Chang, T. H. Phan, J. F. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Pedreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 3.Donnio, P. Y., D. C. Oliveira, N. A. Faria, N. Wilhelm, A. Le Coustumier, and H. de Lencastre. 2005. Partial excision of the chromosomal cassette containing the methicillin resistance determinant results in methicillin-susceptible Staphylococcus aureus. J. Clin. Microbiol. 43:4191-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng, Y., C. J. Chen, L. H. Su, S. Hu, J. Yu, and C. H. Chiu. 2008. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol. Rev. 32:23-37. [DOI] [PubMed] [Google Scholar]
- 5.Goering, R. V., L. K. McDougal, G. E. Fosheim, K. K. Bonnstetter, D. J. Wolter, and F. C. Tenover. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J. Clin. Microbiol. 45:1981-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Highlander, S. K., K. G. Hultén, X. Qin, H. Jiang, S. Yerrapragada, E. O. Mason, Jr., Y. Shang, T. M. Williams, R. M. Fortunov, Y. Liu, O. Igboeli, J. Petrosino, M. Tirumalai, A. Uzman, G. E. Fox, A. M. Cardenas, D. M. Muzny, L. Hemphill, Y. Ding, S. Dugan, P. R. Blyth, C. J. Buhay, H. H. Dinh, A. C. Hawes, M. Holder, C. L. Kovar, S. L. Lee, W. Liu, L. V. Nazareth, Q. Wang, J. Zhou, S. L. Kaplan, and G. M. Weinstock. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazakova, S. V., J. C. Hageman, M. Matava, A. Srinivasan, L. Phelan, B. Garfinkel, T. Boo, S. McAllister, J. Anderson, B. Jensen, D. Dodson, D. Lonsway, L. K. McDougal, M. Arduino, V. J. Fraser, G. Killgore, F. C. Tenover, S. Cody, and D. B. Jernigan. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352:468-475. [DOI] [PubMed] [Google Scholar]
- 9.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 10.McCaskill, M. L., E. O. Mason, Jr., S. L. Kaplan, W. Hammerman, L. B. Lamberth, and K. G. Hultén. 2007. Increase of the USA300 clone among community-acquired methicillin-susceptible Staphylococcus aureus causing invasive infections. Pediatr. Infect. Dis. J. 26:1122-1127. [DOI] [PubMed] [Google Scholar]
- 11.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, L. G., and B. A. Diep. 2008. Colonization, fomites, and virulence: rethinking the pathogenesis of community-acquired methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46:752-760. [DOI] [PubMed] [Google Scholar]
- 13.Orscheln, R. C., D. A. Hunstad, S. A. Fritz, J. A. Loughman, K. Mitchell, E. K. Storch, M. Gaudreault, P. L. Sellenriek, J. R. Armstrong, E. R. Mardis, and G. A. Storch. 2009. Contribution of genetically restricted, methicillin-susceptible strains to the ongoing epidemic of community-acquired Staphylococcus aureus infections. Clin. Infect. Dis. 49:536-542. [DOI] [PMC free article] [PubMed] [Google Scholar]


