Abstract
Molecular assays can provide critical information for malaria diagnosis, speciation, and drug resistance, but their cost and resource requirements limit their application to clinical malaria studies. This study describes the application of a resource-conserving testing algorithm employing sample pooling for real-time PCR assays for malaria in a cohort of 182 pregnant women in Kinshasa. A total of 1,268 peripheral blood samples were collected during the study. Using a real-time PCR assay that detects all Plasmodium species, microscopy-positive samples were amplified individually; the microscopy-negative samples were amplified after pooling the genomic DNA (gDNA) of four samples prior to testing. Of 176 microscopy-positive samples, 74 were positive by the real-time PCR assay; the 1,092 microscopy-negative samples were initially amplified in 293 pools, and subsequently, 35 samples were real-time PCR positive (3%). With the real-time PCR result as the referent standard, microscopy was 67.9% sensitive (95% confidence interval [CI], 58.3% to 76.5%) and 91.2% specific (95% CI, 89.4% to 92.8%) for malaria. In total, we detected 109 parasitemias by real-time PCR and, by pooling samples, obviated over 50% of reactions and halved the cost of testing. Our study highlights both substantial discordance between malaria diagnostics and the utility and parsimony of employing a sample pooling strategy for molecular diagnostics in clinical and epidemiologic malaria studies.
Malaria causes nearly 250 million clinical episodes every year (49), but diagnosis remains challenging (23). Microscopic examination of thin or thick smears of peripheral blood can, under optimal conditions, provide information on infecting species and infection severity, but in many areas of endemicity, the operating characteristics of microscopy are poor (34). Microscopic examination requires highly trained personnel for smear interpretation, and even well-trained microscopists frequently miss mixed-species infections. Furthermore, low-level, “submicroscopic” parasitemia which is below the detectable limit of blood smears is common in settings of acquired host immunity (2) or exposure to antimalarials (27). Though rapid antigen tests detect parasites quickly and cheaply, they cannot provide data relevant to clinical malaria studies of multispecies infections, drug resistance genotypes, and parasite burden (51).
PCR may represent a better alternative for surveillance in some situations. Conventional and real-time PCRs can provide information on parasite density (1), infecting species (41), and drug resistance alleles (50), all with high degrees of sensitivity (42). However, large-scale application of PCR to malaria trials and surveillance has not been implemented, largely because of the cost and human resource requirements of molecular diagnostics.
Pooling samples prior to diagnostic testing for low-prevalence gene targets in a population promises an opportunity to conserve resources without sacrificing diagnostic certainty. First proposed for syphilis screening (14), the technique has been successfully employed to screen blood donors for antibodies to HIV (15), hepatitis B virus (11), and hepatitis C virus (18) and to diagnose acute HIV infections by using PCR (37). In testing algorithms that employ this strategy, the number of samples included in each pool depends on the expected prevalence of the disease under study and the characteristics of the diagnostic test. If a pool tests positive, the individual samples comprising the pool are evaluated in a second round of testing. Depending on the expected prevalence of a target condition, pooling can obviate >90% of individual tests (47), with significant resource savings. Additionally, pooling is usually done robotically, limiting technician time.
Malaria control programs in areas of endemicity are producing significant decreases in clinical malaria episodes and parasite densities (5, 8, 22, 32, 45), and molecular diagnostics may become more critical in detecting parasitemias. Additionally, pooling samples for molecular diagnosis of malaria could make real-time PCR assays more feasible for large, clinical, resource-limited malaria studies. A previous longitudinal cohort of 182 pregnant women in Kinshasa documented high rates of antenatal malaria by blood smear and an association between repeated infection and intrauterine growth retardation (24). We employed a sample pooling/real-time PCR testing strategy for malaria for both quality control and investigation of the impact of submicroscopic parasitemia on birth outcomes. Herein, we report the development and results of this novel molecular testing strategy.
MATERIALS AND METHODS
Patient enrollment and sample collection.
The study was conducted at a single antenatal clinic in Kinshasa, Democratic Republic of the Congo (DRC), to determine the effect of malaria on intrauterine growth retardation. Briefly, all pregnant women >17 years of age with estimated gestational ages of <24 weeks were screened for inclusion; after being screened by ultrasound, women with confirmed gestational ages of <23 weeks were enrolled between May 2005 and 2006 and followed longitudinally until delivery. All women received both intermittent preventive therapy with sulfadoxine-pyrimethamine and an insecticide-treated bed net in accordance with DRC national guidelines. At each scheduled visit, acute visit, and delivery, blood was collected and both prepared as a thick smear and applied to filter paper (Schleicher & Schuell 903 specimen paper). Blood smears were interpreted on-site by a trained microscopist. For quality assurance, a random sample of 10% of the blood smears was reviewed by a second trained microscopist, who was masked to the initial smear results. All women provided written informed consent at enrollment, and the study was approved by the Institutional Review Boards of the University of North Carolina at Chapel Hill and the Kinshasa School of Public Health.
Filter papers with blood spots were placed in individual plastic bags with desiccant and stored at −20°C. They were transported to the United States, where three 0.6-cm-diameter punches were punched from each card and deposited into a single well of a 96-well deep-well plate. Genomic DNA (gDNA) from plates of punches was extracted using a QIAamp 96 DNA blood kit (Qiagen, Germantown, MD) with a vacuum manifold in accordance with the manufacturer's protocol. Genomic DNA was eluted into 150 μl of eluate and stored at 4°C.
Design and validation of the pan-species real-time PCR assay with pooling.
Primer and probe sequences for the block 9 region of the gene encoding the small subunit (18S) of plasmodium rRNA (ribosomal DNA [rDNA]) were modified from a published protocol which detected between 1 and 10 copies of the target DNA and was specific to plasmodium 18S DNA (Table 1 gives all oligonucleotide sequences) (38). The ability of the assay to detect different malaria species and strains was evaluated by testing the assay with gDNA from Plasmodium falciparum strain 3d7 (MR4 no. MRA-102G; ATCC, Manassas, VA); gDNA from Plasmodium vivax strain Nicaragua (MR4 no. MRA-340G); and plasmids containing the 18S rDNA genes from P. falciparum, P. vivax, Plasmodium ovale, and Plasmodium malariae (MR4 no. MRA-177, MRA-178, MRA-179, and MRA-180, respectively). The sensitivity of the assay was determined by evaluating dilutions of P. falciparum gDNA. Real-time PCR was carried out with 25-μl reaction mixtures consisting of 2 μl of DNA, 12.5 μl of 2× TaqMan Universal PCR MasterMix (Applied Biosystems, Foster City, CA), forward and reverse primers at 1,000 nM, and VIC-labeled minor-groove binding (MGB) probe at 200 nM. The cycling conditions for the Applied Biosystems 7300 system were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min.
TABLE 1.
Primer and probe sequences
| Reagent | Sequencec | Reference or source |
|---|---|---|
| Primersa | ||
| Pan-species forward | GTT AAG GGA GTG AAG ACG ATC AGA TA | 38 |
| Pan-species reverse | AAC CCA AAG ACT TTG ATT TCT CAT AAG | 38 |
| P. falciparum 18S rDNA forward | ATT GCT TTT GAG AGG TTT TGT TAC TTT | 46 |
| P. falciparum 18S rDNA reverse | GCT GTA GTA TTC AAA CAC AAT GAA CTC AA | 46 |
| P. ovale 18S rDNA forward | CCG ACT AGG TTT TGG ATG AAA GAT TTT T | 39 |
| P. ovale 18S rDNA reverse | CAA CCC AAA GAC TTT GAT TTC TCA TAA | 46 |
| P. malariae 18S rDNA forward | AGT TAA GGG AGT GAA GAC GAT CAG A | 46 |
| P. malariae 18S rDNA reverse | CAA CCC AAA GAC TTT GAT TTC TCA TAA | 46 |
| Human GAPDH forward | CCT CCC GCT TCG CTC TCT | This study |
| Human GAPDH reverse | GCT GGC GAC GCA AAA GA | This study |
| P. falciparum LDH forward | ACG ATT TGG CTG GAG CAG AT | 36 |
| P. falciparum LDH reverse | TCT CTA TTC CAT TCT TTG TCA CTC TTT C | 36 |
| MGB probesb | ||
| Pan-species | VIC-TCG TAA TCT TAA CCA TAA AC | 38 |
| P. falciparum | FAM-CAT AAC AGA CGG GTA GTC AT | 46 |
| P. ovale | VIC-CGA AAG GAA TTT TCT TAT T | 38 |
| P. malariae | FAM-ATG AGT GTT TCT TTT AGA TAG C | 46 |
| Human GAPDH | VIC-CCT CCT GTT CGA CAG TCA GCC GC | This study |
| TaqMan probeb (P. falciparum LDH) | FAM-GTA ATA GTA ACA GCT GGA TTT ACC AAG GCC CCA-TAMRA | 36 |
Synthesized by MWG/Operon Biotech (High Point, NC) and resuspended in molecular-grade water. LDH, lactate dehydrogenase.
Synthesized by Applied Biosystems (Foster City, CA) and diluted in Tris-EDTA Buffer (FisherBioTech, Fair Lawn, NJ).
“FAM” and “VIC” denote fluorescent dyes.
In order to evaluate the ability of the pan-species assay to diagnose malaria from pooled samples, P. falciparum gDNA was used to create 10 artificial test pools. Each pool was composed of 2 μl from each of 10 components; the components were either water or specified concentrations of parasite gDNA (0.001 ng/μl to 1 ng/μl). Two pools contained only water, and eight pools had a range of gDNA depending on the initial amount of gDNA in each component. The personnel testing the pools were masked to the contents of the components.
All real-time PCRs were run with the Applied Biosystems 7300 system, and all amplification curves were evaluated with ABI 7300 system sequence detection software, version 1.3.
Sample pooling and pan-species detection.
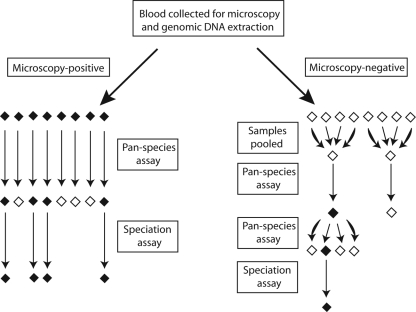
Figure 1 illustrates the sample testing algorithm. Microscopy-positive samples were individually tested in duplicate using 4 μl of extracted DNA. For microscopy-negative samples, the optimal pool size for testing was determined using an online calculator (http://www.bios.unc.edu/∼mhudgens/optimal.pooling.b.htm [last accessed 3 July 2009]), and 10 μl quantities of these samples were combined in pools of 4 original samples. Pools were then amplified in 25-μl reaction mixtures consisting of 8 μl of pooled genomic DNA and reagent concentrations identical to those described above for the pan-species assay. For pools demonstrating amplification, the four individual constituent samples were amplified as described above for the microscopy-positive samples. All reactions were run in duplicate on reaction plates that included positive controls of P. falciparum 3D7 gDNA and negative controls. Pooling and subsequent distribution of samples to reaction plates were facilitated by the use of an epMotion 5070 robot (Eppendorf, Hamburg, Germany).
FIG. 1.
Sample processing and assay work flow schematic. Microscopy-positive samples were amplified directly in the pan-species assay, and positive samples were subsequently tested in the speciation assay. Microscopy-negative samples were first grouped into pools of four and then amplified in the pan-species assay; the individual constituents of positive pools were then retested in the pan-species assay, and positive samples were subsequently tested in the speciation assay.
For quality assurance of the DNA extraction, microscopy-positive samples without amplification in the pan-species assay were subsequently amplified in assays targeting the human RNase P gene and the P. falciparum lactate dehydrogenase gene (Pfldh). The human RNase P assay consisted of 25-μl reaction mixtures with 1 μl genomic DNA, 1.25 μl RNase P reaction mixture (Applied Biosystems), and 12.5 μl TaqMan Universal PCR MasterMix (Applied Biosystems). The Pfldh assay consisted of 25-μl reaction mixtures with 2 μl of genomic DNA, 12.5 μl of TaqMan Universal PCR MasterMix, forward and reverse primers at 300 nM, and 6-carboxyfluorescein (FAM)-6-carboxytetramethylrhodamine (TAMRA) probe at 200 nM (36).
Design and validation of the real-time PCR speciation assay.
PCR primers and probes were adapted from previously published real-time PCR assays for speciation of malaria (39, 46); P. vivax is exceptionally rare in Central Africa (10) and was excluded. The assay was designed to amplify each sample in parallel in two duplex reactions. One reaction mixture consisted of 2 μl of DNA, 12.5 μl of the FastStart high-fidelity PCR system (Roche, Indianapolis, IN), P. falciparum 18S rDNA forward and reverse primers at 300 nM, P. falciparum rDNA FAM-labeled MGB probe at 200 nM, P. ovale 18S rDNA forward and reverse primers at 300 nM, P. ovale rDNA VIC-labeled MGB probe at 200 nM, and molecular-grade water added to give a total reaction volume of 25 μl. The parallel reaction mixture consisted of 2 μl of genomic DNA, 12.5 μl of the FastStart high-fidelity PCR system (Roche), P. malariae 18S rDNA forward and reverse primers at 300 nM, P. malariae rDNA FAM-labeled MGB probe at 200 nM, human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward and reverse primers at 300 nM, GAPDH VIC-labeled MGB probe at 200 nM, and molecular-grade water added to give a total reaction volume of 25 μl. Human GAPDH primers and probes were designed with Primer Express version 3.0 (Applied Biosystems). The cycling conditions for the Applied Biosystems 7300 system were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min.
To evaluate the specificity of the speciation assay, this assay was tested with the same gDNA and plasmid DNA used to evaluate the pan-malaria species assay. Because P. falciparum is the most common species causing malaria in the study region and would be dominant in mixed infections, the assay was tested to determine its ability to diagnose mixed infections with low abundances of P. ovale and P. malariae. Samples that contained 0.01 ng/μl of P. falciparum plasmid and 2.5 ng/μl of human DNA were prepared, in addition to P. ovale or P. malariae DNA ranging from 0.01 ng to 0.000001 ng/μl of plasmid.
Speciation of positive samples.
Samples demonstrating amplification in the pan-species assay were amplified with the speciation real-time PCR assay. All reactions were amplified in duplicate, and all reaction plates included three standards: a positive control consisting of mixed P. falciparum, P. malariae, and human genomic DNA; a positive control consisting of mixed P. falciparum, P. ovale, and human genomic DNA; and a negative control.
Statistical analysis.
Data were entered into Microsoft Excel 2003 (Microsoft, Redmond, WA) and were imported into SAS, version 9.1.3 (SAS, Cary, NC) for analysis. The sensitivity and specificity of peripheral blood microscopy diagnosis were calculated using real-time PCR as the referent standard. Kappa coefficients were used to quantify agreement between diagnostic outcomes. Pearson's correlation coefficient was used to examine the relationship between real-time PCR cycle threshold (CT) values and microscopy-based parasite densities.
RESULTS
Study sample.
A total of 1,111 antenatal clinic attendees were screened for inclusion in the study; 370 women met the screening criteria and, after gestational age was determined by ultrasound, 182 gave consent and were enrolled. Table 2 describes the study cohort. The mean age of the cohort was 27.5 years (standard deviation [SD], 5.3 years), and there were 47 (25.8%) primigravidae and 26 (14.3%) secundigravidae. Five participants were infected with HIV. From the 182 women, 1,268 blood samples were available for both microscopy and molecular analysis. Full details of the study have been published elsewhere (24).
TABLE 2.
Study subject demographics (n = 182)
| Characteristic | Value |
|---|---|
| Mean (SD) age (yr) | 27.5 (5.3) |
| Mean (SD) gestational age (wk) at enrollment | 18.9 (2.8) |
| No. (%) of subjects with gravidity | |
| Primigravid | 47 (25.8) |
| Secundigravid | 26 (14.3) |
| Multigravid | 109 (59.8) |
| No. (%) of subjects HIV infected | 5 (2.7) |
| Mean (SD) total no. of study visits | 6.3 (1.4) |
| Mean (SD) no. of malaria treatments received | 2.5 (0.8) |
| No. (%) of visits to subjects who always slept under an insecticide-treated bed net during the previous 2 wka | 531 (66.6) |
Bed net use was assessed for 797 regular study visits.
Performance of real-time PCR assays.
The pan-species assay reproducibly detected gDNA from stock strains of P. falciparum and P. vivax at a concentration of 0.001 ng/μl (39 parasites per microliter, based on an estimated genome size of 23 Mb), with an average CT value of 32.63 (SD, 0.008).
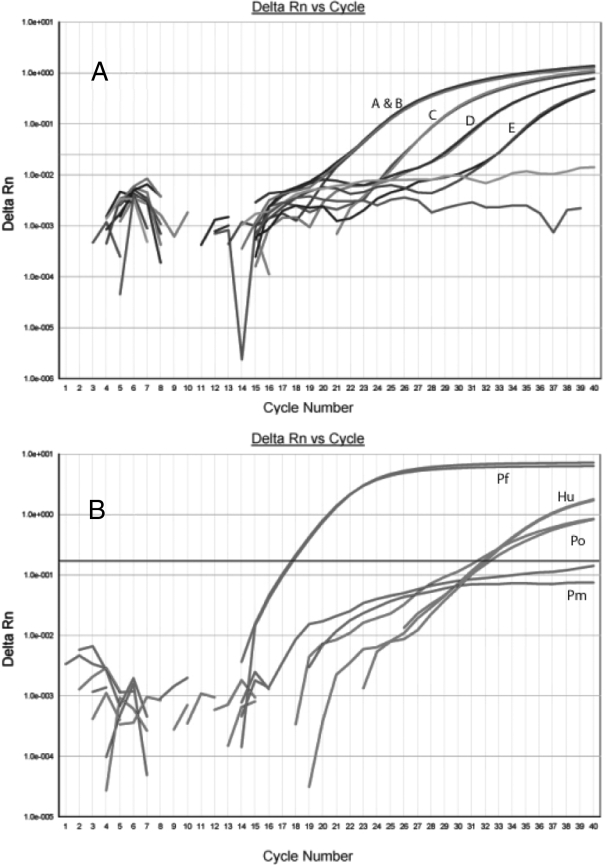
The pooling assay was tested in masked fashion with artificial test pools. The two pools without gDNA failed to amplify, and the eight pools with gDNA successfully amplified with various CT values. The assay successfully detected the pool with the lowest DNA concentration (0.001 ng/μl) (Fig. 2A), which was a single low-concentration sample pooled with nine negative samples; this indicated that the assay was sensitive enough to diagnose a low-level infection, even when tested with a pool of 10 samples. The individual samples from each pool were then tested, and the assay correctly identified all samples with malaria DNA without false-positive results.
FIG. 2.
Real-time PCR output from masked validation of pan-species and speciation assays using artificial test samples. Panel A shows the output of real-time PCR testing with 5 artificial test pools (pools A to E) in the pan-species assay. Each pool contained 2 μl of DNA from 10 samples. Pool A contained a total of 1.111 ng/μl, divided among 4 samples, and pool B contained 1 ng/μl of DNA in a single sample. Pools C, D, and E contained DNA at total concentrations of 0.1, 0.01, and 0.001 ng/μl, respectively. Panel B shows the output observed for the speciation real-time PCR assay when tested on a sample with 0.01 ng/μl of P. falciparum plasmid (Pf), 0.000001 ng/μl of P. ovale plasmid (Po), and 2.5 ng/μl of human DNA (Hu). Delta Rn, fluorescent signal generated relative to background fluorescence.
For the speciation assay, the assay correctly identified samples containing stock and 18S plasmid DNA from each malaria species. The multiplex assay was also compared to each species assay run as a monoplex assay: for each sample, no differences in CT values were observed between the monoplex and multiplex formats, indicating an absence of significant competition in the multiplex format. Additionally, the assay was able to detect the lowest concentration of plasmid accurately for both species (approximately 4 parasites per reaction, based on 6 copies of the gene and a 23-Mb genome) (Fig. 2B).
Comparison of microscopy and real-time PCR.
Overall, 176 of 1,268 blood smears were interpreted as positive (14%) by microscopy. A second microscopist reviewed 140 randomly selected smears and concurred on 11 of the 12 smears initially interpreted as positive and 127 of the 128 smears initially interpreted as negative (κ = 0.91; 95% confidence interval [CI], 0.78 to 1.0). During real-time PCR analysis, 109 DNA samples demonstrated amplification at the plasmodium 18S rDNA locus (9%). Of the 176 microscopy-positive samples, 74 (42%) were positive by direct amplification in the pan-species assay. After pooling and amplification of the 1,092 microscopy-negative samples, 35 additional individual samples were positive in the pan-species assay. All positive controls amplified appropriately; all negative controls failed to amplify. All microscopy-positive/pan-species PCR-negative samples demonstrated amplification in assays targeting the human RNase P gene, confirming successful extraction of DNA from the filter paper. In addition, 2 of the 102 microscopy-positive/pan-species PCR-negative samples amplified at the Pfldh gene, demonstrating that the sensitivity of the assay is not 100%.
With real-time PCR as the referent for malaria diagnosis, the sensitivity of microscopy was low at 67.9% (95% CI, 58.3% to 76.5%) and the specificity was higher at 91.2% (95% CI, 89.4% to 92.8%) (Table 3). Overall, the agreement between the results for microscopy and the real-time PCR assay was moderate (κ = 0.46; 95% CI, 0.39 to 0.54). For the 65 samples that were positive by both microscopy and real-time PCR and for which quantitative microscopy data were available, there was a modest correlation between smear parasite density and the CT value in the pan-species real-time PCR assay (Pearson correlation coefficient, −0.39; P < 0.01).
TABLE 3.
Comparison of results of microscopy and real-time PCR for malaria detection
| Microscopy result | No. of subjects with real-time PCR result |
Total no. of subjects | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 74 | 102 | 176 |
| Negative | 35 | 1,057 | 1,092 |
| Total | 109 | 1,159 | 1,268 |
Speciation by real-time PCR.
All 109 samples demonstrating amplification in the pan-species assay also amplified in the speciation assay. Pure P. falciparum parasitemias accounted for 104 infections, with P. falciparum mixed with P. malariae in 3 additional infections. One infection was purely P. malariae and one purely P. ovale. The human GAPDH gene amplified from all samples, and all positive species controls amplified appropriately.
Efficiency of sample pooling.
With the conservative predictions of a sensitivity of 95%, a specificity of 99%, and a 10% prevalence of parasitemia, a pool size of four samples was determined to achieve the greatest efficiency without sacrificing test sensitivity. Thus, the 1,092 microscopy-negative samples were amplified by the pan-species assay in duplicate in 293 pools of four samples each, resulting in 586 reactions. Because 38 of these pools demonstrated amplification, a further 152 amplifications in duplicate were necessary to identify the 35 positive individual samples (in retrospect, three microscopy-positive samples were inadvertently included in the pooled sample set). In total, 890 amplifications were needed to identify the 35 microscopy-negative/pan-species PCR-positive samples. Because direct amplification of all microscopy-negative samples in duplicate without pooling would have necessitated 2,184 reactions, this represents a reduction of 1,294 total reactions. In total, screening our entire cohort of 1,268 samples with partial pooling required 1,242 reactions, a reduction of 1,294 (51%) from the 2,536 reactions necessary to screen all samples without pooling (excluding control reactions). On the basis of a conservative cost per reaction of approximately $1.25 (including the probe, primers, polymerase, and consumables), directly amplifying all samples would have cost $3,170; our pooling strategy reduced this total to $1,553, for a cost reduction of 51%, or $1,617.
DISCUSSION
This longitudinal study of malaria infection during pregnancy in Kinshasa demonstrates the reliability and parsimony of a sample pooling strategy for detecting parasitemia by real-time PCR. Additionally, this study documents substantial discordance between microscopic and molecular diagnoses of malaria. We believe that these twin findings reinforce both the feasibility and the necessity of incorporating molecular diagnostics for malaria into clinical studies.
Due to the effects of malaria on birth outcomes, prevention of malaria during pregnancy is critical (13). Of the 182 women in our trial, 57 (31%) were parasitemic by PCR either during pregnancy or at delivery, similar to the rates reported in other studies (43), including those involving subjects from Kinshasa (25). Though it can be decreased substantially by intermittent preventive therapy with antimalarials (19), parasitemia is more prevalent during pregnancy (7) and is often asymptomatic (29). Among nonpregnant adults in regions where malaria is endemic, the prevalence of asymptomatic parasitemia is high (3, 17), and the application of PCR diagnoses more cases than standard blood smears (2, 12). Similarly, PCR may provide a more reliable method of screening for parasites in pregnant, semi-immune asymptomatic individuals.
Because asymptomatic populations typically have low parasite burdens (12), a sensitive screening test is essential, and pooling samples can potentially diminish sensitivity of detection of the test target in a pool of samples (47). Prior to application, the described assay was validated for detection of the equivalent of 39 parasites/μl in a single sample pooled with nine parasite-free samples (i.e., ∼4 parasites/μl ultimately); the assay was less intensive in application, with fewer samples pooled and a larger amount of genomic DNA included in each reaction. Current real-time PCR assays can detect <40 parasites/μl (35, 38, 46), a standard with which our pooled assay compares favorably.
Our study demonstrates significant resource savings with pooling samples in comparison with direct testing of individual samples in real-time PCR assays: in the subset of microscopy-negative samples with a predicted low prevalence of parasitemia, the estimated cost saving of pooling, compared with the cost of traditional testing, was over 50%. The efficiency, and therefore the cost saving, associated with sample pooling is primarily dependent on an accurate estimate of the prevalence of the target condition (47). In our study, this saving could have been improved by pooling in greater numbers with more-precise foreknowledge of the low prevalence of parasitemia in the microscopy-negative samples. Pooling-related savings are inversely related to parasite prevalence; given the post-hoc prevalence of PCR positivity of 3% among the microscopy-negative samples, pools of seven would have allowed for an obviation of two-thirds of the reactions without sacrificing test sensitivity. Adaptation of this pooling strategy should take into account the predicted epidemiology of malaria in the study population.
In this study, only 68% of real-time PCR-positive samples were detected by microscopy, despite both the use of a trained and experienced primary microscopist and high levels of interrater agreement with a second interpretation of a random sample of positive and negative smears. This substantial discordance highlights the unreliability of the blood smear for malaria diagnosis in some settings (34). The poor sensitivity of blood smears may be exacerbated by the declines in incident infections and parasite densities in many areas of endemicity (8), and thus, clinical trials of vaccine efficacy, chemoprophylaxis, and antimalarial treatment may require detection methods of greater sensitivity to accurately determine exposures and outcomes (16, 20, 30, 31).
Additionally, the low positive predictive value in our study, in which only 42% of blood smear-positive samples demonstrated amplification in real-time PCR assays, suggests that blood smear use in clinical studies may also be compromised by overdiagnosis and poor specificity. Though the specificity of microscopy in our study was 91% (with real-time PCR as the referent), the application of microscopy in a setting with a low prevalence significantly compromised its positive predictive value. This phenomenon has been observed in other studies (26, 28, 44) and may have several causes: systematic errors in thick smear preparation can produce false-positive interpretations because of red cell fragments, platelets, other blood elements, or environmental particulate contamination (51), and clinical approaches that bias toward overtreatment for malaria may also bias laboratory personnel toward overdiagnosis (9). Declines in parasite prevalence in areas where malaria is endemic (5, 8, 45) may further compromise the positive predictive value of microscopy and limit the utility of microscopy for case detection in clinical studies.
Conversely, this apparently poor specificity of blood smears may actually reflect poor sensitivity on the part of the real-time PCR assay. The sensitivities of PCR-based assays typically differ on the basis of common factors, such as sample quality, real-time system, and cycling conditions, and can be compromised by factors such as low target gene copy number (33), competitive inhibition of amplification by related sequences (6), and target sequence variation (48). Our pan-species screening assay targeted a multicopy gene encoding the small subunit of plasmodium ribosomal DNA, and the primers were adopted from an assay which achieved a lower detection limit of 1 copy per 25-μl reaction mixture (38). Most of the microscopy-positive samples that failed to amplify in the pan-species assay also failed to amplify in a second assay targeting a conserved gene unique to P. falciparum (Pfldh), which is the preponderant species in Kinshasa (21) and the predominant species in pregnancy-associated malaria in Africa (7). Additionally, although blood spot preservation and DNA extraction methods can affect the yield of molecular diagnostics (4), successful real-time amplification of a human gene indicates sufficient DNA preservation and extraction as well as the absence of significant PCR inhibitors. We believe that these represent true false-positive blood smears.
For parasite detection, most clinical malaria studies rely primarily on blood smears, and PCR-based diagnostics have not been widely employed, due to a lack of resources in areas where malaria is endemic. Given the ease and reliability of storing blood on filter paper for downstream molecular analysis (40); the ability to detect parasite species (41), burden (1), and antimalarial susceptibility (50); and the declines in parasite rates observed in areas of endemicity after the implementation of malaria control programs (5, 8, 45), molecular diagnostics may become a more critical case detection method. In this study, we demonstrate both the value-added diagnostic information provided by real-time PCR applied to a clinical study and a resource-saving strategy for its application. We believe that, for clinical malaria studies of prophylaxis, vaccine efficacy, and drug therapy, molecular diagnostics are valuable as both critical and robust measures of exposure and outcome.
Acknowledgments
This work was supported in part and endorsed by the Malaria in Pregnancy Consortium (MiP), which is funded through a grant from the Bill & Melinda Gates Foundation to the Liverpool School of Tropical Medicine. J.J.J. is currently supported by award KL2RR025746 from the National Center for Research Resources.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
We thank Travis Thompson for his assistance with the molecular analyses; Carla Hand for supplying the human GAPDH primers and probes; Daniel Westreich for discussing pooling strategies; and Stephen Rogerson, Alfredo Mayor, and Richard Steketee for their helpful comments on the manuscript. We thank MR4 for the plasmodium genomic DNA and plasmid clones contributed by Peter Zimmerman. Ultimately, we are indebted to the women who participated in the study.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print on 25 November 2009.
REFERENCES
- 1.Andrews, L., R. F. Andersen, D. Webster, S. Dunachie, R. M. Walther, P. Bejon, A. Hunt-Cooke, G. Bergson, F. Sanderson, A. V. Hill, and S. C. Gilbert. 2005. Quantitative real-time polymerase chain reaction for malaria diagnosis and its use in malaria vaccine clinical trials. Am. J. Trop. Med. Hyg. 73:191-198. [PubMed] [Google Scholar]
- 2.Baliraine, F. N., Y. A. Afrane, D. A. Amenya, M. Bonizzoni, D. M. Menge, G. Zhou, D. Zhong, A. M. Vardo-Zalik, A. K. Githeko, and G. Yan. 2009. High prevalence of asymptomatic Plasmodium falciparum infections in a highland area of western Kenya: a cohort study. J. Infect. Dis. 200:66-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bereczky, S., A. Liljander, I. Rooth, L. Faraja, F. Granath, S. M. Montgomery, and A. Farnert. 2007. Multiclonal asymptomatic Plasmodium falciparum infections predict a reduced risk of malaria disease in a Tanzanian population. Microbes Infect. 9:103-110. [DOI] [PubMed] [Google Scholar]
- 4.Bereczky, S., A. Martensson, J. P. Gil, and A. Farnert. 2005. Short report: rapid DNA extraction from archive blood spots on filter paper for genotyping of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 72:249-251. [PubMed] [Google Scholar]
- 5.Bhattarai, A., A. S. Ali, S. P. Kachur, A. Martensson, A. K. Abbas, R. Khatib, A. W. Al-Mafazy, M. Ramsan, G. Rotllant, J. F. Gerstenmaier, F. Molteni, S. Abdulla, S. M. Montgomery, A. Kaneko, and A. Bjorkman. 2007. Impact of artemisinin-based combination therapy and insecticide-treated nets on malaria burden in Zanzibar. PLoS Med. 4:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bialasiewicz, S., D. M. Whiley, M. D. Nissen, and T. P. Sloots. 2007. Impact of competitive inhibition and sequence variation upon the sensitivity of malaria PCR. J. Clin. Microbiol. 45:1621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brabin, B. J. 1983. An analysis of malaria in pregnancy in Africa. Bull. World Health Organ. 61:1005-1016. [PMC free article] [PubMed] [Google Scholar]
- 8.Ceesay, S. J., C. Casals-Pascual, J. Erskine, S. E. Anya, N. O. Duah, A. J. Fulford, S. S. Sesay, I. Abubakar, S. Dunyo, O. Sey, A. Palmer, M. Fofana, T. Corrah, K. A. Bojang, H. C. Whittle, B. M. Greenwood, and D. J. Conway. 2008. Changes in malaria indices between 1999 and 2007 in The Gambia: a retrospective analysis. Lancet 372:1545-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandler, C. I., C. Jones, G. Boniface, K. Juma, H. Reyburn, and C. J. Whitty. 2008. Guidelines and mindlines: why do clinical staff over-diagnose malaria in Tanzania? A qualitative study. Malar. J. 7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Culleton, R. L., T. Mita, M. Ndounga, H. Unger, P. V. Cravo, G. M. Paganotti, N. Takahashi, A. Kaneko, H. Eto, H. Tinto, C. Karema, U. D'Alessandro, V. do Rosario, T. Kobayakawa, F. Ntoumi, R. Carter, and K. Tanabe. 2008. Failure to detect Plasmodium vivax in West and Central Africa by PCR species typing. Malar. J. 7:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham, R., J. L. Northwood, C. D. Kelly, E. H. Boxall, and N. J. Andrews. 1998. Routine antenatal screening for hepatitis B using pooled sera: validation and review of 10 years experience. J. Clin. Pathol. 51:392-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dal-Bianco, M. P., K. B. Koster, U. D. Kombila, J. F. Kun, M. P. Grobusch, G. M. Ngoma, P. B. Matsiegui, C. Supan, C. L. Salazar, M. A. Missinou, S. Issifou, B. Lell, and P. Kremsner. 2007. High prevalence of asymptomatic Plasmodium falciparum infection in Gabonese adults. Am. J. Trop. Med. Hyg. 77:939-942. [PubMed] [Google Scholar]
- 13.Desai, M., F. O. ter Kuile, F. Nosten, R. McGready, K. Asamoa, B. Brabin, and R. D. Newman. 2007. Epidemiology and burden of malaria in pregnancy. Lancet Infect. Dis. 7:93-104. [DOI] [PubMed] [Google Scholar]
- 14.Dorfman, R. 1943. The detection of defective members of large populations. Ann. Math. Stat. 14:436-440. [Google Scholar]
- 15.Emmanuel, J. C., M. T. Bassett, H. J. Smith, and J. A. Jacobs. 1988. Pooling of sera for human immunodeficiency virus (HIV) testing: an economical method for use in developing countries. J. Clin. Pathol. 41:582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felger, I., B. Genton, T. Smith, M. Tanner, and H. P. Beck. 2003. Molecular monitoring in malaria vaccine trials. Trends Parasitol. 19:60-63. [DOI] [PubMed] [Google Scholar]
- 17.Franks, S., K. A. Koram, G. E. Wagner, K. Tetteh, D. McGuinness, J. G. Wheeler, F. Nkrumah, L. Ranford-Cartwright, and E. M. Riley. 2001. Frequent and persistent, asymptomatic Plasmodium falciparum infections in African infants, characterized by multilocus genotyping. J. Infect. Dis. 183:796-804. [DOI] [PubMed] [Google Scholar]
- 18.García, Z., L. Taylor, A. Ruano, L. Pavón, E. Ayerdis, R. B. Luftig, and K. A. Visoná. 1996. Evaluation of a pooling method for routine anti-HCV screening of blood donors to lower the cost burden on blood banks in countries under development. J. Med. Virol. 49:218-222. [DOI] [PubMed] [Google Scholar]
- 19.Garner, P., and A. M. Gulmezoglu. 2006. Drugs for preventing malaria in pregnant women. Cochrane Database Syst. Rev. 4:CD000169.. [DOI] [PubMed] [Google Scholar]
- 20.Imoukhuede, E. B., L. Andrews, P. Milligan, T. Berthoud, K. Bojang, D. Nwakanma, J. Ismaili, C. Buckee, F. Njie, S. Keita, M. Sowe, T. Lang, S. C. Gilbert, B. M. Greenwood, and A. V. Hill. 2007. Low-level malaria infections detected by a sensitive polymerase chain reaction assay and use of this technique in the evaluation of malaria vaccines in an endemic area. Am. J. Trop. Med. Hyg. 76:486-493. [PMC free article] [PubMed] [Google Scholar]
- 21.Kazadi, W., J. D. Sexton, M. Bigonsa, B. W'Okanga, and M. Way. 2004. Malaria in primary school children and infants in Kinshasa, Democratic Republic of the Congo: surveys from the 1980s and 2000. Am. J. Trop. Med. Hyg. 71:97-102. [PubMed] [Google Scholar]
- 22.Kleinschmidt, I., C. Schwabe, L. Benavente, M. Torrez, F. C. Ridl, J. L. Segura, P. Ehmer, and G. N. Nchama. 2009. Marked increase in child survival after four years of intensive malaria control. Am. J. Trop. Med. Hyg. 80:882-888. [PMC free article] [PubMed] [Google Scholar]
- 23.Koram, K. A., and M. E. Molyneux. 2007. When is “malaria” malaria? The different burdens of malaria infection, malaria disease, and malaria-like illnesses. Am. J. Trop. Med. Hyg. 77:1-5. [PubMed] [Google Scholar]
- 24.Landis, S. H., V. Lokomba, C. V. Ananth, J. Atibu, R. W. Ryder, K. E. Hartmann, J. M. Thorp, A. Tshefu, and S. R. Meshnick. 2009. Impact of maternal malaria and under-nutrition on intrauterine growth restriction: a prospective ultrasound study in Democratic Republic of Congo. Epidemiol. Infect. 137:294-304. [DOI] [PubMed] [Google Scholar]
- 25.Lukuka, K. A., O. S. Fumie, M. R. Mulumbu, B. J. Lokombe, and T. J. Muyembe. 2006. Malaria prevalence at delivery in four maternity hospitals of Kinshasa City, Democratic Republic of Congo. Bull. Soc. Pathol. Exot. 99:200-201. [PubMed] [Google Scholar]
- 26.McKenzie, F. E., J. Sirichaisinthop, R. S. Miller, R. A. Gasser, and C. Wongsrichanalai. 2003. Dependence of malaria detection and species diagnosis by microscopy on parasite density. Am. J. Trop. Med. Hyg. 69:372-376. [PMC free article] [PubMed] [Google Scholar]
- 27.Mockenhaupt, F. P., B. Rong, H. Till, T. A. Eggelte, S. Beck, C. Gyasi-Sarpong, W. N. Thompson, and U. Bienzle. 2000. Submicroscopic Plasmodium falciparum infections in pregnancy in Ghana. Trop. Med. Int. Health 5:167-173. [DOI] [PubMed] [Google Scholar]
- 28.Nankabirwa, J., D. Zurovac, J. N. Njogu, J. B. Rwakimari, H. Counihan, R. W. Snow, and J. K. Tibenderana. 2009. Malaria misdiagnosis in Uganda—implications for policy change. Malar. J. 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nwagha, U. I., V. O. Ugwu, T. U. Nwagha, and B. U. Anyaehie. 2009. Asymptomatic Plasmodium parasitaemia in pregnant Nigerian women: almost a decade after Roll Back Malaria. Trans. R. Soc. Trop. Med. Hyg. 103:16-20. [DOI] [PubMed] [Google Scholar]
- 30.Ohrt, C., Purnomo, A. Sutamihardja, D. Tang, and K. C. Kain. 2002. Impact of microscopy error on estimates of protective efficacy in malaria-prevention trials. J. Infect. Dis. 186:540-546. [DOI] [PubMed] [Google Scholar]
- 31.O'Meara, W., F. Hall, and E. McKenzie. 2007. Malaria vaccine efficacy: the difficulty of detecting and diagnosing malaria. Malar. J. 6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otten, M., M. Aregawi, W. Were, C. Karema, A. Medin, W. Bekele, D. Jima, K. Gausi, R. Komatsu, E. Korenromp, D. Low-Beer, and M. Grabowsky. 2009. Initial evidence of reduction of malaria cases and deaths in Rwanda and Ethiopia due to rapid scale-up of malaria prevention and treatment. Malar. J. 8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oyedeji, S. I., H. O. Awobode, G. C. Monday, E. Kendjo, P. G. Kremsner, and J. F. Kun. 2007. Comparison of PCR-based detection of Plasmodium falciparum infections based on single and multicopy genes. Malar. J. 6:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne, D. 1988. Use and limitations of light microscopy for diagnosing malaria at the primary health care level. Bull. World Health Organ. 66:621-626. [PMC free article] [PubMed] [Google Scholar]
- 35.Perandin, F., N. Manca, A. Calderaro, G. Piccolo, L. Galati, L. Ricci, M. C. Medici, M. C. Arcangeletti, G. Snounou, G. Dettori, and C. Chezzi. 2004. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J. Clin. Microbiol. 42:1214-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickard, A. L., C. Wongsrichanalai, A. Purfield, D. Kamwendo, K. Emery, C. Zalewski, F. Kawamoto, R. S. Miller, and S. R. Meshnick. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilcher, C. D., S. A. Fiscus, T. Q. Nguyen, E. Foust, L. Wolf, D. Williams, R. Ashby, J. O. O'Dowd, J. T. McPherson, B. Stalzer, L. Hightow, W. C. Miller, J. J. Eron, Jr., M. S. Cohen, and P. A. Leone. 2005. Detection of acute infections during HIV testing in North Carolina. N. Engl. J. Med. 352:1873-1883. [DOI] [PubMed] [Google Scholar]
- 38.Rougemont, M., M. Van Saanen, R. Sahli, H. P. Hinrikson, J. Bille, and K. Jaton. 2004. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J. Clin. Microbiol. 42:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shokoples, S., M. Ndao, K. Kowalewska-Grochowska, and S. Yanow. 2009. Multiplexed real-time PCR assay for discrimination of Plasmodium species with improved sensitivity for mixed infections. J. Clin. Microbiol. 47:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, B., J. Cox-Singh, A. O. Miller, M. S. Abdullah, G. Snounou, and H. A. Rahman. 1996. Detection of malaria in Malaysia by nested polymerase chain reaction amplification of dried blood spots on filter papers. Trans. R. Soc. Trop. Med. Hyg. 90:519-521. [DOI] [PubMed] [Google Scholar]
- 41.Snounou, G., S. Viriyakosol, W. Jarra, S. Thaithong, and K. N. Brown. 1993. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol. Biochem. Parasitol. 58:283-292. [DOI] [PubMed] [Google Scholar]
- 42.Snounou, G., S. Viriyakosol, X. P. Zhu, W. Jarra, L. Pinheiro, V. E. do Rosario, S. Thaithong, and K. N. Brown. 1993. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol. Biochem. Parasitol. 61:315-320. [DOI] [PubMed] [Google Scholar]
- 43.Steketee, R. W., B. L. Nahlen, M. E. Parise, and C. Menendez. 2001. The burden of malaria in pregnancy in malaria-endemic areas. Am. J. Trop. Med. Hyg. 64:28-35. [DOI] [PubMed] [Google Scholar]
- 44.Stow, N. W., J. K. Torrens, and J. Walker. 1999. An assessment of the accuracy of clinical diagnosis, local microscopy and a rapid immunochromatographic card test in comparison with expert microscopy in the diagnosis of malaria in rural Kenya. Trans. R. Soc. Trop. Med. Hyg. 93:519-520. [DOI] [PubMed] [Google Scholar]
- 45.Teklehaimanot, H. D., A. Teklehaimanot, A. Kiszewski, H. S. Rampao, and J. D. Sachs. 2009. Malaria in Sao Tome and Principe: on the brink of elimination after three years of effective antimalarial measures. Am. J. Trop. Med. Hyg. 80:133-140. [PubMed] [Google Scholar]
- 46.Veron, V., S. Simon, and B. Carme. 2009. Multiplex real-time PCR detection of P. falciparum, P. vivax and P. malariae in human blood samples. Exp. Parasitol. 121:346-351. [DOI] [PubMed] [Google Scholar]
- 47.Westreich, D., M. Hudgens, S. Fiscus, and C. Pilcher. 2008. Optimizing screening for acute human immunodeficiency virus infection with pooled nucleic acid amplification tests. J. Clin. Microbiol. 46:1785-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiley, D. M., S. B. Lambert, S. Bialasiewicz, N. Goire, M. D. Nissen, and T. P. Sloots. 2008. False-negative results in nucleic acid amplification tests—do we need to routinely use two genetic targets in all assays to overcome problems caused by sequence variation? Crit. Rev. Microbiol. 34:71-76. [DOI] [PubMed] [Google Scholar]
- 49.WHO. 2008. World malaria report 2008. WHO, Geneva, Switzerland.
- 50.Wilson, P. E., A. P. Alker, and S. R. Meshnick. 2005. Real-time PCR methods for monitoring antimalarial drug resistance. Trends Parasitol. 21:278-283. [DOI] [PubMed] [Google Scholar]
- 51.Wongsrichanalai, C., M. J. Barcus, S. Muth, A. Sutamihardja, and W. H. Wernsdorfer. 2007. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am. J. Trop. Med. Hyg. 77:119-127. [PubMed] [Google Scholar]