Abstract
Rapid detection and identification of Ehrlichia species improves clinical outcome for patients suspected of ehrlichiosis. We describe an assay that employs multilocus PCR and electrospray ionization mass spectrometry (PCR/ESI-MS) to detect and identify Ehrlichia species directly from blood specimens. The results were compared to those of a colorimetric microtiter PCR enzyme immunoassay (PCR-EIA) used as a diagnostic assay. Among 213 whole-blood samples collected from patients who were clinically suspected of ehrlichiosis from 1 May to 1 August 2008 at Vanderbilt University Hospital, 40 were positive for an Ehrlichia species by PCR/ESI-MS, giving a positive rate of 18.8%. In comparison to the PCR-EIA, PCR/ESI-MS possessed a sensitivity, a specificity, and positive and negative predictive values of 95.0%, 98.8%, 95.0%, and 98.8%, respectively. The 38 specimens that were positive for Ehrlichia by both PCR/ESI-MS and the PCR-EIA were further characterized to the species level, with 100% agreement between the two assays. In addition, Rickettsia rickettsii was detected by PCR/ESI-MS from four specimens that were confirmed retrospectively by serology and PCR-EIA. In three specimens, the PCR/ESI-MS assay identified Pseudomonas aeruginosa, Neisseria meningitidis, and Staphylococcus aureus; these were confirmed by culture and/or clinical diagnosis as being clinically relevant. From specimen processing to result reporting, the PCR/ESI-MS assay can be completed within 6 h, providing another laboratory tool for the diagnosis of ehrlichiosis. Moreover, this system may provide rapid detection and identification of additional pathogens directly from blood specimens.
Ehrlichiosis patients typically present with fever, headache, and other nonspecific symptoms, such as myalgias, arthralgias, and chills several days following a bite by an infected tick (5, 8, 25, 27). These infections are difficult to diagnose clinically, due to the nonspecific nature of the symptoms, and may be confused with other diseases (8). Ehrlichiosis can be diagnosed in the laboratory by direct identification of intracytoplasmic morulae in leukocytes upon examination of stained blood smears (18). Cell culture is difficult at best for Ehrlichia chaffeensis and has not been achieved for Ehrlichia ewingii (10, 11). Serological tests have been used as the tests of choice, but antibody titers typically do not develop in the first week of acute clinical ehrlichiosis (6), when treatment is critical to the patient's prognosis (36).
Several PCR-based diagnostic assays have been developed and used for the laboratory diagnosis of ehrlichiosis. In one assay, PCR primers specific to E. chaffeensis, along with a radiolabeled probe, are used to detect the 16S ribosomal gene from this species (24). Another study evaluated the detection of Ehrlichia in the whole blood of patients by using a nested-PCR approach that targets the 16S rRNA gene. These researchers detected E. chaffeensis in all seven blood samples tested (31). Bell and Patel employed a real-time PCR assay targeting groEL to detect and differentiate E. chaffeensis, E. ewingii, and Anaplasma phagocytophilum in the blood of patients (3). Recently, a quantitative multicolor TaqMan real-time PCR assay targeting the dsb gene was reported to detect and differentiate medically important Ehrlichia species (9). We have developed a two-step, colorimetric microtiter plate PCR enzyme immunoassay (PCR-EIA) capable of detecting and differentiating medically important Ehrlichia species (33, 35). The first step involves PCR amplification of a genus-specific region in the 16S rRNA gene. In the second step, the PCR amplification product is identified as either E. chaffeensis, E. ewingii, or A. phagocytophilum by using three species-specific probes in an EIA format (9, 26). This assay has been implemented at Vanderbilt University Hospital for routine laboratory diagnosis of ehrlichiosis.
We report here the development of a multilocus PCR and electrospray ionization mass spectrometry (PCR/ESI-MS) method for the detection of bacterial tick-borne pathogens, including Ehrlichia and Anaplasma species. The assay employs 16 primer pairs; 4 target the bacterial 16S and 23S ribosomal genes and can broadly detect bacteria, while the others are targeted to broadly conserved genes encoding housekeeping proteins that differentiate known tick-borne bacteria. Following PCR, the amplicons are analyzed in an electrospray ionization mass spectrometer (Ibis Biosciences, Carlsbad, CA). From the masses of the amplicons, the numbers of A's, G's, C's, and T's in each amplicon are determined. By analysis of the base compositions of amplicons from all primer pairs, the organisms present in the sample can be identified and quantified (12, 15, 20, 29). In this study we explored the use of this assay to directly detect and identify tick-borne pathogens in blood specimens for the laboratory diagnosis of ehrlichiosis. Clinical samples of whole blood, plasma, or cerebrospinal fluid (CSF) from clinically suspected ehrlichiosis patients were sent to Vanderbilt University Hospital for the detection of Ehrlichia species using the PCR-EIA. Blinded DNA extracts from these specimens were subsequently screened by the PCR/ESI-MS assay. The PCR/ESI-MS assay detected E. chaffeensis and E. ewingii with 95.0% sensitivity and 98.8% specificity relative to the PCR-EIA results.
(This study was presented in part at the 109th General Meeting of the American Society for Microbiology, Philadelphia, PA, 17 to 21 May 2009.)
MATERIALS AND METHODS
Clinical samples and processing.
Whole-blood specimens were collected consecutively from 1 May to 1 August 2008 and were submitted to the Molecular Infectious Disease Laboratory at the Vanderbilt University Hospital for Ehrlichia species testing by the PCR-EIA. Total nucleic acids were extracted using a NucliSens easyMAG system (bioMerieux, Durham, NC). Briefly, 200 μl of whole blood was mixed with 200 μl of phosphate-buffered saline. After a thorough vortexing, the default extraction protocol was used (34). Total nucleic acids were eluted in 55 μl of elution buffer (bioMerieux). The sample extracts were blinded prior to analysis at Ibis Biosciences.
PCR/ESI-MS primers and calibrant.
Sixteen broad-range primer pairs (Table 1) targeting the conserved regions of DNA that border variable regions were employed. All primers used in this study had a thymine nucleotide at the 5′ end to minimize the addition of nontemplated adenosines during amplification using Taq polymerase (4). An internal positive control (calibrant) made from synthetic DNA (Blue Heron Biotechnology, Bothell, WA) was included in each PCR at 100 and 16 copies per PCR for the ribosomal and nonribosomal primer targets, respectively. The calibrant sequence contains a 5-bp deletion within the amplicon so that the calibrant amplicons could be readily resolved and distinguished from the bacterium-derived amplicon.
TABLE 1.
PCR/ESI-MS primers, gene targets, and bacterial targets
| Primer pair | Primer IDa | Primer sequence | Target | Target clade/genus |
|---|---|---|---|---|
| BCT3517 | BCT8241F | TGCTGAAGAGCTTGGAATGCA | Flagellin | All Borrelia spp. |
| BCT8242R | TACAGCAATTGCTTCATCTTGATTTGC | |||
| BCT3515 | BCT8237F | TCCACAAGGTGGTGGTGAAGG | rplB | All Spirochaetes |
| BCT8238R | TCGGCTGTCCCCAAGGAG | |||
| BCT1083 | BCT2764F | TAAGAGCGCACCGGTAAGTTGG | RNaseP | All Rickettsia spp. |
| BCT2763R | TCAAGCGATCTACCCGCATTACAA | |||
| BCT1084 | BCT2765F | TCCACCAAGAGCAAGATCAAATAGGC | RNaseP | All Rickettsia spp. |
| BCT2763R | TCAAGCGATCTACCCGCATTACAA | |||
| BCT3569 | BCT8334F | TGCATGCAGATCATGAACAAAATGC | gltA | Bartonella, Anaplasma, and Ehrlichia |
| BCT8335R | TCCATGTGCTGGTCCCCA | |||
| BCT3575 | BCT8346F | TGCATCACTTGGTTGATGATAAGATACATGC | rpoB | Bartonella, Anaplasma, and Ehrlichia |
| BCT8347R | TCACCAAAACGCTGACCACCAAA | |||
| BCT3570 | BCT8336F | TGCATGCAGATCATGAACAGAATGC | GLTA | Bartonella, Anaplasma, and Ehrlichia |
| BCT8337R | TCCACCATGAGCTGGTCCCCA | |||
| BCT3571 | BCT8338F | TAAGGTTGGTGGATCTAGTGAAGTTGA | groEL | Anaplasma and Ehrlichia |
| BCT8339R | TACACCTTCCTCAACAGCAGC | |||
| BCT3573 | BCT8342F | TGTGGAAGGTGAAGCTTTGGCAAC | groEL | Bartonella |
| BCT8343R | TAACATGGCTTTACGGCGATCACC | |||
| BCT3574 | BCT8344F | TTCTGACTATGACCGTGAGAAATTGCAAG | groEL | Bartonella and Anaplasma |
| BCT8345R | TCACCAACACGGATAACAGCAACACC | |||
| BCT2328 | BCT5602F | TGAGGGTTTTATGCTTAAAGTTGGTTTTATTGGTT | asd | Francisella tularensis |
| BCT5603R | TGATTCGATCATACGAGACATTAAAACTGAG | |||
| BCT1079 | BCT2717F | TCGCCGTGGAAAAATCCTACGCT | icd | Coxiella burnetii |
| BCT2718R | TAGCCTTTTCTCCGGCGTAGATCT | |||
| BCT346 | BCT1366F | TAGAACACCGATGGCGAAGGC | 16S rRNA gene | All bacteria |
| BCT1367R | TCGTGGACTACCAGGGTATCTA | |||
| BCT348 | BCT1393F | TTTCGATGCAACGCGAAGAACCT | 16S rRNA gene | All bacteria |
| BCT1370R | TACGAGCTGACGACAGCCATG | |||
| BCT360 | BCT1386F | TCTGTTCTTAGTACGAGAGGACC | 23S rRNA gene | All bacteria |
| BCT1402R | TTTCGTGCTTAGATGCTTTCAG | |||
| BCT361 | BCT1396F | TTTAAGTCCCGCAACGAGCGCAA | 16S rRNA gene | All bacteria |
| BCT1403R | TTGACGTCATCCCCACCTTCCTC |
ID, identification.
PCR amplification.
Each sample was added to 16 wells on a 96-well plate already containing primer pairs and a reagent. The PCR mixture consisted of 1 U of Immolase Taq polymerase (Bioline USA, Randolph, MA), 20 mM Tris (pH 8.3), 75 mM KCl, 1.5 mM MgCl2, 0.4 M betaine, 200 μM (each) dATP, dCTP, and dTTP (Bioline USA, Randolph, MA), 200 μM 13C-enriched dGTP (Cambridge Isotope Laboratories, Andover, MA), and 250 nM primer. The following PCR cycling conditions were used on a 96-well MJ Dyad thermocycler (Bio-Rad Inc., Hercules, CA): 95°C for 10 min, followed by 8 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 30 s, with the 48°C annealing temperature increasing 0.9°C for each cycle. The PCR was then continued for 37 additional cycles of 95°C for 15 s, 56°C for 20 s, and 72°C for 20 s. The PCR cycle ended with a final extension of 2 min at 72°C, followed by a 4°C hold.
Mass spectrometry and base composition analysis.
After amplification, 30-μl aliquots of each PCR product were desalted and purified by using a weak anion-exchange protocol as described elsewhere (12). Accurate-mass (±20 ppm), high-resolution (mass/delta mass, >8,000 full width at half maximum [FWHM]) mass spectra were acquired for each sample using high-throughput ESI-MS protocols, described previously (21). For each sample, approximately 1.5 μl of analyte solution was consumed during the 34-s spectral acquisition. Raw mass spectra were postcalibrated with an internal mass standard and deconvolved to monoisotopic molecular masses. Unambiguous base compositions were derived from the exact-mass measurements of the complementary single-stranded oligonucleotides (23). Quantitative results were obtained by comparing the peak heights with the calibrant present in every PCR well at 100 molecules (21).
Data analysis and interpretation.
The broad-range PCR/ESI-MS assay employs primers targeting protein genes for a broad group of bacteria and 16S and 23S ribosomal genes that can detect all bacteria. A specimen was considered to be positive if at least two of the protein gene-targeting primers detected the organism. Identifications made using the ribosomal gene-targeting primer required that all four primer pairs detect the organism. Identification of a specimen as Ehrlichia positive by the PCR/ESI-MS assay required detection by at least two of the following protein gene-targeting primer pairs: BCT3569, BCT3575, and BCT3570 (Table 2). The base composition signatures for E. chaffeensis and E. ewingii with these three primers are unique and are not known to be shared with any other bacteria. Additionally, primer pairs BCT3569, BCT3575, BCT3570, BCT3571, BCT346, BCT348, BCT360, and BCT361 (Tables 1 and 2) may prime Ehrlichia in the samples, depending on the levels of Ehrlichia DNA in the extract and the levels of background DNA. The Rickettsia rickettsii-positive specimens were detected with two broad-range Rickettsia primers (BCT1083 and BCT1084). In addition, R. rickettsii may also be detected by primer pair BCT3570 and the four broad-range ribosomal primers.
TABLE 2.
Base composition signatures for bacteria detected in this study
| Primer pair | No. of each base (A, G, C, T) in the PCR amplicon |
||||||
|---|---|---|---|---|---|---|---|
| E. chaffeensis | E. ewingii | R. rickettsii | Bacteroides sp. | N. meningitidis | P. aeruginosa | S. aureus | |
| BCT1083 | DNPa | DNP | 40, 34, 30, 31 | DNP | DNP | DNP | DNP |
| BCT1084 | DNP | DNP | 25, 22, 21, 23 | DNP | DNP | DNP | DNP |
| BCT3569 | 29, 32, 22, 39 | 34, 29, 24, 35 | DNP | DNP | DNP | DNP | DNP |
| BCT3575 | 28, 30, 17, 37 | 29, 30, 17, 36 | DNP | DNP | DNP | DNP | DNP |
| BCT3570 | 27, 35, 22, 41 | 32, 32, 24, 37 | 28, 30, 32, 35 | DNP | DNP | DNP | DNP |
| BCT3571 | 30, 30, 11, 26 | DNP | DNP | DNP | DNP | DNP | DNP |
| BCT346 | 28, 32, 21, 18 | 28, 32, 21, 18 | 28, 31, 24, 16 | 30, 28, 22, 19 | 29, 28, 26, 16 | 30, 31, 23, 15 | 27, 30, 21, 21 |
| BCT348 | 26, 31, 30, 32 | 27, 30, 29, 33 | 25, 32, 32, 31 | 29, 31, 27, 28 | 26, 34, 30, 26 | 26, 32, 29, 29 | 30, 29, 30, 29 |
| BCT360 | 35, 32, 23, 32 | 35, 32, 23, 32 | 33, 38, 27, 24 | 27, 37, 26, 32 | 34, 37, 25, 26 | 31, 36, 27, 28 | 31, 38, 24, 29 |
| BCT361 | 33, 29, 26, 22 | 33, 29, 26, 22 | 31, 32, 25, 22 | 29, 31, 24, 26 | 27, 31, 26, 24 | 27, 33, 29, 20 | 29, 30, 25, 24 |
DNP, does not prime; no PCR amplicon was generated using the specified primer pair.
PCR-EIA for Ehrlichia and R. rickettsii.
A two-step colorimetric microtiter plate PCR assay capable of detecting and differentiating medically important Ehrlichia species was used (33, 35). The first step involved PCR amplification of a genus-specific region in the 16S rRNA gene that has been described elsewhere (9, 26). In the second step, the PCR amplification product was identified and differentiated to E. chaffeensis, E. ewingii, or A. phagocytophilum using three species-specific probes (5′-CAA TTG CTT ATA ACC TTT TGG TTA T-3′, 5′-TCG AAC GAA CAA TTC CTA AA-3′, and 5′-CGG ATT ATT CTT TAT AGC TTG CTA T-3′) in a colorimetric microtiter plate format (33). The same PCR-EIA format and procedure were used to detect R. rickettsii by targeting a 154-bp fragment of the R. rickettsii-specific rickettsial outer membrane protein A (rOmpA) gene. RR190.547F, RR190.701R, and RR190.588F were used as the forward primer, reverse primer, and hybridization probe, respectively, as described previously (16).
RESULTS
PCR/ESI-MS assay.
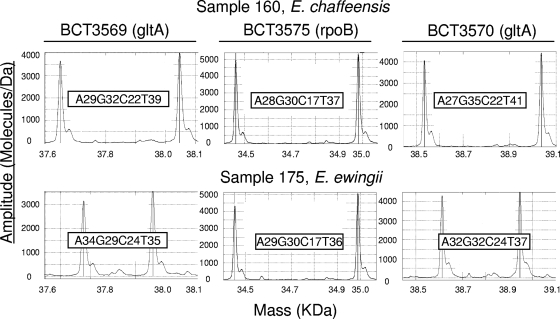
The PCR/ESI-MS assay employs 16 primer pairs and is designed for use with the T5000 PCR mass spectrometry system (Ibis Biosciences, Carlsbad, CA). The panel includes four broad-range primer pairs targeting the 16S and 23S genes of all bacteria. The other primers were selected based on their coverage of groups of known tick-borne pathogens, including members of the Spirochaetales (e.g., Borrelia spp.), Alphaproteobacteria (e.g., Ehrlichia, Anaplasma, Bartonella, and Rickettsia spp.), and Gammaproteobacteria (e.g., Francisella and Coxiella spp.). The 16 primer sequences and gene targets are shown in Table 1. All of the primers perform under uniform PCR cycling conditions and thus are used together arrayed on a 96-well plate. Each PCR mixture contains an internal positive control called the calibrant. This calibrant is quantified as the optical density at 260 nm (OD260) and is added at 100 or 16 copies/PCR for the ribosomal or nonribosomal primer targets, respectively. These low calibrant levels are consistently detected in negative specimens and thus serve as a PCR positive control. Figure 1 shows the representative mass spectra obtained from specimens containing E. chaffeensis or E. ewingii with three primer pairs. The whole procedure of the PCR/ESI-MS assay, from specimen processing to result reporting, can be completed within 6 h.
FIG. 1.
Mass spectra from PCR/ESI-MS analysis of patient specimens. The spectra show the masses of the forward and reverse strands of the amplicons generated using primer pairs BCT3569, BCT3575, and BCT3570 on sample 160, containing E. chaffeensis (top frames), or sample 175, containing E. ewingii (bottom frames). For each spectrum shown, the reverse strand corresponds to the left peak and the forward strand corresponds to the right peak. The base compositions of the amplicons are indicated. E. chaffeensis and E. ewingii can be readily distinguished from each other with any of these three primer pairs by the base composition of the PCR amplicons.
Analytical sensitivity.
The Ibis PCR/ESI-MS assay employs a quantified calibrant template in each PCR. For the Ehrlichia-positive samples, the amount of Ehrlichia genomic DNA ranged from 10 to >1,000 genomes/PCR, as shown in Table S1 in the supplemental material. This corresponds to a starting concentration of 2.75 × 103 to >2.75 × 105 Ehrlichia genomes per milliliter of clinical specimen, assuming 100% nucleic acid extraction efficiency. For the R. rickettsii-positive samples, the numbers of genomes ranged from 30 to >1,000 copies per PCR, corresponding to 8.25 × 103 to 2.75 × 105 Rickettsia genomes per ml of whole blood. Similar levels were observed in the samples that tested positive for Pseudomonas aeruginosa, Neisseria meningitidis, Bacteroides spp., and Staphylococcus aureus (see Table 4).
TABLE 4.
Detection and identification of additional bacterial pathogens from whole-blood specimens
| Specimen(s) | Gender | Age (yr) | T5000 result | No. of genomes/ml of blood | Clinical diagnosis | Laboratory confirmation |
|---|---|---|---|---|---|---|
| 61 | Female | 11 | R. rickettsii | 8.3 × 103 | Likely tick-borne illness with dehydration and myalgia | Serum collected at acute phase positive for rickettsial IgM and negative for IgG. R. rickettsii-specific PCR-EIA was positive. |
| 73 | Male | 49 | R. rickettsii | ≥2.8 × 105 | Clinical findings consistent with RMSF | Serum rickettsial IgG titer of 1:512. R. rickettsii-specific PCR-EIA was positive. |
| 83, 81 | Female | <1 (9 mo) | R. rickettsii | 9.9 × 104, 3.0 × 104 | Sepsis with multiorgan failure secondary to RMSF | Acute-phase serum was negative, and convalescent-phase serum was positive, for rickettsial IgG and IgM. R. rickettsii-specific PCR-EIA was positive. |
| 58 | Male | 71 | P. aeruginosa | ≥6.9 × 104 | Bacteremia, aspergillosis | Pseudomonas aeruginosa recovered from blood culture |
| 9 | Female | 19 | N. meningitidis | ≥6.9 × 104 | Bacterial meningitis, sepsis | Neisseria meningitidis recovered from blood culture |
| 2 | Male | 47 | Bacteroides spp. | >3.9 × 104 | Diverticulitis, retroperitoneal abscess | Blood culture was negative |
| 138 | Male | 2 | S. aureus | 1.7 × 104 | Sepsis, septic arthritis, osteomyelitis | MRSAa recovered from blood culture |
MRSA, methicillin-resistant S. aureus.
Diagnostic sensitivity and specificity and predictive values.
A total of 213 clinical specimens, including 197 (92.5%) whole-blood, 1 (0.5%) plasma, and 15 (7.0%) CSF specimens, were collected during the study period. Forty specimens were positive for an Ehrlichia species by the reference PCR-EIA, giving a positive rate of 18.8%. Of these 40 specimens, 38 were positive by the PCR/ESI-MS assay, with an average of 4.7 primers yielding positive detections. Two samples were positive by the PCR/ESI-MS assay but negative by the PCR-EIAs. All three E. ewingii samples were positive by both assays. In comparison to the PCR-EIA, the sensitivity, specificity, and positive and negative predictive values of the PCR/ESI-MS assay for Ehrlichia species detection were 95.0%, 98.8%, 95.0%, and 98.8%, respectively (Table 3). The individual specimens, their origins, and the results by both assays are shown in Table S1 in the supplemental material.
TABLE 3.
Diagnostic sensitivity and specificity and predictive values of the PCR/ESI-MS assay for detection of Ehrlichia species in blood
| Pathogen | No. of samplesa: |
Diagnostic sensitivity (%) | Diagnostic specificity (%) | Positive predictive value (%) | Negative predictive value (%) | |||
|---|---|---|---|---|---|---|---|---|
| S+ T+ (true positive) | S+ T− (false negative) | S− T+ (false positive) | S− T− (true negative) | |||||
| E. chaffeensis | 35 | 2 | 2 | 174 | 94.6 | 98.9 | 94.6 | 98.9 |
| E. ewingii | 3 | 0 | 0 | 210 | 100.0 | 100.0 | 100.0 | 100.0 |
| Ehrlichia species | 38 | 2 | 2 | 171 | 95.0 | 98.8 | 95.0 | 98.8 |
S, PCR-EIA; T, PCR/ESI-MS; +, positive; −, negative.
Detection of Rickettsia rickettsii in whole blood by PCR/ESI-MS.
In four of the specimens, the PCR/ESI-MS assay identified Rickettsia rickettsii, the causative agent of Rocky Mountain spotted fever (RMSF) (Table 4). The four positive samples came from three patients who had clinical diagnoses consistent with RMSF. For the 9-month-old patient (specimens 81 and 83), the acute-phase serum sample was negative for rickettsial IgM and IgG but the convalescent-phase serum was positive for both. The acute-phase serum sample from the patient corresponding to specimen 61 was weakly positive for rickettsial IgM and negative for IgG. The third Rickettsia rickettsii-positive patient (specimen 73) had a rickettsial IgG titer consistent with a diagnosis of RMSF (Table 4). All three specimens tested positive retrospectively by the R. rickettsii PCR-EIA.
Detection of non-tick-borne pathogens in whole blood by PCR/ESI-MS.
Four specimens tested positive for non-tick-borne pathogens. The organisms found in these four specimens were identified using the broad-range ribosomal primers; the base composition signatures observed are shown in Table 2. Specimens 58, 9, and 138 tested positive for Pseudomonas aeruginosa, Neisseria meningitidis, and Staphylococcus aureus, respectively. Each of these patients was diagnosed with sepsis or bacteremia, and the corresponding organisms were detected in the blood culture (Table 4). Specimen 2 tested positive for a Bacteroides sp. While the detection of Bacteroides correlated with the clinical diagnosis of diverticulitis and retroperitoneal abscess, the blood culture for this patient remained negative.
DISCUSSION
Human monocytotropic ehrlichiosis (HME) is caused by E. chaffeensis or E. ewingii (1, 2), and the symptoms, such as fever, headache, myalgias, arthralgias, and chills, are usually nonspecific (5, 8, 25). These generic symptoms can be mistaken for those of other infectious diseases. The PCR/ESI-MS assay described in this report and performed on the Ibis T5000 biosensor was able to detect Ehrlichia spp. and identify them to species level with a sensitivity of 95% and a specificity of 98.8% compared to the reference PCR-EIA, providing another rapid, early, and accurate laboratory diagnostic tool for ehrlichiosis.
Broad-range PCR and electrospray ionization mass spectrometry for diagnostic applications have several advantages over organism-specific nucleic acid-based detection assays. First, a small set of PCRs can be used to detect and distinguish among a wider range of organisms (15, 29). PCR/ESI-MS can also be used to detect mixtures of bacteria or viruses in a single sample (14, 17, 30). Ticks carry numerous pathogens, including coinfections with different pathogens (7, 32). The Ibis T5000 system, along with the panel of broad-range PCR primers described, has the ability to improve the treatment of tick-borne illnesses by the rapid and correct diagnosis of the infectious agents. This assay has been used to screen questing Ixodes, Amblyomma, and Dermacentor ticks from various parts of the United States and Europe. Efforts are under way to add primers covering the Babesia and flavivirus groups of tick-borne pathogens to the current PCR/ESI-MS assay in order to increase the breadth of coverage of the assay. Such an assay, when applied to clinical specimens, would enable the health care provider to better treat tick-borne illnesses, including coinfections with multiple pathogens, by knowing the optimal antimicrobial therapy to employ.
The Ibis PCR/ESI-MS assay detected Rickettsia rickettsii in four of the blinded samples. These infections were likely the result of tick bites. Specimens 81 and 83 tested positive for Rickettsia rickettsii and came from a single 9-month-old patient whose acute-phase serum sample tested negative for rickettsial IgG and IgM but whose convalescent-phase serum sample was IgG and IgM positive. Similarly, an acute-phase serum sample from the patient associated with specimen 61 was only weakly positive for rickettsial IgM and was negative for rickettsial IgG. These results exemplify the advantage of a direct nucleic acid-based diagnostic test over a serological test: Serological analyses are typically negative during the critical time when health care providers need to make treatment decisions. Providers are also dependent on serologically based diagnostics for other important tick-borne diseases, such as Lyme disease, for which early antibiotic therapy can greatly improve prognosis (36).
Twelve of the 16 primers used in the PCR/ESI-MS assay target groups of known tick-borne pathogens: Spirochaetes, Rickettsia, Francisella, Coxiella, Bartonella, Anaplasma, and Ehrlichia spp. The remaining four primers target the 16S and 23S rRNA genes and are designed to amplify these regions from all bacteria. The ribosomal primers detected pathogens in four samples that were negative for Ehrlichia or Rickettsia spp.; these pathogens were likely responsible for febrile illness and incidental to the tick bite. Specimens 58, 9, and 138 tested positive for Pseudomonas aeruginosa, Neisseria meningitidis, and Staphylococcus aureus, respectively, and these detections were confirmed both by the clinical diagnosis and by blood cultures. Specimen 2 tested positive for a Bacteroides sp. closely related to Bacteroides vulgatus. Extended, well-designed studies are needed to determine whether this PCR/ESI-MS assay can be used in place of current blood culture methodologies to diagnose sepsis.
The Ibis PCR/ESI-MS assay employs a synthetic calibrant DNA molecule that can be used to quantify the amount of the pathogen genome in the sample. The calibrant levels are intentionally set at a relatively low level per PCR so that the calibrant amplicons do not compete with the amplicons from the organism(s) of interest. For the target genes encoding housekeeping proteins, which are typically present at 1 copy per genome, the calibrant is set at 16 copies per well, just above the stochastic limits of PCR. Since ribosomal genes are typically present in multiple copies per genome, with the exception of a few organisms, such as Ehrlichia spp. (22), the ribosomal calibrants are set at the higher number of 100 copies per PCR. The ratio of the calibrant to the target amplicon can then be used to estimate the amount of target template in the sample extracts (13, 15). The number of Ehrlichia genomes per milliliter of specimen ranged from 2,750 to >2.75 × 105, the maximal quantification limit of the assay; the actual genome levels could be much higher. Similarly high bacterial genome levels were observed in specimens from patients infected with Rickettsia rickettsii, Pseudomonas aeruginosa, Neisseria meningitidis, Staphylococcus aureus, and Bacteroides.
The high-throughput PCR/ESI-MS assay run on the Ibis T5000 system allows universal bacterial pathogen identification directly from clinical samples without culture, enabling the health care provider to make informed treatment decisions without waiting days for culture results, provided that the pathogen is culturable. Immediate treatment is critical for a positive prognosis for many tick-borne diseases, such as Rocky Mountain spotted fever and ehrlichiosis (19, 28). All of these diseases typically begin with febrile illness. In this study, we demonstrated the ability of our PCR/ESI-MS assay to correctly diagnose the pathogen responsible for ehrlichiosis, identifying it to the species level. The assay also detected Rickettsia rickettsii, Neisseria meningitidis, and Staphylococcus aureus. The assay is rapid; it can provide results from extracted specimens in 4 to 5 h, and a single system can process 90 samples a day. Thus, this assay may potentially provide a system for the rapid detection and identification of a broad range of bacterial pathogens directly from crude blood specimens.
Supplementary Material
Acknowledgments
We thank Criziel Quinn, Jessica Kilby, Anna Raymer, Sharon Smith, Mary Beth Underwood, Melinda McCormac, Rebekah Glisson, Korrin Crossno, and Steve Lawson for assistance with the collection and processing of clinical specimens.
We also acknowledge the financial support of the Lyme Disease Association, the Tami Fund, and the National Institute of Allergy and Infectious Diseases (R43-AI-077156).
Footnotes
Published ahead of print on 2 December 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Anderson, B. E., J. E. Dawson, D. C. Jones, and K. H. Wilson. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 29:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, B. E., C. E. Greene, D. C. Jones, and J. E. Dawson. 1992. Ehrlichia ewingii sp. nov., the etiologic agent of canine granulocytic ehrlichiosis. Int. J. Syst. Bacteriol. 42:299-302. [DOI] [PubMed] [Google Scholar]
- 3.Bell, C. A., and R. Patel. 2005. A real-time combined polymerase chain reaction assay for the rapid detection and differentiation of Anaplasma phagocytophilum, Ehrlichia chaffeensis, and Ehrlichia ewingii. Diagn. Microbiol. Infect. Dis. 53:301-306. [DOI] [PubMed] [Google Scholar]
- 4.Brownstein, M. J., J. D. Carpten, and J. R. Smith. 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004-1010. [DOI] [PubMed] [Google Scholar]
- 5.Buller, R. S., M. Arens, S. P. Hmiel, C. D. Paddock, J. W. Sumner, Y. Rikhisa, A. Unver, M. Gaudreault-Keener, F. A. Manian, A. M. Liddell, N. Schmulewitz, and G. A. Storch. 1999. Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N. Engl. J. Med. 341:148-155. [DOI] [PubMed] [Google Scholar]
- 6.Childs, J. E., J. W. Sumner, W. L. Nicholson, R. F. Massung, S. M. Standaert, and C. D. Paddock. 1999. Outcome of diagnostic tests using samples from patients with culture-proven human monocytic ehrlichiosis: implications for surveillance. J. Clin. Microbiol. 37:2997-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clay, K., O. Klyachko, N. Grindle, D. Civitello, D. Oleske, and C. Fuqua. 2008. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol. Ecol. 17:4371-4381. [DOI] [PubMed] [Google Scholar]
- 8.Dawson, J. E., S. A. Ewing, W. R. Davidson, J. E. Childs, S. E. Little, and S. M. Standaert. 2005. Human monocytotropic ehrlichiosis, p. 239-257. In J. L. Goodman, D. T. Dennis, and D. E. Sonenshine (ed.), Tick-borne diseases of humans. ASM Press, Washington, DC.
- 9.Doyle, C. K., M. B. Labruna, E. B. Breitschwerdt, Y. W. Tang, R. E. Corstvet, B. C. Hegarty, K. C. Bloch, P. Li, D. H. Walker, and J. W. McBride. 2005. Detection of medically important Ehrlichia by quantitative multicolor TaqMan real-time polymerase chain reaction of the dsb gene. J. Mol. Diagn. 7:504-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumler, J. S. 2005. Anaplasma and Ehrlichia infection. Ann. N. Y. Acad. Sci. 1063:361-373. [DOI] [PubMed] [Google Scholar]
- 11.Dumler, J. S., J. E. Madigan, N. Pusterla, and J. S. Bakken. 2007. Ehrlichioses in humans: epidemiology, clinical presentation, diagnosis, and treatment. Clin. Infect. Dis. 45(Suppl. 1):S45-S51. [DOI] [PubMed] [Google Scholar]
- 12.Ecker, D. J., J. Drader, J. Gutierrez, A. Gutierrez, J. Hannis, A. Schink, R. Sampath, J. A. Ecker, L. B. Blyn, M. W. Eshoo, T. A. Hall, M. Tobarmosquera, Y. Jiang, K. Sannes-Lowery, L. Cummins, B. Libby, D. J. Walcott, C. Massire, R. Ranken, S. M. Manalili, C. Ivy, R. Melton, H. Levene, V. Harpin, F. Li, N. White, M. Pear, V. Samant, D. Knize, D. Robbins, K. Rudnick, F. Hajjar, and S. A. Hofstadler. 2006. The Ibis T5000 universal biosensor: an automated platform for pathogen identification and strain typing. JALA 11:341-351. [Google Scholar]
- 13.Ecker, D. J., J. A. Ecker, T. A. Hall, C. Massire, L. B. Blyn, S. A. Hofstadler, M. Eshoo, R. Sampath, and P. Scott. 2006. Rapid, high-throughput bacterial genotyping to reduce healthcare-associated infections, p. 97. In LabAutomation 2006, 10th Anniversary Conference and Exhibition, Final Program & Abstracts. Association for Laboratory Automation, Geneva, IL. http://www.labautomation.org/conference/pdfs/LA06_final_program.pdf.
- 14.Ecker, D. J., R. Sampath, L. B. Blyn, M. W. Eshoo, C. Ivy, J. A. Ecker, B. Libby, V. Samant, K. Sannes-Lowery, R. E. Melton, K. Russell, N. Freed, C. Barrozo, J. Wu, K. Rudnick, A. Desai, E. Moradi, D. J. Knize, D. W. Robbins, J. C. Hannis, P. M. Harrell, C. Massire, T. A. Hall, Y. Jiang, R. Ranken, J. J. Drader, N. White, J. A. McNeil, S. T. Crooke, and S. A. Hofstadler. 2005. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl. Acad. Sci. U. S. A. 102:8012-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ecker, D. J., R. Sampath, C. Massire, L. B. Blyn, T. A. Hall, M. W. Eshoo, and S. A. Hofstadler. 2008. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 6:553-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eremeeva, M. E., G. A. Dasch, and D. J. Silverman. 2003. Evaluation of a PCR assay for quantitation of Rickettsia rickettsii and closely related spotted fever group rickettsiae. J. Clin. Microbiol. 41:5466-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eshoo, M. W., C. A. Whitehouse, S. T. Zoll, C. Massire, T. T. Pennella, L. B. Blyn, R. Sampath, T. A. Hall, J. A. Ecker, A. Desai, L. P. Wasieloski, F. Li, M. J. Turell, A. Schink, K. Rudnick, G. Otero, S. C. Weaver, G. V. Ludwig, S. A. Hofstadler, and D. J. Ecker. 2007. Direct broad-range detection of alphaviruses in mosquito extracts. Virology 368:286-295. [DOI] [PubMed] [Google Scholar]
- 18.Fishbein, D. B., J. E. Dawson, and L. E. Robinson. 1994. Human ehrlichiosis in the United States, 1985 to 1990. Ann. Intern. Med. 120:736-743. [DOI] [PubMed] [Google Scholar]
- 19.Hamburg, B. J., G. A. Storch, S. T. Micek, and M. H. Kollef. 2008. The importance of early treatment with doxycycline in human ehrlichiosis. Medicine (Baltimore) 87:53-60. [DOI] [PubMed] [Google Scholar]
- 20.Hofstadler, S. A., K. L. Hari, R. Sampath, L. B. Blyn, M. W. Eshoo, and D. J. Ecker. 2005. Detection of microbial agents using broad-range PCR with detection by mass spectrometry: the TIGER concept, p. 287-307. In M. J. Miller (ed.), Encyclopedia of rapid microbiological methods, vol. 3. Parenteral Drug Association, Bethesda, MD. [Google Scholar]
- 21.Hofstadler, S. A., R. Sampath, L. B. Blyn, M. W. Eshoo, T. A. Hall, Y. Jiang, J. J. Drader, J. C. Hannis, K. A. Sannes-Lowery, L. L. Cummins, B. Libby, D. J. Walcott, A. Schink, C. Massire, R. Ranken, J. Gutierrez, S. Manalili, C. Ivy, R. Melton, H. Levene, G. Barrett-Wilt, F. Li, V. Zapp, N. White, V. Samant, J. A. McNeil, D. Knize, D. Robbins, K. Rudnik, A. Desai, E. Moradi, and D. J. Ecker. 2005. TIGER: the universal biosensor. Int. J. Mass Spectrom. 242:23-41. [Google Scholar]
- 22.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. A. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muddiman, D. C., G. A. Anderson, S. A. Hofstadler, and R. D. Smith. 1997. Length and base composition of PCR-amplified nucleic acids using mass measurements from electrospray ionization mass spectrometry. Anal. Chem. 69:1543-1549. [DOI] [PubMed] [Google Scholar]
- 24.Olano, J. P., W. Hogrefe, B. Seaton, and D. H. Walker. 2003. Clinical manifestations, epidemiology, and laboratory diagnosis of human monocytotropic ehrlichiosis in a commercial laboratory setting. Clin. Diagn. Lab. Immunol. 10:891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paddock, C. D., and J. E. Childs. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16:37-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paddock, C. D., S. M. Folk, G. M. Shore, L. J. Machado, M. M. Huycke, L. N. Slater, A. M. Liddell, R. S. Buller, G. A. Storch, T. P. Monson, D. Rimland, J. W. Sumner, J. Singleton, K. C. Bloch, Y. W. Tang, S. M. Standaert, and J. E. Childs. 2001. Infections with Ehrlichia chaffeensis and Ehrlichia ewingii in persons coinfected with human immunodeficiency virus. Clin. Infect. Dis. 33:1586-1594. [DOI] [PubMed] [Google Scholar]
- 27.Paddock, C. D., A. M. Liddell, and G. A. Storch. 2005. Other causes of tick-borne ehrlichioses, including Ehrlichia ewingii, p. 258-267. In J. L. Goodman, D. T. Dennis, and D. E. Sonenshine (ed.), Tick-borne diseases of humans. ASM Press, Washington, DC.
- 28.Prince, L. K., A. A. Shah, L. J. Martinez, and K. A. Moran. 2007. Ehrlichiosis: making the diagnosis in the acute setting. South. Med. J. 100:825-828. [DOI] [PubMed] [Google Scholar]
- 29.Sampath, R., T. A. Hall, C. Massire, F. Li, L. B. Blyn, M. W. Eshoo, S. A. Hofstadler, and D. J. Ecker. 2007. Rapid identification of emerging infectious agents using PCR and electrospray ionization mass spectrometry. Ann. N. Y. Acad. Sci. 1102:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampath, R., K. L. Russell, C. Massire, M. W. Eshoo, V. Harpin, L. B. Blyn, R. Melton, C. Ivy, T. T. Pennella, F. Li, H. Levene, T. Hall, B. Libby, N. Fan, D. J. Walcott, R. Ranken, M. Pear, A. Schink, J. Gutierrez, J. Drader, D. Moore, D. Metzgar, L. Addington, R. Rothman, C. A. Gaydos, S. Yang, K. St. George, M. E. Fuschino, A. B. Dean, D. Stallknecht, G. Goekjian, S. Yingst, M. Monteville, M. D. Saad, C. A. Whitehouse, C. Baldwin, K. H. Rudnick, S. A. Hofstadler, S. M. Lemon, and D. J. Ecker. 2007. Global surveillance of emerging influenza virus genotypes by mass spectrometry. PLoS One 2:e489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Standaert, S. M., T. Yu, M. A. Scott, J. E. Childs, C. D. Paddock, W. L. Nicholson, J. Singleton, Jr., and M. J. Blaser. 2000. Primary isolation of Ehrlichia chaffeensis from patients with febrile illnesses: clinical and molecular characteristics. J. Infect. Dis. 181:1082-1088. [DOI] [PubMed] [Google Scholar]
- 32.Steiner, F. E., R. R. Pinger, C. N. Vann, N. Grindle, D. Civitello, K. Clay, and C. Fuqua. 2008. Infection and co-infection rates of Anaplasma phagocytophilum variants, Babesia spp., Borrelia burgdorferi, and the rickettsial endosymbiont in Ixodes scapularis (Acari: Ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J. Med. Entomol. 45:289-297. [DOI] [PubMed] [Google Scholar]
- 33.Tang, Y. W., P. N. Rys, B. J. Rutledge, P. S. Mitchell, T. F. Smith, and D. H. Persing. 1998. Comparative evaluation of colorimetric microtiter plate systems for detection of herpes simplex virus in cerebrospinal fluid. J. Clin. Microbiol. 36:2714-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, Y. W., S. E. Sefers, H. Li, D. J. Kohn, and G. W. Procop. 2005. Comparative evaluation of three commercial systems for nucleic acid extraction from urine specimens. J. Clin. Microbiol. 43:4830-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas, L. D., I. Hongo, K. C. Bloch, Y. W. Tang, and S. Dummer. 2007. Human ehrlichiosis in transplant recipients. Am. J. Transplant. 7:1641-1647. [DOI] [PubMed] [Google Scholar]
- 36.Wormser, G. P., R. J. Dattwyler, E. D. Shapiro, J. J. Halperin, A. C. Steere, M. S. Klempner, P. J. Krause, J. S. Bakken, F. Strle, G. Stanek, L. Bockenstedt, D. Fish, J. S. Dumler, and R. B. Nadelman. 2006. The clinical assessment, treatment, and prevention of Lyme disease, human granulocytic anaplasmosis, and babesiosis: clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 43:1089-1134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.