Contact: pawel@wzw.tum.de
Supplementary information: Supplementary data are available at Bioinformatics online.
Data mining is a cornerstone of modern bioinformatics. Techniques such as feature selection or data-driven model (classifier) building are used broadly in many fields including gene expression data analysis (Wood et al., 2007), proteomics (Barla et al., 2008), secondary and tertiary protein structure prediction (Heringa, 2000; Kryshtafovych et al., 2007), prediction of experimental behavior of proteins or other molecules (Liao et al., 2006; Smialowski et al., 2007) and medical science (Patel and Goyal, 2007). It is widely accepted that any model building requires stringent evaluation of its performance and generalization capabilities using well-established methods such as k-fold cross-validation, leave-one-out cross-validation, bootstrap and others (Frank et al., 2004).
The growing complexity of the data and the urge to improve available methods have led to the development of procedures combining feature selection and model building. Feature selection is often used to limit the amount and dimensionality of the data or to select features that correlate well with the target class. Feature selection methods can be subdivided into those that are unsupervised, i.e. unaware of class attributes [e.g. removal of a feature with the same constant values throughout the whole dataset, PCA (principal component analysis), MF (matrix factorization)] and those that are supervised, i.e. driven by class information. The latter group includes filter methods using, e.g. information gain as well as the Wrapper approach (Witten and Frank, 2005).
Special consideration is required when supervised feature selection is used to construct input data for model building (classification). In order to correctly evaluate classifiers built on such projected data, the entire procedure including feature selection and model training has to be evaluated against independent data. In other words, the test set must not be used for supervised feature selection (or more generally, supervised preprocessing). Otherwise, estimates of the classifiers’ performance will be over optimistic (Ambroise and McLachlan, 2002; Efron, 2005; Kohavi and John, 1997; Molinaro et al., 2005; Reunane, 2004). Any model building method integrated with feature selection must be externally evaluated. Evaluation must include both: supervised feature selection and classification and should not be limited to classification only (Fig. 1).
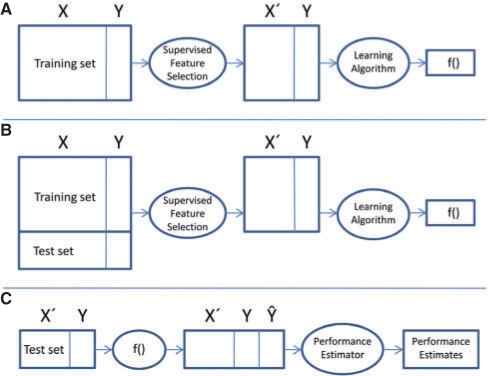
Fig. 1.
The correct [(A) followed (C)] and the incorrect [(B) followed by (C)] procedure for combining supervised feature selection and learning a classifier. In the figure, processes and products are depicted by ellipses and rectangles, respectively. Training and test sets consist of features X and a target attribute Y (to be predicted). X′ is a subset of features reduced by supervised feature selection, f() is a classifier and Ŷ contains the prediction of Y values by this function. (A and B) show the workflows for the correct and incorrect application of supervised feature selection, and (C) holds the evaluation workflow (more description in the text).
The correct procedure is depicted in Figure 1 and consists of the workflow from Figure 1A for training and Figure 1C for testing (evaluation). In a preprocessing step, supervised feature selection reduces the set of features X to a subset X′ (Y is the target attribute). Subsequently, the reduced training set is used to infer a classifier f(). During testing (Fig. 1C), the trained classifier f() is evaluated using an independent test set with the feature space reduced to X′ according to the feature selection derived in the previous step (Fig. 1A). The classifier predicts Ŷ for each instance. Various performance measures can then be calculated by comparing the predictions Ŷ with the true values for Y.
In contrast with this procedure, the most common mistake (according to our observation) of machine learning applications in the life sciences is illustrated in Figure 1B: both training and test sets are used for supervised feature selection. After that, a classifier f() is learned from the reduced training set as before. During testing, this classifier is applied to the test set as described above (Fig. 1C). However, as evident from the illustration, information from the test set has already been used for the inference of the classifier, by choosing an appropriate subset of features for the learning algorithm (Fig. 1B). Therefore, class information from the test set has leaked to the training phase. This analysis remains true for classification and regression and regardless whether training and test sets are created as part of a k-fold cross-validation or any other method. Recently, it has repeatedly come to our attention while reading or reviewing manuscripts submitted to Bioinformatics and other journals that often no proper external evaluations of the whole method consisting of supervised feature selection and subsequent classification is provided. In this brief note we would like to demonstrate the consequences of this type of a mistake for classification performance estimation using data where attribute values are generated randomly as well as data with random class assignments. We also estimate the consequences of such erroneous overoptimism as a function of the dataset size. For simplicity, we focus only on classification and leave out regression methods.
Three types of datasets were used in this study. All have 21 attributes: 20 frequencies of amino acids and one class attribute assigning each instance to one of two classes. Datasets of the first type (randomly generated attributes) contain real random attribute values resembling natural frequency distributions of 20 amino acids. To generate randomly perturbated frequencies of the 20 amino acids, we used Gaussian random numbers with peak maximums at their natural occurrence frequencies (as provided at http://prowl.rockefeller.edu/aainfo/struct.htm) and standard deviation (SD) equal to 0.25 of this value. All negative values were set to 0 and the sum of 20 frequencies for a given instance was not allowed to be greater than 100.
Datasets of the second type (randomize class data) consist of instances picked randomly from a dataset of 1000 proteins of which a half showed good solubility upon heterologous expression in Escherichia coli, while the other half was notoriously insoluble. Classes of these instances—soluble or non-soluble—were assigned randomly. Third type of datasets (real data) was same as second except that real class labels were preserved.
Quantum Random Bit Generator Service (QRBG; Stevanovic et al., 2008) was used as a source of seeds for random numbers. We maintained even class distribution at all times.
For selecting the best features, the following methods were used: Wrapper (Kohavi and John, 1997), Relief Attribute Evaluation with Ranker (Witten and Frank, 2005) and PCA (Pearson, 1901). The Wrapper method takes into account class information by evaluating feature sets based on the performance of the classifier. Hence, the resulting feature set is tailored to a given classification method. In our comparison, the Wrapper method is the most ‘aggressive’ feature selection method. It was setup to use Naive Bayes for classification and Best First for attribute space search (Witten and Frank, 2005). The Relief method is also supervised, but does not optimize feature sets directly for classifier performance. Thus, it takes into account class information in a ‘less aggressive’ manner than the Wrapper method. The threshold of Ranker coupled to the Relief Attribute Evaluation method was set to zero. PCA (Jolliffe, 2002) is an unsupervised feature selection method and hence does not take into account class information at all. PCA dimensionality reduction was accomplished by the following steps: data normalization, calculation of orthonormal vectors [PC (principal components)], sorting PC according to decreasing variance. We save PC accounting cumulatively for 95% of the data variance (Jolliffe, 2002). Naive Bayes and nearest neighbor IB1 (Aha and Kibler, 1991) algorithms were used for classification, because they are among the simplest and most fundamental classification methods. Classifiers were trained and evaluated using 10-fold cross-validation. MCC (Matthew's correlation coefficient) and AUROC (area under receiver operating curve) were calculated to measure classifier performance. For each dataset size, feature selection and classification algorithm the whole procedure starting from data construction was repeated 30 times. Overfitting (overoptimism) was measured as the difference in classification performance (denoted by ΔAUROC in the following) on data after and before feature selection, averaged over 30 trials.
Information density is defined as the number of instances per number of attributes. For datasets resulting from feature selection, we calculate the information density ratio (ID ratio, the factor by which the number of features is reduced) to measure loss of information.
We generated a range of datasets containing between 10 and 1000 instances. For both types of randomize datasets, half of the data were tagged with the ‘no’ class and the rest with ‘yes’ to simulate a two class problem with an even class distribution.
We found that using supervised feature selection with dataset containing both training and test sets leads to a significant overoptimistic assessment of classifier performance. This holds true for datasets with randomly generated attribute values, randomized class data (Fig. 2) and real data (Supplementary Fig. 1, Supplementary Table 1). The extent of overfitting depends on the dataset size. The strongest overfitting (AUROC increase ∼0.5) was observed for the smallest real datasets with random class labels using the Wrapper method. Classification on randomly generated attribute data exhibits a slightly smaller increase (∼0.4) (data with real class labels also reached 0.4). These values are extremely high considering that the AUROC ranges from 0.5 for random guessing to 1.0 for the best theoretically possible model. Improper use of feature selection also distorts classifier performance when larger datasets are used. Even for datasets with 1000 instances we observed slight albeit statistically significant increase in AUROC values (0.04) (whiskers in Fig. 2 mark 95% confidence intervals). As seen in Figure 2, the use of both the Wrapper and the Relief feature selection algorithms results in falsified measures of classifier performance. Application of PCA which is not supervised did not lead to substantial overfitting on any tested dataset.
Fig. 2.
Relation between the number of instances and the extent of overfitting caused by feature selection as measured by AUROC growth. (A) Randomly generated attribute values, (B) randomly tagged real data. Three different feature selection algorithms were used: Wrapper (hashed bars), Relief Attribute Evaluation (white bars), PCA (black bars). Whiskers mark 95% confidence intervals.
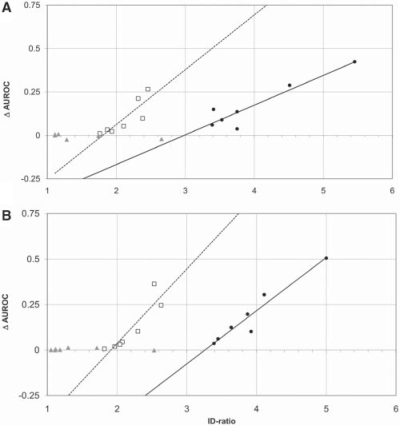
Feature selection reduces the amount of information by lowering the number of attributes and thus leads to an increase of the ratio between the number of instances and the number of attributes. We assessed the extent of information loss by calculating the ID ratio between datasets before and after feature selection as described above. A higher ID ratio means that a higher percentage of features was removed. For example, ID ratio = 2, means that after feature selection the number of attributes is reduced by two. For each of the feature selection method, we found that the more features got removed, the stronger were the effects of overfitting (Fig. 3). Interestingly, there are also differences between attribute selection algorithms. Wrapper seems to be less prone to overfitting compared with the Relief method. By almost the same magnitude of overfitting (increase in AUROC 0.25), it reduces the information content more effectively (4.5 times) compared with Relief (2.5 times). Wrappers reduce the number of features stronger than Relief method possibly because it comprehensively evaluates also combinations of features, selecting features based on not only their own relevancy but also redundancy (Kohavi and John, 1997). Moreover, Wrapper efficiency in feature selection depends on internally employed classifier and search algorithms. Data with randomly generated attribute values showed higher resistance against classification bias permitting a greater decrease in the feature number compared with real data with randomized class tags and real data. PCA did not cause overfitting while still being able to slightly reduce the number of attributes of all data types. Similar results were obtained when MCC was used instead of AUROC and also when the whole analysis was repeated using the IB1 nearest neighbor classification method (Supplementary Table 1, Data not shown).
Fig. 3.
Relation between information loss and overfitting measured by ΔAUROC growth. (A) Randomly generated attribute values, (B) randomly tagged real data. Three feature selection methods were examined: Wrapper (black circles), Relief Attribute Evaluation (open squares) and PCA (gray triangles). Lines were fitted by linear regression: solid lines to Wrapper and dashed to Relief Attribute Evaluation data points. ID ratio is the information density ratio.
In summary, we show that supervised feature selection coupled with or integral to model building (classification) requires external evaluation. Failure in setting up evaluation with external data (Fig. 1B) leads to overoptimism in the assessment of classifier performance. It is reasonable to expect that overfitting will be at least in the same order of magnitude as what is observed on randomly generated data for most of the real life classification tasks, where amino acid composition is used as input. Real data may be more prone to overfitting because they are likely to contain more patterns than randomly generated data. Even if patterns are orthogonal to the problem under consideration overfitting can still occur. For this reason, whenever model building (classification) is integrated with supervised attribute selection, it is crucial to evaluate classifiers with the data not used for attribute selection (Fig. 1 A and C).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Aha D, Kibler D. Instance-based learning algorithms. Mach. Learn. 1991;6:37–66. [Google Scholar]
- Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl Acad. Sci. USA. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barla A, et al. Machine learning methods for predictive proteomics. Brief Bioinform. 2008;9:119–128. doi: 10.1093/bib/bbn008. [DOI] [PubMed] [Google Scholar]
- Efron B. 22nd International Conference on Machine learning. Bonn: ACM; 2005. Reducing overfitting in process model induction; pp. 81–88. [Google Scholar]
- Frank E, et al. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- Heringa J. Computational methods for protein secondary structure prediction using multiple sequence alignments. Curr. Protein Pept. Sci. 2000;1:273–301. doi: 10.2174/1389203003381324. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT. Principal Component Analysis. NY: Springer; 2002. [Google Scholar]
- Kohavi R, John G. Wrappers for feature subset selection. Artif. Intell. 1997;97:273–324. [Google Scholar]
- Kryshtafovych A, et al. Progress from CASP6 to CASP7. Proteins. 2007;69(Suppl. 8):194–207. doi: 10.1002/prot.21769. [DOI] [PubMed] [Google Scholar]
- Liao Q, et al. SVM approach for predicting LogP. Mol. Divers. 2006;10:301–309. doi: 10.1007/s11030-006-9036-2. [DOI] [PubMed] [Google Scholar]
- Molinaro AM, et al. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21:3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- Patel JL, Goyal RK. Applications of artificial neural networks in medical science. Curr. Clin. Pharmacol. 2007;2:217–226. doi: 10.2174/157488407781668811. [DOI] [PubMed] [Google Scholar]
- Pearson K. On lines and planes of closest fit to systems of points in space. Phil. Mag. 1901;2:559–572. [Google Scholar]
- Reunane J. Workshop on Pattern Recognition in Information Systems Proceedings. Portugal: Porto; 2004. A pitfall in determining the optimal feature subset size; pp. 176–185. April'04. [Google Scholar]
- Smialowski P, et al. Protein solubility: sequence based prediction and experimental verification. Bioinformatics. 2007;23:2536–2542. doi: 10.1093/bioinformatics/btl623. [DOI] [PubMed] [Google Scholar]
- Stevanovic R, et al. Quantum random bit generator service for Monte Carlo and other stochastic simulations. In: Lirkov I, et al., editors. Lecture Notes in Computer Science. Berlin, Heidelberg: Springer; 2008. pp. 508–515. [Google Scholar]
- Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- Wood IA, et al. Classification based upon gene expression data: bias and precision of error rates. Bioinformatics. 2007;23:1363–1370. doi: 10.1093/bioinformatics/btm117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.