Abstract
Summary: Representing models of cellular processes or pathways in a graphically rich form facilitates interpretation of biological observations and generation of new hypotheses. Solving biological problems using large pathway datasets requires software that can combine data mapping, querying and visualization as well as providing access to diverse data resources on the Internet. ChiBE is an open source software application that features user-friendly multi-view display, navigation and manipulation of pathway models in BioPAX format. Pathway views are rendered in a feature-rich format, and may be laid out and edited with state-of-the-art visualization methods, including compound or nested structures for visualizing cellular compartments and molecular complexes. Users can easily query and visualize pathways through an integrated Pathway Commons query tool and analyze molecular profiles in pathway context.
Availability: http://www.bilkent.edu.tr/%7Ebcbi/chibe.html
Contact: ugur@cs.bilkent.edu.tr
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Our knowledge of cellular processes has been increasing at a dramatic rate. Efficient construction and use of a knowledge base of cellular pathways depends highly on a strong representation (i.e. a structured semantic encoding of knowledge). Such a knowledge base can then act as a blueprint for simulations and other analysis methods, improving our ability to understand and predict the behavior of cells (Demir et al., 2004).
Computational biologists have advanced pathway knowledge representation, created standards and formats (http://www.biopax.org; http://sbml.org; Caspi et al., 2008; Demir et al., 2004); Kanehisa et al., 2007; Matthews et al., 2009), and built more than 300 pathway and interaction databases (Bader et al., 2006) in recent years. However, current bioinformatics infrastructure is still lacking in software tools for visualizing and analyzing pathways. A main obstacle in this direction has been the fragmented, incomplete and incompatible nature of pathway knowledge, making representation and integration of pathways extremely difficult.
A number of interesting pathway visualization tools (Elliott et al., 2008; Funahashi et al., 2008; Hu et al., 2008; Yeung et al., 2008) have been developed over the past decade, with diverse analysis focus, from analyzing gene expression profiles to effective database querying, to discovering graph-theoretic properties in biological networks. Such tools can benefit substantially from standardization of knowledge representation, pathway-specific layout algorithms and representation of compound graphs.
Knowledge representation: BioPAX has made great progress in developing a standard exchange format for biological pathway data, as a result of several years of community effort. Pathway Commons (PC), based on BioPAX, was developed as an integrated single point of access to publicly available pathway information. PC covers major pathway databases (http://www.pathwaycommons.org) and already provides integration at the level of molecular identifiers. Therefore, the community now has an emerging platform for building software tools and services without worrying about compatibility and fragmentation issues.
Pathway layout: general graph layout algorithms do not address the specific needs and established conventions of pathway graphs. So far, work on pathway layout algorithms (Becker and Rojas, 2001; Genc and Dogrusoz, 2006; Karp and Paley, 1994; Schreiber, 2002; Wegner and Kummer, 2005) has primarily focused on biochemical pathways. Thus, the need for layout of complex pathways, such as signal transduction, remains to be addressed.
Compound graphs: an extension of graph-based representation, namely hierarchically structured or compound graphs, in which a member of a biological network may recursively contain a sub-network of other pathway elements, can be used for representing subpathways, molecular complexes and subcellular location. Compound graphs also help in managing complexity by interactively decomposing a pathway into distinct components or modules (Demir et al., 2004; Fukuda and Takagi, 2001). The recently introduced visualization standard SBGN (Le Novere et al., 2009) also uses compound graphs extensively.
ChiBE is an open source visualization tool, which for the first time, brings together compound graph-based BioPAX visualization, seamless PC access, pathway-specific layout and strong visualization and data analysis capabilities. A comparison of ChiBE with similar visualization tools can be found in Section 3 of Supplementary Material.
2 DESCRIPTION
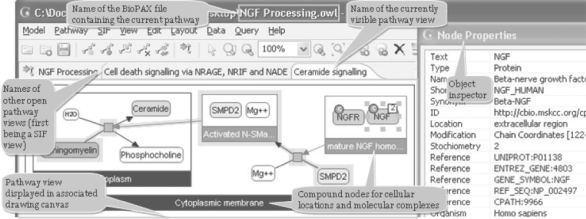
ChiBE accepts data in BioPAX Level 2 format. Section 1 of Supplementary Material summarizes how BioPAX models are interpreted by ChiBE. We call the entire set of biological information loaded from a BioPAX file a pathway model. As defined by BioPAX, a pathway is a set or series of interactions, often forming a network, which biologists have assembled for organizational, historic, biophysical or other reasons. We use pathways to determine the boundaries of a coherent view. Each loaded pathway is displayed in a separate canvas, organized with tabs (Fig. 1). A pathway model may be expanded by merging it with another BioPAX file or PC query.
Fig. 1.
ChiBE views are organized in canvasses, each displaying one or more BioPAX pathways.
A pathway view is composed of pathway objects and their interactions. Compound nodes are exclusively used to represent molecular complexes and cellular compartments (Fig. 1). Our notation is similar to that of PATIKA (Demir et al., 2004). ChiBE has context-sensitive pop-up menus associated with pathway objects, providing fast access to popular operations for the associated pathway object. All kinds of nodes and edges in a pathway view have distinct properties and UIs (User Interfaces). These properties can be changed by using inspectors for each pathway object.
Viewing and editing pathways: the user has various mechanisms for navigating and editing the topology as well as the geometry of pathway views. These mechanisms range from standard zoom/scroll and highlight operations to modifying the UI associated with each pathway object and to automatic layout of the pathway view.
Pathway operations: any subset of available pathways in a model may be displayed as a separate view, and may be saved as an image or printed. Each subset may then be modified as desired. Also, new pathway views may be created by duplicating, cropping or capturing a neighborhood of a view.
Querying PC: PC is a convenient point of access to biological pathway information collected from public pathway databases, which one can browse or search. ChiBE provides a graphical user interface to search this knowledge base to find pathways that contain a specified molecule (using its UniProt or Entrez Gene ID), and present the results in a visual form. The resulting view may contain either the whole pathway or only the immediate neighborhood of the specified molecule in all the pathways in which it appears.
SIF operations: SIF (simple interaction format) is a format introduced by Cytoscape (Yeung et al., 2008) for describing interactions in a biological network. ChiBE can reduce BioPAX pathways to SIF using a customizable set of rules to obtain a simpler view. Pathways can also be saved in SIF.
Visualizing high-throughput data: multiple types of high-throughput data, such as gene expression or proteomics profiles, copy number variation and mutation data, can be loaded into ChiBE, and overlaid onto pathway views using color coding or displayed in tables that can be searched and filtered (Section 2 of Supplementary Material).
Availability and components: ChiBE is a free (EPL v1.0) Java application that runs on Windows, Mac OS and Linux. It was built using Chisio 1.0 (http://www.cs.bilkent.edu.tr/%7Eivis/chisio.html) and Eclipse GEF 3.1 (http://www.eclipse.org/gef) for graph visualization, Paxtools (http://www.biopax.org/paxtools) for accessing and manipulating BioPAX files and PATIKAmad (Babur et al., 2008) for high-throughput data visualization.
3 CONCLUSIONS
ChiBE can be a valuable tool for visualizing BioPAX models and high-throughput data in the context of pathways, and as a front-end for PC, making pathway knowledge more accessible and usable for biologists. We aim to enrich editing and querying support in ChiBE, provide support for the recently released BioPAX Level 3 ontology and format, and for the graphical notation standard SBGN.
Funding: The Scientific and Technological Research Council of Turkey (grant 104E049); US National Institutes of Health (grant P41HG004118).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Babur O, et al. PATIKAmad: putting microarray data into pathway context. Proteomics. 2008;8:2196–2198. doi: 10.1002/pmic.200700769. [DOI] [PubMed] [Google Scholar]
- Bader GD, et al. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MY, Rojas I. A graph layout algorithm for drawing metabolic pathways. Bioinformatics. 2001;17:461–467. doi: 10.1093/bioinformatics/17.5.461. [DOI] [PubMed] [Google Scholar]
- Caspi R, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E, et al. An ontology for collaborative construction and analysis of cellular pathways. Bioinformatics. 2004;20:349–356. doi: 10.1093/bioinformatics/btg416. [DOI] [PubMed] [Google Scholar]
- Elliott B, et al. PathCase: pathways database system. Bioinformatics. 2008;24:2526–2533. doi: 10.1093/bioinformatics/btn459. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Takagi T. Knowledge representation of signal transduction pathways. Bioinformatics. 2001;17:829–837. doi: 10.1093/bioinformatics/17.9.829. [DOI] [PubMed] [Google Scholar]
- Funahashi A, et al. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc. IEEE. 2008;96:1254–1265. [Google Scholar]
- Genc B, Dogrusoz U. A layout algorithm for signaling pathways. Inf. Sci. 2006;176:135–149. [Google Scholar]
- Hu Z, et al. VisANT: an integrative framework for networks in systems biology. Brief. Bioinform. 2008;9:317–325. doi: 10.1093/bib/bbn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Paley S. Third International Conference on Bioinformatics and Genome Research. Tallahassee, Florida: 1994. Automated drawing of metabolic pathways; pp. 225–238. [Google Scholar]
- Le Novere N, et al. The systems biology graphical notation. Nat. Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- Matthews L, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber F. High quality visualization of biochemical pathways in BioPath. In Silico Biol. 2002;2:59–73. [PubMed] [Google Scholar]
- Wegner K, Kummer U. A new dynamical layout algorithm for complex biochemical reaction networks. BMC Bioinformatics. 2005;6:212. doi: 10.1186/1471-2105-6-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung N, et al. Exploring biological networks with Cytoscape software. Curr. Protoc. Bioinformatics. 2008 doi: 10.1002/0471250953.bi0813s23. Chapter 8Unit 8.13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.