Abstract
Summary: Tablet is a lightweight, high-performance graphical viewer for next-generation sequence assemblies and alignments. Supporting a range of input assembly formats, Tablet provides high-quality visualizations showing data in packed or stacked views, allowing instant access and navigation to any region of interest, and whole contig overviews and data summaries. Tablet is both multi-core aware and memory efficient, allowing it to handle assemblies containing millions of reads, even on a 32-bit desktop machine.
Availability: Tablet is freely available for Microsoft Windows, Apple Mac OS X, Linux and Solaris. Fully bundled installers can be downloaded from http://bioinf.scri.ac.uk/tablet in 32- and 64-bit versions.
Contact: tablet@scri.ac.uk
1 INTRODUCTION
The advent of next-generation sequencing (NGS) technologies such as Roche 454 (Margulies et al., 2005) and Illumina Solexa (http://www.illumina.com/sequencing) has brought about a need for fast, efficient and user-friendly tools for analyzing the outcome of sequencing runs. This includes visualization software for viewing the resultant assemblies or alignments, for example, Consed (Gordon et al., 1998), Hawkeye (Schatz et al., 2007), EagleView (Huang and Marth, 2008), MapView (Bao et al., 2009), SAMtools' tview (Li et al., 2009) and Maqview (http://maq.sourceforge.net/maqview.shtml).
All assembly visualization packages face the following challenges when dealing with NGS data: processing a very large number of reads, providing high-quality rendering and navigation of assembled reads, and supporting a widening range of assembly formats. Additionally, as analysis and interpretation of the data moves from large-genome centers to smaller laboratories, there is an increasing need for biologist-friendly software that has an intuitive user interface, is available for a range of common desktop platforms and has no complicated installation dependencies.
With these features in mind, we have developed Tablet, a lightweight, high-performance and memory efficient assembly viewer. Tablet is aimed at users of all abilities and combines simple installation on a desktop machine with ease of use and a visually rich interface. The application supports both single and multi-core processor architectures and will scale its performance according to the number of processor cores available.
2 FEATURES
Tablet can import data from ACE, AFG, MAQ and SOAP assembly formats (with preliminary support for SAM), and can handle both 454 and Solexa data. Its visualizations are split into several areas; the main display provides a view of a single contig at a time, with reads aligned against their consensus sequence. Reads are colored according to nucleotide type and subtle use of gradients and color choice allow visual structure to be maintained even when fully zoomed out. Tablet will lay out the data in either packed (showing as many reads per line as possible without overlap) or stacked (showing one read per line) formats, and allows the user to switch instantly between them. A sortable list, containing all available contigs shows contig lengths as well as numbers of reads and annotation features, and can be dynamically filtered by any of its fields. Continuous zooming of the entire contig in real time is supported by means of a slider, and there is also an option for varying the contrast between variant and non-variant nucleotides which adjusts the brightness used to display read bases that differ from the consensus, thus aiding identification of potential single nucleotide polymorphisms (SNPs) or sequencing errors.
An overview window located above the consensus can display either a scaled-to-fit summary of all the reads in a contig, or a coverage graph which shows the read coverage along the entire length of the contig independent of the current zoom level.
Navigation within a contig is catered for in several ways. First, the current view point is controlled by manipulating the scroll bars to move in either direction around the display or by dragging with the mouse directly on the canvas itself. We also provide a page-at-a-time navigation option that will move the view left or right by the number of bases that are currently visible. High-speed navigation to any area of the view is also available by clicking and dragging on the overview window, which always displays a bounding rectangle representing the portion of the overall data currently visible within the main display.
Protein translations are optionally provided for all six reading frames of the consensus sequence. Annotation features such as SNPs and indels can either be imported with the assembly file itself or separately in GFF3 format, and are then listed on a separate tab attached to the contig list.
Information on a given read is provided as a graphical overlay as soon as the mouse is moved over it. Tablet will display its name, start and end positions (optionally with unpadded consensus values), the sequence length, whether it is complemented or not, and also provide a scaled-to-fit graphical representation of the bases within the read. All of the raw data can be copied to the clipboard at any time.
3 IMPLEMENTATION AND PERFORMANCE
Tablet is written in Java and is compatible with any Java-enabled system with a runtime level of ≥1.6. We provide installable versions that include everything required to run the application, including a suitable Java runtime. The installers are available for Windows, Mac OS X, Linux and Solaris, in both 32- and 64-bit versions. Once installed and running, Tablet will also monitor our server for new versions and will prompt, download and update quickly and easily whenever a new release is available, along with redirecting the user to a web page describing the new features that have been added.
A prime requisite during development of Tablet has been computing efficiency and speed. The two main approaches to handling assembly data in viewers are either memory-based, where all the data are loaded into memory, or disk-cached, where the data reside on disk with only the currently visible segment of the dataset held in memory. Memory-based applications are faster for viewing and navigation (after an initial delay while loading the data) and can provide whole dataset overviews and statistical summaries, but the size of dataset they can handle is limited by the amount of available memory. In contrast, cache-based applications can display views from much larger datasets using a minimum of memory, but access to the data can be orders of magnitude slower (which then affects navigation and rendering), and the feature sets available are often limited.
With Tablet, we have chosen a hybrid solution that provides us with advantages from both approaches. We hold a ‘skeleton’ layout of the reads in memory, with data on each read limited to just an internal ID, its position against the consensus or reference sequence and its length. The nucleotide data itself (efficiently compressed so it can be read as quickly as possible), along with other supplementary information—such as the read's name and its orientation—is held in an indexed disk-cache and is only accessed (via the read's ID) when required. Tablet also allocates memory on a per-contig basis, including information for features such as how to pack the data for display, coverage calculations, padded-to-unpadded mappings, etc. These data are calculated and stored before each contig is rendered and discarded again after display. This approach allows us to provide maximum functionality—instant access to any portion of the data; extremely fast and high-quality rendering; entire dataset overviews—yet memory usage is kept relatively low.
Comparing data indexing/loading times and memory consumption across a range of tools for an assembly file containing ∼2.9 million Illumina Solexa reads of length 51, we found that the cache-based viewers (Maqview, MapView, tview) were fairly constant in memory usage (between 35 MB and 70 MB while viewing), with indexing times varying from 10 s to 50 s, although memory consumption during indexing did peak as high as 350 MB with MapView. For the memory-based viewers, we compared Hawkeye (5500 MB; 107 s), Consed (2600 MB; 73 s) and EagleView (2450 MB; 98 s). Tablet, being a hybrid, loads the data in 25 s, and uses just 175 MB of memory.
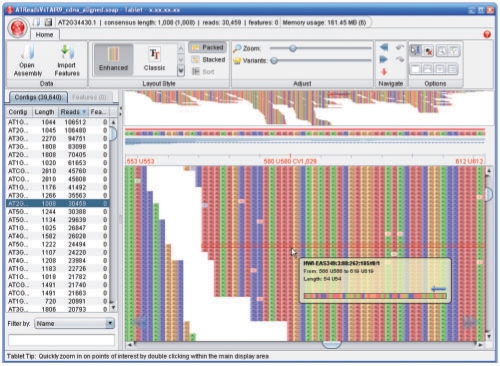
Fig. 1.
Tablet showing Illumina Solexa reads against an Arabidopsis thaliana cDNA reference sequence (additional screenshots can be seen online at http://bioinf.scri.ac.uk/tablet/screenshots.shtml).
4 FUTURE WORK
Work is in progress to support paired-end sequence data, and to enhance Tablet's visualization of annotation data. We also plan to further reduce Tablet's memory requirements by cutting down on the amount of reference/consensus information held at any time. Experiments have shown that further reductions should be possible without compromising data access times, graphical rendering speed or visualization quality.
ACKNOWLEDGEMENTS
We would like to thank colleagues within the Genetics, Pathology and BioSS Programmes at SCRI for their input to this project.
Funding: Scottish Government (RERAD, Programme 1); the Scottish Funding Council and Scottish Enterprise through the Scottish Bioinformatics Research Network (SBRN) project.
Conflict of Interest: none declared.
REFERENCES
- Bao H, et al. MapView: visualization of short reads alignment on a desktop computer. Bioinformatics. 2009;25:1554–1555. doi: 10.1093/bioinformatics/btp255. [DOI] [PubMed] [Google Scholar]
- Gordon D, et al. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Huang W, Marth G. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–1543. doi: 10.1101/gr.076067.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMTools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, et al. Hawkeye: an interactive visual analytics tool for genome assemblies. Genome Biol. 2007;8:R34. doi: 10.1186/gb-2007-8-3-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]