The crystal structure of uracil-DNA N-glycosylase from Vibrio cholerae has been determined at 1.5 Å, and used for mapping temperature adaptation.
Keywords: cold adaptation, mutational analysis, uracil-DNA N-glycosylase, Aliivibrio salmonicida, Vibrio cholerae
Abstract
The crystal structure of Vibrio cholerae uracil-DNA N-glycosylase (vcUNG) has been determined to 1.5 Å resolution. Based on this structure, a homology model of Aliivibrio salmonicida uracil-DNA N-glycosylase (asUNG) was built. A previous study demonstrated that asUNG possesses typical cold-adapted features compared with vcUNG, such as a higher catalytic efficiency owing to increased substrate affinity. Specific amino-acid substitutions in asUNG were suggested to be responsible for the increased substrate affinity and the elevated catalytic efficiency by increasing the positive surface charge in the DNA-binding region. The temperature adaptation of these enzymes has been investigated using structural and mutational analyses, in which mutations of vcUNG demonstrated an increased substrate affinity that more resembled that of asUNG. Visualization of surface potentials revealed a more positive potential for asUNG compared with vcUNG; a modelled double mutant of vcUNG had a potential around the substrate-binding region that was more like that of asUNG, thus rationalizing the results obtained from the kinetic studies.
1. Introduction
Cold-adapted enzymes are generally characterized by a higher catalytic efficiency, a lower thermal stability and a lower temperature optimum compared with their mesophilic counterparts. The cold-adapted enzymes studied to date utilize different adaptive strategies to perform more efficient catalysis compared with their mesophilic counterparts (Smalås et al., 2000 ▶; Russell, 2000 ▶; D’Amico et al., 2002 ▶; Sheridan et al., 2000 ▶). The main theory, which has not been proven, proposes that an increase in the structural flexibility of psychrophilic enzymes also increases their catalytic efficiency (Hochachka & Somero, 1984 ▶). Increased flexibility is not necessarily observed as an overall increase, as in many cases increased flexibility in localized areas seems to be one of the strategies used to maintain high catalytic activity at lower temperatures.
An optimization of electrostatic surface potentials near the active sites, which might favour binding to substrates, has also been observed for some cold-adapted enzymes and could be another strategy for cold adaptation. This is particularly useful for enzymes that interact with charged substrates; salmon trypsin, for example, has been reported to have a more negatively charged surface in the substrate-binding region compared with a mesophilic homologue (Gorfe et al., 2000 ▶). This increase in negative potential is believed to cause an increased substrate affinity for positively charged substrates. For cold-adapted Atlantic cod uracil-DNA N-glycosylase, both increased local flexibility (Olufsen et al., 2005 ▶) and optimization of electrostatic surface potential have been suggested as strategies for cold adaptation (Leiros et al., 2003 ▶; Moe et al., 2004 ▶).
Uracil DNA glycosylases (UDGs) have been identified in most organisms (Kavli et al., 2002 ▶) and several classes of UDGs have been described. The major cellular form of UDG is uracil-DNA N-glycosylase (UNG), encoded by the UNG gene (Slupphaug et al., 1995 ▶). UNG is highly conserved in evolution and high sequence identity has been shown between the catalytic domains of UNG from different species (Krokan et al., 1997 ▶). UNG is the primary enzyme responsible for the removal of uracil from DNA catalysing the first step in the uracil-excision repair pathway. The biological function of UNG is the specific removal of the RNA base uracil from DNA, liberating free uracil and generating apyrimidinic sites. Uracil in DNA arises either as a result of the deamination of cytosine or via misincorporation of dUTP during DNA synthesis (Lindahl, 1974 ▶).
Structures of UNG are known from various species and include human UNG (hUNG; Mol et al., 1995 ▶), cod UNG (cUNG; Leiros et al., 2001 ▶, 2003 ▶), Escherichia coli UNG (ecUNG; Ravishankar et al., 1998 ▶), Deinococcus radiodurans UNG (drUNG; Leiros et al., 2005 ▶) and herpes simplex virus 1 (HSV-1) UNG (Savva et al., 1995 ▶). Structures of UNG in complex with the uracil DNA-glycosylase inhibitor protein (Ugi) have also been determined (Géoui et al., 2007 ▶; Kaushal et al., 2008 ▶). The UNG structures have a classic single-domain α/β-fold, with a central four-stranded parallel β-sheet surrounded by a variable number of α-helices. The N- and C-terminal ends are on opposite sides of the central β-sheet. The active site is located at the C-terminal side of the central four-stranded β-sheet and in this region there is a conical cleft shaped to bind double-stranded DNA. In the middle of this cleft there is a cavity that accommodates a uracil base which is flipped-out of the double-stranded DNA (Slupphaug et al., 1996 ▶). Four loops are involved in the detection and removal of uracil in DNA. These are the water-activating loop (145-DPYH-148), the 4-Pro loop (165-PPPPS-169), the Gly-Ser loop (246-GS-247) and the Leu272 loop (268-HPSPLSVY-275) (hUNG numbering). Some of the most important residues for catalysis are the conserved His268 and Asp145. By movement of the Leu272 loop upon substrate binding, His268 is brought to within hydrogen-bonding distance of the uracil O2 and a uracil-recognition pocket is formed (Parikh et al., 1999 ▶). The glycosidic bond (linking the ribose and uracil functional groups) is proposed to be cleaved in a stepwise mechanism, yielding an oxocarbenium cation and a uracil-anion intermediate (Werner & Stivers, 2000 ▶; Dinner et al., 2001 ▶; Jiang, Ichikawa et al., 2002 ▶; Jiang, Drohat et al., 2002 ▶; Jiang et al., 2003 ▶). Subsequent attack by a water molecule and transfer of a proton to Asp145 results in the products. The side chains of both Asp145 and His268 contribute to lowering the activation energy by stabilizing the transition state.
UNG from the psychrophile Aliivibrio salmonicida (asUNG) has previously been characterized and compared with the mesophilic Vibrio cholerae UNG (vcUNG; Raeder et al., 2008 ▶). Biochemical characterization revealed typical cold-adapted features of asUNG such as low temperature stability, a lower temperature optimum and an increased catalytic efficiency (k cat/K m) compared with vcUNG. The increased catalytic efficiency arose from lowered K m values (increased substrate affinity) compared with vcUNG and not a higher k cat as often found for cold-adapted enzymes. Further analyses and comparisons of these and other UNG sequences resulted in a hypothesis suggesting that substitutions of specific amino acids in the asUNG sequence could account for increased positive surface charges in the DNA-binding area, leading to increased affinity for the negatively charged substrate DNA. The aim of the present study was to further investigate the effect of charge distributions in the DNA-interaction regions of these enzymes through structural and mutational studies.
2. Materials and methods
2.1. Recombinant expression and purification of vcUNG
The gene encoding vcUNG (accession No. Q9KPK8) was cloned, recombinantly expressed using Gateway technology (Invitrogen, Carlsbad, California, USA) and purified as described previously (Raeder et al., 2008 ▶). In brief, E. coli BL21-CodonPlus (DE3) strain (Stratagene, Cedar Creek, Texas, USA) was transformed with the pDEST14 vector containing the vcUNG gene. The resulting transformant was used in large-scale expression in 1 l cultures. The protein extract (50 ml) was applied onto a HisTrap HP Column (1 ml; GE Healthcare, London, England) and equilibrated with buffer A1 (50 mM Tris–HCl pH 7.5, 150 mM NaCl and 5 mM β-mercaptoethanol). The protein was eluted from the column using a gradient of 0–100% buffer B1 (50 mM Tris–HCl, 150 mM NaCl, 5 mM β-mercaptoethanol and 500 mM imidazole). Fractions containing UNG activity were diluted with buffer A1, applied onto a HisTrap HP Column (1 ml) once more and eluted using the same conditions as in the first step. Fractions containing UNG activity were pooled, concentrated to 150 µl with an Amicon Ultra-4 spin column (Millipore, Billerica, Massachusetts, USA) and loaded onto a Superdex 75 (10/300) gel-filtration column (GE Healthcare) equilibrated in buffer A2 (50 mM Tris–HCl pH 7.5, 5 mM β-mercaptoethanol). The pooled Superdex 75 fractions containing the enzyme were concentrated to 15 mg ml−1 using an Amicon Ultra-4 spin column (Millipore).
2.2. Crystallization of vcUNG
vcUNG was crystallized using the hanging-drop vapour-diffusion method. Crystals were obtained by mixing 1 µl 15 mg ml−1 protein solution with a precipitant solution containing 50 mM Na MES pH 6.0 and 28%(w/v) polyethylene glycol (PEG) 3350 (Sigma Low Ionic Kit; Sigma–Aldrich, St Louis, Missouri, USA) on siliconized cover slides and equilibrating against a reservoir containing 1.0 ml precipitant solution. Crystals generally grew within one week at room temperature (about 294 K).
2.3. Data collection, structure determination and refinement of vcUNG
Crystals with overall dimensions of about 200 × 200 × 150 µm were rapidly soaked in reservoir solution with 20%(v/v) glycerol added. This solution sufficed to cryoprotect the crystals for flash-cooling in liquid nitrogen. X-ray data were collected from a single crystal in a nitrogen cold stream (Oxford Instruments) at 100 K. This crystal diffracted to 1.5 Å resolution on beamline 14.1 at BESSY (Berlin, Germany). Data-collection characteristics are listed in Table 1 ▶. The intensity data were indexed, integrated and scaled using XDS (Kabsch, 1993 ▶) before being converted to structure factors using the CCP4 program TRUNCATE (Collaborative Computational Project, Number 4, 1994 ▶). The crystals were orthorhombic, with unit-cell parameters a = 60.14, b = 60.44, c = 61.33 Å, α = β = γ = 90°. Systematic absences in the collected data identified twofold screws along all three axes, leaving P212121 as the only possible choice of space group. This choice of crystal symmetry and absences were also indicated by analysis of the intensity data using the Bruker–Nonius program XPREP. The solvent content was estimated to be around 44%, with a Matthews coefficient of 2.19 Å3 Da−1, assuming the presence of one protein molecule in the asymmetric unit. The crystal structure of vcUNG was determined by the molecular-replacement method using MOLREP (Collaborative Computational Project, Number 4, 1994 ▶), with the crystal structure of ecUNG (Xiao et al., 1999 ▶), which has a sequence identity of 69.9%, as a search model. The automated program functions of MOLREP were utilized in order to obtain the search model with the presumably best fit to the search model, i.e. the side chains of the model were stripped to fit the vcUNG sequence. Using a high-resolution cutoff of 3 Å, one well resolved solution was found with a correlation coefficient of 0.487 and an R factor of 43.9% (the second solution had a correlation coefficient of 0.103 and an R factor of 58.3%). A round of refinement in REFMAC5 (Murshudov et al., 1999 ▶) followed by automated model building using ARP/wARP (Perrakis et al., 1999 ▶) using all available reflections built 216 of the possible 226 amino-acid residues. Subsequent refinement was performed by alternating cycles of manual adjustments with O (Jones et al., 1991 ▶) followed by refinement with REFMAC5 (Murshudov et al., 1999 ▶). The final R work and R free values were 17.5% and 21.0%, respectively, with acceptable protein stereochemistry. The final model of vcUNG consisted of 223 amino-acid residues in a single polypeptide chain comprising residues 4–226 of the amino-acid sequence; no electron density could be observed for the three N-terminal residues. One chloride ion and 327 water molecules were also added during the course of the refinement. The data-collection and refinement statistics are summarized in Table 1 ▶. The final model and structure-factor file of vcUNG have been deposited in the Protein Data Bank under accession number 2jhq.
Table 1. Data-collection and refinement summary.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Resolution range (Å) | 12–1.50 (1.58–1.50) |
| No. of unique reflections | 36324 |
| Redundancy | 3.9 (3.9) |
| Rmerge† (%) | 7.6 (47.8) |
| Completeness (%) | 99.7 (100.0) |
| Mean I/σ(I) | 12.5 (2.9) |
| Wilson B factor (Å2) | 11.8 |
| Refinement statistics | |
| R value (%) | 17.5 |
| Free R value (%) | 21.0 |
| Deviations from ideal geometry | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.329 |
| ESU‡ (Å) | 0.052 |
| Average B values (Å2) | |
| Main-chain atoms | 9.9 |
| Side-chain atoms | 11.5 |
| Protein atoms | 10.7 |
| Chloride (1) | 11.9 |
| Water molecules (327) | 23.3 |
| All atoms | 12.7 |
| Ramachandran plot (%) | |
| Most favoured | 92.0 |
| Additionally allowed | 7.4 |
| Generously allowed | 0.5 |
R
merge = 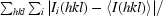
 , where Ii(hkl) is the ith measurement of reflection hkl and 〈I(hkl)〉 is the weighted mean of all measurements of reflection hkl.
, where Ii(hkl) is the ith measurement of reflection hkl and 〈I(hkl)〉 is the weighted mean of all measurements of reflection hkl.
Estimated overall coordinate error from REFMAC5 based on maximum likelihood.
2.4. Model building of UNG from A. salmonicida
A sequence alignment was created using the ESPript web server (Gouet et al., 1999 ▶). A homology model of asUNG was made by aligning the sequence of asUNG onto the crystal structure of vcUNG using the tools available in Swiss-PdbViewer (Guex & Peitsch, 1997 ▶). Side-chain geometries were manually adjusted (in order to optimize the ion-pair and hydrogen-bond interactions with neighbouring amino-acid residues) using the molecular-graphics program O (Jones et al., 1991 ▶). Visualization of the mutations considered was performed with the same program. Molecular surfaces were made using PyMOL (DeLano, 2002 ▶) using an electrostatic surface potential imported from DelPhi (Rocchia et al., 2002 ▶)
2.5. Construction of mutants
All mutants were constructed using the QuikChange site-directed mutagenesis kit following the manufacturer’s instructions (Stratagene). The primers used for the vcUNGV90R mutant were the forward primer FPvcUNGV90R (5′-GCCCTCGTTAAGAAATATTTATAAAGAGT-3′) and the reverse primer RPvcUNGV90R (5′-ACTCTTTATAAATATTTCTTAACGAGGGC-3′). For the asUNGR88V mutant the primers were the forward primer FPasUNGR88V (5′-GATCCCGCCTTCTCTGGTGAATATGTATAAAG-3′) and the reverse primer RPasUNGR88V (5′-CTTTATACATATTCACCAGAGAAGGCGGGATC-3′).
For the vcUNGH194R mutant the primers were the forward primer FPvcUNGH194R (5′-GCTTTCGGCGCGCCGCGGCTTTTTAGGATG-3′) and the reverse primer RPvcUNGH194R (5′-CATCCTAAAAAGCCGCGGCGCGCCGAAAGC-3′). The primers used for the asUNGR192H mutant were the forward primer FPasUNGR192H (5′-CTTTATCGGCACATCGCGGTTTTTTGGTTTG-3′) and the reverse primer RPasUNGR192H (5′-CAACCAAAAAAACCGCGATGTGCCGATAAAG-3′). The mutations were confirmed by DNA sequencing. The vcUNG mutants were expressed and purified as described for vcUNG and the asUNG mutants were expressed and purified as described previously (Raeder et al., 2008 ▶), but with a large-scale expression volume of 10 l in a Techfors S fermenter (Infors, Bottmingen, Switzerland).
2.6. Activity measurements and kinetics
Preparation of the substrate ([3H]-dUMP DNA made by nick translation) and measurements of UNG activity was performed as described previously (Lanes et al., 2000 ▶). One unit is defined as the amount of enzyme required to release 1 nmol of acid-soluble uracil per minute at 310 K. K m and k cat were measured in the presence of eight different [3H]-dUMP substrate concentrations in the range 0.56–4.5 µM at 310 K. The amounts of UNG used were different for the two mutants, giving values of between 500 and 5000 counts min−1. Assays were performed in 25 mM Tris–HCl pH 7.5 at 310 K, 25 mM NaCl, 1 mM EDTA and 100 µg ml−1 BSA. Each reaction was run in triplicate. Calculation of the kinetic constants was performed using the enzyme-kinetics module of the SigmaPlot software (Systat Software, San Jose, California, USA).
3. Results and discussion
3.1. Overall structure
The crystal structure of vcUNG was solved to 1.5 Å resolution by molecular replacement using the crystal structure of ecUNG, which has a sequence identity of 69.9%, as a search model. The overall dimensions of the protein were approximately 35 × 38 × 48 Å. The overall topology of vcUNG is that of a typical α/β protein, as described previously for hUNG (Slupphaug et al., 1996 ▶). The central part of the structure is a four-stranded parallel β-sheet with strand order 2-1-3-4 surrounded by a total of eight α-helices (Figs. 1 ▶ and 2 ▶). The two termini are situated on opposite sides of the β-sheet. Except for the three N-terminal amino-acid residues, which are flexible, the rest of the protein is well defined in electron density. The overall structure of vcUNG is similar to that of ecUNG, with a root-mean-square (r.m.s.) deviation of 0.57 Å for the 222 Cα atoms that could be structurally aligned.
Figure 1.
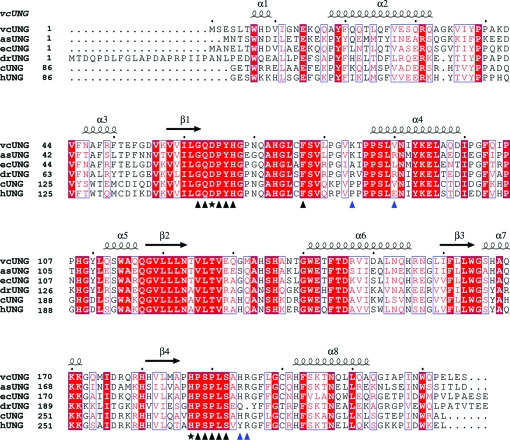
Sequence alignment of the sequences of V. cholerae, A. salmonicida, E. coli and D. radiodurans UNGs and the catalytic domains of cod and human UNGs. The secondary-structure elements of vcUNG are indicated above the alignment. α-Helices are indicated as spirals and β-strands as arrows. Identical residues are indicated as white letters on a red background. The residues marked with black triangles below the alignment are important for substrate binding and catalysis, while the residues marked with stars are directly involved in catalysis. Residues that are proposed to be important for substrate affinity are indicated by blue triangles below the alignment. 85 N-terminal residues of the regulatory domain of cUNG and hUNG were not included in the alignment; the numbering starts from amino acid 86 for these.
Figure 2.
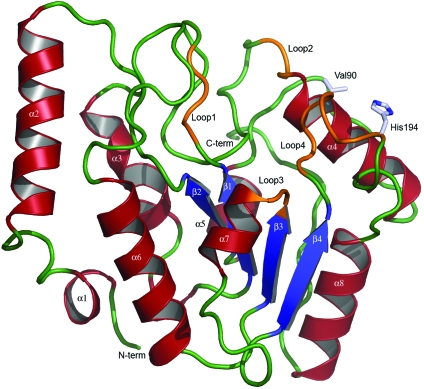
Ribbon illustration of the crystal structure of vcUNG. α-Helices are illustrated in red, β-strands in blue and loop regions in orange. Loops involved in the detection and removal of uracil in DNA are marked. Loop1 is the water-activating loop (64-DPYH-67), Loop2 is the Pro-rich loop (84-KTPPS-88), Loop3 is the Gly-Ser loop (165-GS-166) and Loop4 is the Leu loop (187-HPSPLSAH-194). The sites of the mutations Val90 and His194 are also shown.
3.2. Comparsion of vcUNG and asUNG
3.2.1. Sequence
As a first approach to identifying the amino-acid residues that are responsible for the lower substrate affinity (higher K m) and higher stability of vcUNG compared with asUNG, their sequences were aligned and compared (Fig. 1 ▶). The sequences of ecUNG, drUNG, cUNG and hUNG were also included in the alignment. These represent both psychrophilic and mesophilic UNGs and both cUNG and drUNG demonstrated stronger substrate-binding affinity compared with hUNG (Moe et al., 2004 ▶; Leiros et al., 2005 ▶). Comparison of the sequences of vcUNG and asUNG shows that there are 88 amino-acid substitutions in vcUNG compared with asUNG. Residues that differ between the two enzymes and that are unique to asUNG are of particular interest because they might be involved in the cold-adaptation of asUNG. The sequence identity between vcUNG (226 amino acids) and asUNG (224 amino acids) is 60%, while the similarity is 76%. The most striking difference in the amino-acid composition of vcUNG and asUNG is the low number of strictly charged residues (Arg, Lys, Asp and Glu) in vcUNG (34) compared with asUNG (49). The distribution of positively and negatively charged residues is 17 positive and 17 negative in vcUNG and 20 positive and 29 negative in asUNG. The difference in negative charges is mainly caused by eight specific substitutions from Gln to Glu in asUNG. In addition, the differences can be observed as an increased content of arginine (eight versus five) and proline (15 versus 12) residues, a decreased content of serine (12 versus 17) residues and a higher Arg/(Arg+Lys) ratio (0.47 versus 0.25) when comparing vcUNG with asUNG. These are all amino-acid differences that have been reported for several cold-adapted enzymes when compared with their mesophilic homologues (D’Amico et al., 2002 ▶; Russell, 2000 ▶; Smalås et al., 2000 ▶).
3.2.2. Electrostatics
The observed increase in the substrate-binding affinity of asUNG compared with vcUNG could be caused in part by increased positive charge close to the active site of the enzyme (Raeder et al., 2008 ▶). This part of the enzyme is mainly involved in interactions with the negatively charged DNA substrate.
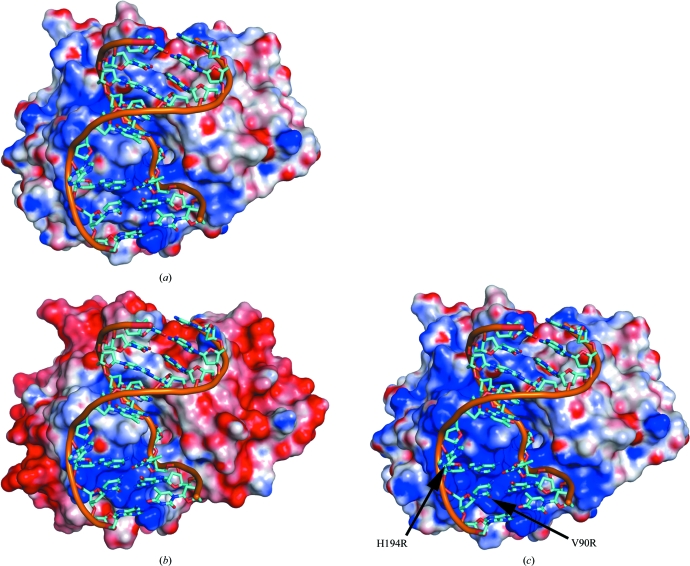
The difference in the distribution of charged residues in the two enzymes was investigated by visualization of the surface potentials of the proteins using PyMOL (DeLano, 2002 ▶). Although in an overall view vcUNG (Fig. 3 ▶ a) clearly has a more positive surface potential than asUNG (Fig. 3 ▶ b), asUNG appears to possess a more positive potential close to the active site. In particular, the charge of the residue in position 90 seems to be important in this regard. When comparing the residues corresponding to position 90 in the various aligned enzymes (Fig. 1 ▶), there seems to be a tendency for this residue to change from having negative charge in some species to being neutral in others and to having a positive charge in a few. As examples, the negatively charged Glu is observed at this position in mesophilic mammalian UNGs such as hUNG. Neutral amino acids are observed in several bacterial UNGs such as ecUNG (Leu), Mycobacterium tuberculosis UNG (Ala), vcUNG (Val) and in the cold-adapted cUNG (Val). Positive charges are observed as an Arg residue as in the UNGs from various cold-adapted Vibrio species (unpublished results) and in drUNG, HSV-1 UNG and Epstein–Barr virus UNG (Géoui et al., 2007 ▶) or as a Lys residue as observed in some other bacterial UNGs and vaccinia virus UNG. This tendency seems to correlate with the increasing substrate affinities observed for the species characterized within these categories (see below). A similar tendency is observed for the residue at position 84 (Fig. 1 ▶), which is the first residue of the 4-Pro loop; most species possess a neutral amino acid at this position, while some possess positively charged amino acids as in V. cholerae, A. salmonicida (Lys) and M. tuberculosis (Arg) (Kaushal et al., 2008 ▶) and some possess a negative charge in this position (e.g vaccinia virus UNG).
Figure 3.
Estimated electrostatic surface potentials of the crystal structure of vcUNG (a), the homology model of asUNG (b) and the modelled double mutant V90R/H194R of vcUNG (c). The dsDNA from the crystal structure of the complex of hUDG with DNA (PDB code 1emh; Parikh et al., 2000 ▶) has been modelled into all surfaces in order to illustrate the protein–DNA interaction area. The surface potentials are contoured at ±5kT/e, where red describes a negative potential and blue a positive potential. The figure was made by PyMOL (DeLano, 2002 ▶) using an electrostatic surface potential imported from DelPhi (Rocchia et al., 2002 ▶).
An observed increase in the substrate-binding affinity of drUNG compared with hUNG was suggested to reflect the positive charges in the substrate-binding site, including the residues in these two positions (Leiros et al., 2005 ▶). The cold-adapted cUNG also demonstrates increased substrate-binding affinity when compared with hUNG (Moe et al., 2004 ▶), caused by a substitution (Val171Glu) in a position that is structurally conserved as position 90 in vcUNG. The charge of this residue was demonstrated by structural and mutational analysis to have a pronounced effect on the binding affinity and was proposed to explain the cold-adaptation of cUNG.
To investigate the possible effects arising from the charge of the residue at position 90 (Fig. 1 ▶) of asUNG and vcUNG, mutational analysis was performed. Based on these assumptions, two mutants were constructed: one of asUNG (asUNGR88V) and one of the corresponding residue in vcUNG (vcUNGV90R). Only the vcUNG mutant was expressed in amounts that gave sufficient pure protein for successful characterization experiments. The kinetic constants of the vcUNG mutant vcUNGV90R (and also another vcUNG mutant; see below) were determined at 310 K. The residues were substituted by the corresponding asUNG residues and the results were compared with the data obtained previously for vcUNG and asUNG (Table 2 ▶). The kinetic studies of vcUNGV90R demonstrated an increased substrate affinity that more resembled the features of asUNG, further indicating the importance of this residue in substrate binding and reflecting the change from a neutral to a positive charge.
Table 2. Kinetic constants determined at 310 K for asUNG, vcUNG (Raeder et al., 2008 ▶) and the vcUNG mutants vcV90R and vcH194R.
| T (K) | Vmax (U mg−1) | kcat (min−1) | Km (µM) | kcat/Km (min−1 µM−1) | |
|---|---|---|---|---|---|
| asUNG | 310 | 6872 ± 352 | 174 | 0.40 ± 0.09 | 471 |
| vcUNG | 310 | 11590 ± 1728 | 293 | 2.80 ± 0.78 | 105 |
| vcV90R | 310 | 6077 ± 791 | 154 | 1.10 ± 0.35 | 141 |
| vcH194R | 310 | 32347 ± 5478 | 821 | 1.80 ± 0.57 | 454 |
The characterized enzymes possessing positive or neutral charges at both positions 84 and 90 (Fig. 1 ▶) showed the highest affinity for substrate (lowest K m values; Table 3 ▶), whereas those with neutral/negative or negative charges at these positions showed lower substrate-binding affinity. The charge at position 90 (Fig. 1 ▶) seems to be of particular importance in this regard, as confirmed by mutational analysis of the corresponding residues in both cUNG and hUNG (Moe et al., 2004 ▶) and in vcUNG.
Table 3. Charges of positions 84 and 90 (or residues corresponding to these positions) and observed substrate affinities (K m values; see also Fig. 1 ▶).
The Leu272 loop is another catalytically important area in UNG. Upon substrate binding, the catalytically important His268 in hUNG is brought to within hydrogen-bonding distance of uracil O2 by movement of this loop and a uracil-recognition pocket is formed as a consequence (Parikh et al., 1998 ▶). Residues in this area have been demonstrated to be important for uracil detection and substrate binding (Parikh et al., 1998 ▶). Comparison of this area of vcUNG and asUNG shows that asUNG possesses a positively charged Arg at position 194 (corresponding to Tyr275 in hUNG), whereas vcUNG possesses a His (see also Fig. 1 ▶). An Arg at this position is apparently unique to asUNG.
To investigate the effect of arginine at position 194, mutational analysis was performed. One asUNG mutant (asUNGR192H) and one mutation of the corresponding residue in vcUNG (vcUNGH194R) were again constructed in this case, but again only the vcUNG mutant could be successfully used in characterization experiments owing to low yields of the asUNG mutant. The kinetic constants obtained are given in Table 2 ▶. An increased substrate affinity that more resembles the features of asUNG indicates that this residue is also important in substrate binding. However, the effect on k cat differed for the two vcUNG mutants characterized. The mutant vcUNGV90R displayed a reduced k cat compared with vcUNG, while the mutant vcUNGH194R displayed an increased k cat value.
To further evaluate whether the increased positive surface potential at the active site is linked to the V90R and H194R substitutions, a double mutant of vcUNG with these substitutions was modelled and compared with the surface potentials obtained for vcUNG and asUNG (Fig. 3 ▶ c). The resulting surface potential around the substrate-binding site more closely resembles that seen for asUNG, thus rationalizing the results obtained from biochemical characterization of the individual mutants.
From the above, it appears that optimization of charge distributions, with the presence of residues giving rise to a more positive electrostatic potential of asUNG in the substrate-interacting regions, can explain the increased substrate affinity and enhanced catalytic efficiency of asUNG compared with vcUNG.
From Fig. 3 ▶, it can also be seen that apart from the substrate-binding region, the overall surface potential of asUNG is generally more negatively charged than that of vcUNG. These observations support previous studies (Smalås et al., 2000 ▶) proposing that the hydrophilicity of the surfaces of some cold-adapted enzymes is higher than those of their mesophilic homologues and that this is caused by an increased number of charged and polar residues (especially Asp and Glu, which have a negative potential) on the exterior of the enzymes. Indeed, a significantly higher number of Asp and Glu residues can be found in the psychrophilic asUNG (29) compared with the mesophilic vcUNG (17), which also results in a much lower theoretical pI for asUNG than for vcUNG (5.48 and 7.24, respectively). The overall highly negatively charged surface of asUNG (Fig. 3 ▶ b) might also cause a more systematic orientation of the enzyme against the substrate. An increase in surface charge and particularly a negative charge has been described for several cold-adapted enzymes (Siddiqui & Cavicchioli, 2006 ▶). This may enable proper solvation of the proteins and could be important for maintaining flexibility at lower temperatures (Siddiqui & Cavicchioli, 2006 ▶).
4. Conclusions
As for previously characterized UNGs, electrostatic interactions have been demonstrated to be important for substrate binding in vcUNG and asUNG. The electrostatic interactions are important for the increased catalytic efficiency of asUNG compared with vcUNG, indicating that the charge distribution in the substrate-binding region is a strategy for environmental adaptation of these enzymes. Mutational analyses suggested specific amino acids in the sequences to be of particular importance in this regard and mutations in vcUNG have demonstrated increased affinity for substrate, more resembling the features of asUNG. Visualization of surface potentials around the substrate-binding region of the enzymes have demonstrated a more positive potential for asUNG compared with vcUNG; for a modelled double mutant of vcUNG the potential in the substrate-binding area is more like that observed for asUNG, thus rationalizing the results obtained from the kinetic studies of the mutants.
Supplementary Material
PDB reference: uracil-DNA N-glycosylase, 2jhq
Acknowledgments
This work was supported by the Norwegian Research Council (project No. 146613/431). Financial support was also received from The University of Tromsø, Norway. The Norwegian Structural Biology Centre (NorStruct) is funded by the National Programme for Research in Functional Genomics in Norway (FUGE). We also gratefully acknowledge Gry Evjen and Stefan Hauglid at the Norwegian Structural Biology Centre (NorStruct), Department of Chemistry, University of Tromsø, Norway for technical assistance. Provision of synchrotron beamtime at BESSY is gratefully acknowledged.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- D’Amico, S., Claverie, P., Collins, T., Georlette, D., Gratia, E., Hoyoux, A., Meuwis, M. A., Feller, G. & Gerday, C. (2002). Philos. Trans. R. Soc. Lond. B Biol. Sci.357, 917–925. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL Molecular Viewer. DeLano Scientific, San Carlos, California, USA. http://www.pymol.org.
- Dinner, A. R., Blackburn, G. M. & Karplus, M. (2001). Nature (London), 413, 752–755. [DOI] [PubMed]
- Géoui, T., Buisson, M., Tarbouriech, N. & Burmeister, W. (2007). J. Mol. Biol.366, 117–131. [DOI] [PubMed]
- Gorfe, A. A., Brandsdal, B. O., Leiros, H. K., Helland, R. & Smalås, A. O. (2000). Proteins, 40, 207–217. [PubMed]
- Gouet, P., Courcelle, E., Stuart, D. I. & Métoz, F. (1999). Bioinformatics, 15, 305–308. [DOI] [PubMed]
- Guex, N. & Peitsch, M. C. (1997). Electrophoresis, 18, 2714–2723. [DOI] [PubMed]
- Hochachka, P. W. & Somero, G. N. (1984). Biochemical Adaptation. Princeton, USA: Princeton University Press.
- Jiang, Y. L., Drohat, A. C., Ichikawa, Y. & Stivers, J. T. (2002). J. Biol. Chem.277, 15385–15392. [DOI] [PubMed]
- Jiang, Y. L., Ichikawa, Y., Song, F. & Stivers, J. T. (2003). Biochemistry, 42, 1922–1929. [DOI] [PubMed]
- Jiang, Y. L., Ichikawa, Y. & Stivers, J. T. (2002). Biochemistry, 41, 7116–7124. [DOI] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kaushal, P. S., Talawar, R. K., Krishna, P. D. V., Varshney, U. & Vijayan, M. (2008). Acta Cryst. D64, 551–560. [DOI] [PubMed]
- Kavli, B., Sundheim, O., Akbari, M., Otterlei, M., Nilsen, H., Skorpen, F., Aas, P. A., Hagen, L., Krokan, H. E. & Slupphaug, G. (2002). J. Biol. Chem.277, 39926–39936. [DOI] [PubMed]
- Krokan, H. E., Standal, R. & Slupphaug, G. (1997). Biochem. J.325, 1–16. [DOI] [PMC free article] [PubMed]
- Lanes, O., Guddal, P. H., Gjellesvik, D. R. & Willassen, N. P. (2000). Comp. Biochem. Physiol. B, 127, 399–410. [DOI] [PubMed]
- Leiros, I., Lanes, O., Sundheim, O., Helland, R., Smalås, A. O. & Willassen, N. P. (2001). Acta Cryst. D57, 1706–1708. [DOI] [PubMed]
- Leiros, I., Moe, E., Lanes, O., Smalås, A. O. & Willassen, N. P. (2003). Acta Cryst. D59, 1357–1365. [DOI] [PubMed]
- Leiros, I., Moe, E., Smalås, A. O. & McSweeney, S. (2005). Acta Cryst. D61, 1049–1056. [DOI] [PubMed]
- Lindahl, T. (1974). Proc. Natl Acad. Sci. USA, 71, 3649–3653. [DOI] [PMC free article] [PubMed]
- Moe, E., Leiros, I., Riise, E. K., Olufsen, M., Lanes, O., Smalås, A. O. & Willassen, N. P. (2004). J. Mol. Biol.343, 1221–1230. [DOI] [PubMed]
- Mol, C. D., Arvai, A. S., Slupphaug, G., Kavli, B., Alseth, I., Krokan, H. E. & Tainer, J. A. (1995). Cell, 80, 869–878. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999). Acta Cryst. D55, 247–255. [DOI] [PubMed]
- Olufsen, M., Smalås, A. O., Moe, E. & Brandsdal, B. O. (2005). J. Biol. Chem.280, 18042–18048. [DOI] [PubMed]
- Parikh, S. S., Mol, C. D., Hosfield, D. J. & Tainer, J. A. (1999). Curr. Opin. Struct. Biol.9, 37–47. [DOI] [PubMed]
- Parikh, S. S., Mol, C. D., Slupphaug, G., Bharati, S., Krokan, H. E. & Tainer, J. A. (1998). EMBO J.17, 5214–5226. [DOI] [PMC free article] [PubMed]
- Parikh, S. S., Walcher, G., Jones, G. D., Slupphaug, G., Krokan, H. E., Blackburn, G. M. & Tainer, J. A. (2000). Proc. Natl Acad. Sci. USA, 97, 5083–5088. [DOI] [PMC free article] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed]
- Raeder, I. L. U., Leiros, I., Willassen, N. P., Smalås, A. O. & Moe, E. (2008). Enzyme Microb. Technol.42, 594–600.
- Ravishankar, R., Bidya Sagar, M., Roy, S., Purnapatre, K., Handa, P., Varshney, U. & Vijayan, M. (1998). Nucleic Acids Res.26, 4880–4887. [DOI] [PMC free article] [PubMed]
- Rocchia, W., Sridharan, S., Nicholls, A., Alexov, E., Chiabrera, A. & Honig, B. (2002). J. Comput. Chem.23, 128–137. [DOI] [PubMed]
- Russell, N. J. (2000). Extremophiles, 4, 83–90. [DOI] [PubMed]
- Savva, R., McAuley-Hecht, K., Brown, T. & Pearl, L. (1995). Nature (London), 373, 487–493. [DOI] [PubMed]
- Sheridan, P. P., Panasik, N., Coombs, J. M. & Brenchley, J. E. (2000). Biochim. Biophys. Acta, 1543, 417–433. [DOI] [PubMed]
- Siddiqui, K. S. & Cavicchioli, R. (2006). Annu. Rev. Biochem.75, 403–433. [DOI] [PubMed]
- Slupphaug, G., Eftedal, I., Kavli, B., Bharati, S., Helle, N. M., Haug, T., Levine, D. W. & Krokan, H. E. (1995). Biochemistry, 34, 128–138. [DOI] [PubMed]
- Slupphaug, G., Mol, C. D., Kavli, B., Arvai, A. S., Krokan, H. E. & Tainer, J. A. (1996). Nature (London), 384, 87–92. [DOI] [PubMed]
- Smalås, A. O., Leiros, H. K., Os, V. & Willassen, N. P. (2000). Biotechnol. Annu. Rev.6, 1–57. [DOI] [PubMed]
- Werner, R. M. & Stivers, J. T. (2000). Biochemistry, 39, 14054–14064. [DOI] [PubMed]
- Xiao, G., Tordova, M., Jagadeesh, J., Drohat, A. C., Stivers, J. T. & Gilliland, G. L. (1999). Proteins, 35, 13–24. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: uracil-DNA N-glycosylase, 2jhq
PDB reference: uracil-DNA N-glycosylase, 2jhq