Recombinant glycine decarboxylase from Synechocystis sp. PCC 6803 was expressed in E. coli and purified to homogeneity. Crystals were obtained that diffracted to 2.1 Å resolution using synchrotron radiation.
Keywords: glycine decarboxylases, P-proteins
Abstract
Glycine decarboxylase, or P-protein, is a major enzyme that is involved in the C1 metabolism of all organisms and in the photorespiratory pathway of plants and cyanobacteria. The protein from Synechocystis sp. PCC 6803 is a homodimer with a mass of 215 kDa. Recombinant glycine decarboxylase was expressed in Escherichia coli and purified by metal-affinity, ion-exchange and gel-filtration chromatography. Crystals of P-protein that diffracted to a resolution of 2.1 Å were obtained using the hanging-drop vapour-diffusion method at 291 K. X-ray diffraction data were collected from cryocooled crystals using synchrotron radiation. The crystals belonged to space group P212121, with unit-cell parameters a = 96.30, b = 135.81, c = 179.08 Å.
1. Introduction
Glycine decarboxylase (EC 1.4.4.2), or P-protein, is part of the glycine-cleavage system (GCS), which is widely distributed in bacteria, plants and animals. The GCS cooperates with serine hydroxymethyltransferase in the reversible glycine–serine interconversion (Douce et al., 2001 ▶) and catalyses the oxidative cleavage of glycine to CO2, NH4 + and a methylene group (–CH2–) in a multistep reaction. The methylene group is accepted by tetrahydrofolate to yield methylene tetrahydrofolate, a major cofactor in one-carbon metabolism. Hence, it is not surprising that defects in the GCS result in serious metabolic disturbances in eukaryotes. In humans, absence of glycine decarboxylase activity results in an enhanced accumulation of glycine in the blood, leading to nonketotic hyperglycinaemia. This disease is often caused by mutations in the P-protein subunit (Kume et al., 1988 ▶; Kure et al., 1998 ▶). The GCS is also essential in the plant photorespiratory C2 cycle, which salvages 2-phosphoglycolate resulting from the oxygenation reaction catalysed by ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) under atmospheric conditions. In Arabidopsis thaliana lack of P-protein also causes lethality under nonphotorespiratory conditions, underlining the fact that GSC activity is also essential for plant C1 metabolism (Engel et al., 2007 ▶). Cyanobacteria possess a carbon-concentrating mechanism that reduces but does not abolish photorespiration (Eisenhut et al., 2008 ▶). In Synechocystis sp. PCC 6803 GCS deletion leads to glycine accumulation. However, because of its greater metabolic flexibility this organism can survive without a GCS (Hagemann et al., 2005 ▶; Eisenhut et al., 2006 ▶).
The complete GCS reaction cycle requires the cooperation of three different enzymes (P-protein, T-protein and L-protein) and the small heat-stable H-protein. The latter acts as an electron acceptor and an aminomethyl carrier and functions as a mobile substrate of P-protein, T-protein and L-protein. P-protein is the actual glycine-decarboxylating enzyme and uses pyridoxal 5′-phosphate (PLP) as a cofactor; CO2 is released in the reaction and the residual aminomethyl group is bound to the oxidized lipoamide arm of H-protein. Next, T-protein or aminomethyltransferase (EC 2.1.2.10) catalyzes the transfer of the methylene group from the H-protein-bound aminomethyl moiety to tetrahydrofolate. T-protein releases NH3 and regenerates the fully reduced H-protein. In a final step, H-protein is reoxidized by L-protein or dihydrolipoamide dehydrogenase (EC 1.8.1.4). This last step is accompanied by the formation of NADH.
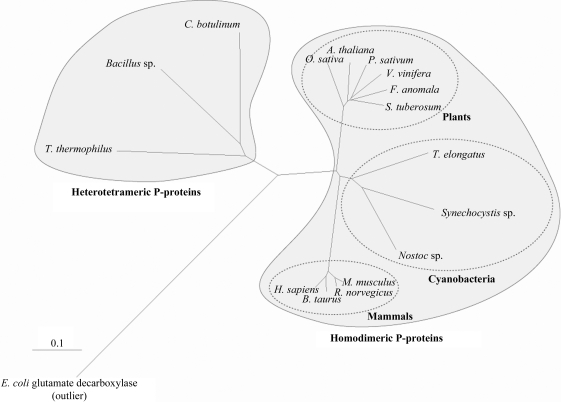
P-protein is a PLP-dependent enzyme with a molecular mass of about 215 kDa. Depending on their subunit composition, P-proteins can be classified into two types (Fig. 1 ▶). The majority of P-proteins, including those from humans (Kume et al., 1991 ▶), pea (Turner et al., 1992 ▶), Escherichia coli (Okamura-Ikeda et al., 1993 ▶) and Synechocystis (Hasse et al., 2007 ▶), are α2 homodimers. The P-proteins from Eubacterium acidaminophilum (Freudenberg & Andreesen, 1989 ▶) and Thermus thermophilus (Nakai et al., 2005 ▶) are heterotetrameric (αNβC)2, in which the single subunits of the homodimeric P-proteins are split into α and β subunits. The three-dimensional structure of the heterotetrameric P-protein from T. thermophilus (Nakai et al., 2005 ▶) is known, whereas structures of P-proteins of the homodimeric type are not yet available.
Figure 1.
Rooted tree of glycine decarboxylase proteins constructed from a ClustalX (Larkin et al., 2007 ▶) alignment of homodimeric and heterotetrameric glycine decarboxylase enzymes. The phylogenetic tree was constructed with the program TreeView (Page, 1996 ▶). To enable the alignment of enzymes with different oligomeric compositions, subunit 2 of the heterotetrameric P-proteins was fused to the carboxy-terminus of subunit 1. The tree shows that the Synechocystis P-protein clusters more closely with eukaryotic P-proteins than any of the heterotetrameric enzymes, including the T. thermophilus enzyme. E. coli glutamate decarboxylase (NP_287662) was defined as an outlier. P-protein sequences from the following eukaryotic species and bacterial isolates were used: Synechocystis sp. PCC 6803 (P74416), Thermosynechococcus elongatus Bp-1 (NP_682393), Nostoc sp. PCC 7120 (NP_488647), Oryza sativa (AAQ24377), Arabidopsis thaliana (BAE98954), Pisum sativum (P26969), Solanum tuberosum (O49954), Flaveria anomala (O49850), Vitis vinifera (XP_002279590), Mus musculus (NP_613061), Bos taurus (XP_869239), Homo sapiens (NP_000161), Rattus norvegicus (NP_001101053), Thermus thermophilus HB-8 (subunit 1, YP_143791; subunit 2, YP_143792), Bacillus sp. B14905 (subunit 1, ZP_01725792; subunit 2, ZP_01725793), Clostridium botulinum A strain ATCC 3502 (subunit 1, YP_001253239; subunit 2, YP_001253240). The scale bar shows the distance equal to 0.1 amino-acid substitutions per sequence position.
As the ancestors of plant chloroplasts, cyanobacteria have delivered many genes to plant genomes (Martin et al., 2002 ▶). In contrast to that of T. thermophilus, cyanobacterial P-proteins have a homodimeric structure and are thus more closely related to eukaryotic homologues (Fig. 1 ▶). A three-dimensional structure of a homodimeric enzymatically active glycine decarboxylase would therefore be important and could be used to model the human P-protein. However, it would also provide insight into the reaction mechanism of P-proteins and the structural architecture of the enzyme complexes. Recently, we presented the biochemical characterization of the enzymatically active recombinant P-protein from Synechocystis sp. PCC 6803 (Hasse et al., 2007 ▶). Here, we report the crystallization and preliminary X-ray analyses of the Synechocystis P-protein.
2. Materials and methods
2.1. Protein expression
The P-protein gene slr0293 from Synechocystis sp. PCC 6803 was cloned as described elsewhere (Hasse et al., 2007 ▶). For gene expression, cells of Escherichia coli strain LMG194 harbouring plasmid pBAD-slr0293 were cultivated in 2YT medium at 310 K to an optical density (OD600) of 0.6. After cooling the culture to 291 K, gene expression was induced by adding 0.02% arabinose. Cells were harvested after a further 16 h of incubation at 291 K.
2.2. Protein purification
Unless stated otherwise, all purification steps were performed at 277 K and within 1 d. After gene expression, cells were harvested by centrifugation and suspended in homogenization buffer (20 mM sodium phosphate pH 7.8, 500 mM sodium chloride, 40 mM imidazole and 15 mM 2-mercaptoethanol). After cell disruption by sonication, the lysate was centrifuged (Sorvall RC6, 20 000 rev min−1, SS34 rotor) for 40 min at 277 K. Metal-affinity chromatography was carried out using Ni2+-Sepharose (GE Healthcare, Sweden) and Poly-Prep gravity-flow columns (Bio-Rad). The protein was washed using two column volumes (CVs) of washing buffer (20 mM sodium phosphate pH 7.8, 500 mM sodium chloride, 80 mM imidazole and 15 mM 2-mercaptoethanol) and eluted with 3 ml elution buffer (20 mM sodium phosphate pH 7.8, 500 mM sodium chloride, 500 mM imidazole and 15 mM 2-mercaptoethanol). Before loading onto a MonoQ column (8 ml; GE Healthcare, Sweden) equilibrated in buffer A (20 mM Tris–HCl pH 7.8, 50 mM sodium chloride, 5 mM 2-mercaptoethanol), fractions were pooled and rebuffered in buffer A using PD-10 desalting columns (GE Healthcare, Sweden). The protein was eluted with a linear gradient of 50–500 mM sodium chloride in 20 CVs. Fractions containing the target protein were pooled, concentrated (Vivaspin 20, GE Healthcare, Sweden) and loaded onto a HiLoad 16/60 Superdex 200 prep-grade column (GE Healthcare, Sweden) equilibrated with buffer B (20 mM Tris–HCl pH 7.8, 50 mM sodium chloride, 10 mM 2-mercaptoethanol) and eluted with the same buffer. The peak fractions were collected in a buffer containing 10 mM 2-mercaptoethanol, concentrated and stored at 277 K. After each step, fractions were analysed by SDS–PAGE using the PhastSystem (GE Healthcare, Sweden). The kinetic constants of the purified protein were measured according to Hasse et al. (2007 ▶) with slight modifications (Hasse et al., 2009 ▶).
2.3. Crystallization of Synechocystis P-protein
Initial crystallization screening of native P-protein was performed by the sitting-drop vapour-diffusion method using commercially available kits. Microcrystals were obtained using PEG/Ion Screen HT (Hampton Research, USA) at a protein concentration of 20 mg ml−1 and a temperature of 293 K. The initial drop size was 2 µl, comprising 1 µl each of the protein and reservoir solutions. The first three-dimensional crystals were obtained at 100 mM Tris–HCl pH 8, 15% PEG 3350, 0.15 M CsCl and 10 mM 2-mercaptoethanol using the hanging-drop vapour-diffusion method. The size and quality of the crystals could be improved by tweaking the crystallization conditions (the final condition was 100 mM Tris–HCl pH 7.75, 15–25% PEG 3350, 0.15–0.3 M CsCl or LiCl and 10 mM 2-mercaptoethanol) and by streak-seeding. A further increase in crystal size was achieved by increasing the drop size to 4 µl (containing equal volumes of protein at 40 mg ml−1 in buffer B and reservoir solution) and placing each droplet over 1 ml reservoir solution. Prism-shaped crystals (50 × 25 × 100 µm) were obtained 1–3 d after streak-seeding at 293 K.
2.4. Data collection
X-ray diffraction data were collected using a CCD detector (ADSC Q315r) on beamline ID14-4 at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) at a wavelength of 0.98 Å. The crystal was soaked for a few seconds in reservoir solution containing 20%(v/v) ethylene glycol as a cryoprotectant, transferred to a nylon CryoLoop (Hampton Research, USA) and then flash-cooled in liquid nitrogen and maintained at 100 K for data collection. The crystal-to-detector distance was 289.4 mm. The crystal diffracted to 2.1 Å resolution. 360 frames were collected with 0.5° oscillation per frame. The exposure time per frame was 0.5 s. The data were processed with DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
To enable the production of crystallization-grade P-protein, a purification protocol was established that involved sequential anion-exchange (MonoQ) and gel-filtration chromatography. Large-scale production of P-protein was then carried out; the yield was about 8 mg of protein per litre of culture. The molecular mass of the overexpressed recombinant protein was consistent with the predicted mass from the cloned DNA sequence (Fig. 2 ▶). As previously reported (Hasse et al., 2007 ▶), the overexpressed Synechocystis P-protein is biologically active in vitro. The kinetic constants were determined by glycine–bicarbonate exchange. The estimated V max was 5.53 µmol mg−1 min−1. This is identical to the value determined recently (Hasse et al., 2009 ▶), but is about 70 times higher than the originally measured value for the protein obtained after Ni2+ metal-affinity chromatography (Hasse et al., 2007 ▶). The reason for this increase is not clear at present, but presumably the additional use of a reducing agent (2-mercaptoethanol) and/or the addition of PLP to the purification and storage buffers leads to higher enzyme activity. The K m (H-protein) was 1.57 µM, which is similar to the previously determined value. Ion-exchange chromatography using a MonoQ column revealed charge inhomogeneity of the purified protein, resulting in two major peaks. This could be overcome by switching from Luria–Bertani (LB) to 2YT expression medium, which resulted in the major fraction of the protein residing in one peak (not shown).
Figure 2.
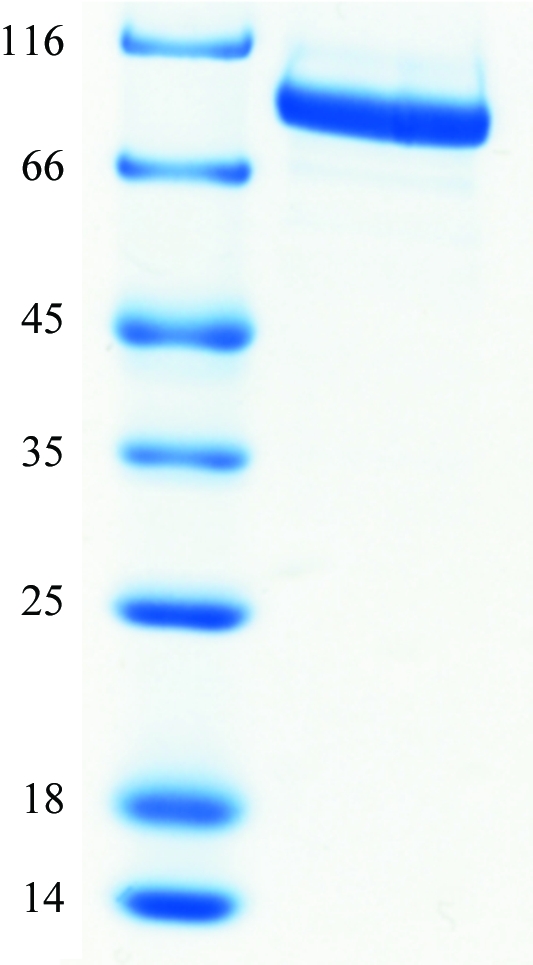
12.5% SDS–PAGE of the purified Synechocystis P-protein after metal-affinity chromatography. Left lane, molecular-mass markers (kDa); right lane, P-protein.
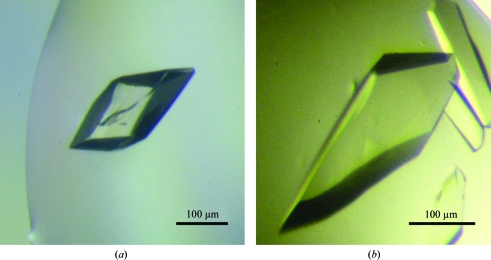
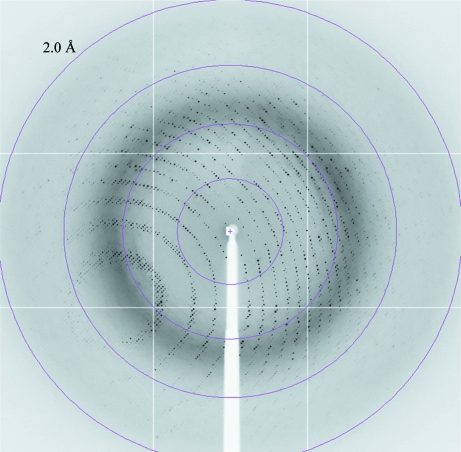
Several crystal forms were obtained in the search for crystallization conditions. The most promising condition, 15% PEG 3350, 0.15 M CsCl, was used as a starting point to produce large homogenous crystals. The same crystal form could be obtained using LiCl instead of CsCl (Fig. 3 ▶), but the crystals obtained from the caesium-containing conditions usually diffracted to higher resolution. Crystals of the native Synechocystis P-protein were obtained by streak-seeding into drops prepared under optimized conditions. This procedure gave prism-shaped crystals (Fig. 3 ▶) that diffracted to 2.1 Å resolution (Fig. 4 ▶; for further details, see §2).
Figure 3.
Crystals of recombinant P-protein. The crystal in (a) was grown in 100 mM Tris–HCl pH 7.75, 0.25 M CsCl, 17% PEG 3350 and 10 mM 2-mercaptoethanol. The crystal in (b) was grown in 100 mM Tris–HCl pH 7.75, 0.25 M LiCl, 17% PEG 3350 and 10 mM 2-mercaptoethanol. The diffraction data were collected from a crystal grown using conditions similar those used to obtain the crystal in (a).
Figure 4.
X-ray diffraction image of the apo P-protein crystal used for data collection. The diffraction data were collected on ESRF beamline ID14-4; the oscillation range was 0.5°. The edge of the detector corresponds to a resolution of 2.0 Å.
X-ray diffraction data were collected to 2.1 Å resolution from a native P-protein crystal using synchrotron radiation. The crystal belonged to the orthorhombic system, space group P222, with unit-cell parameters a = 96.30, b = 135.81, c = 179.08 Å, α = β = γ = 90°. Based on the pattern of systematic absences and the results of initial molecular-replacement trials, the most likely space group is P212121. The data-collection statistics are shown in Table 1 ▶. Cryocooled crystals were sufficiently stable in the X-ray beam to allow the collection of a complete data set from one single crystal. Calculation of the Matthews coefficient (Matthews, 1968 ▶) predicted the presence of one homodimer per asymmetric unit, assuming a molecular mass of 215 kDa. The Synechocystis P-protein sequence shows 35% identity to that of the T. thermophilus P-protein (BLAST2). We have initiated attempts to solve the structure by molecular replacement using the T. thermophilus P-protein (PDB code 1wyt; Nakai et al., 2005 ▶) as a search model.
Table 1. Data-collection statistics.
Values in parentheses are for the outer resolution shell.
| Wavelength (Å) | 0.9795 |
| Beamline | ID14-4, ESRF |
| Resolution (Å) | 50–2.1 (2.18–2.10) |
| Space group | P222 |
| Unit-cell parameters | |
| a (Å) | 96.30 |
| b (Å) | 135.81 |
| c (Å) | 179.08 |
| α (°) | 90 |
| β (°) | 90 |
| γ (°) | 90 |
| No. of observed reflections | 967636 |
| No. of unique reflections | 136940 |
| Mosaicity (°) | 0.44 |
| Redundancy | 7.1 (7.2) |
| Completeness (%) | 99.8 (100) |
| Rmerge† | 0.079 (0.353) |
| Average I/σ(I) | 29.90 (4.49) |
R
merge = 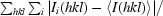
 , where 〈I(hkl)〉 is the average intensity of symmetry-equivalent reflections.
, where 〈I(hkl)〉 is the average intensity of symmetry-equivalent reflections.
Acknowledgments
The authors wish to thank the ESRF and the EMBL Outstation in Grenoble for providing beam time and data-collection facilities and the staff of ID14-4, ESRF for outstanding support. This research was financially supported by the DAAD (Deutscher Akademischer Austausch Dienst) and Landesgraduiertenstipendium Mecklenburg-Vorpommern and an excellence grant to IA from the Swedish University of Agricultural Sciences.
References
- Douce, R., Bourguignon, J., Neuburger, M. & Rebeille, F. (2001). Trends Plant Sci.6, 167–176. [DOI] [PubMed]
- Eisenhut, M., Kahlon, S., Hasse, D., Ewald, R., Lieman-Hurwitz, J., Ogawa, T., Ruth, W., Bauwe, H., Kaplan, A. & Hagemann, M. (2006). Plant Physiol.142, 333–342. [DOI] [PMC free article] [PubMed]
- Eisenhut, M., Ruth, W., Haimovich, M., Bauwe, H., Kaplan, A. & Hagemann, M. (2008). Proc. Natl Acad. Sci. USA, 105, 17199–17204. [DOI] [PMC free article] [PubMed]
- Engel, N., van den Daele, K., Kolukisaoglu, U., Morgenthal, K., Weckwerth, W., Parnik, T., Keerberg, O. & Bauwe, H. (2007). Plant Physiol.144, 1328–1335. [DOI] [PMC free article] [PubMed]
- Freudenberg, W. & Andreesen, J. R. (1989). J. Bacteriol.171, 2209–2215. [DOI] [PMC free article] [PubMed]
- Hagemann, M., Vinnemeier, J., Oberpichler, I., Boldt, R. & Bauwe, H. (2005). Plant Biol.7, 15–22. [DOI] [PubMed]
- Hasse, D., Mikkat, S., Hagemann, M. & Bauwe, H. (2009). FEBS J.276, 6985–6991. [DOI] [PubMed]
- Hasse, D., Mikkat, S., Thrun, H. A., Hagemann, M. & Bauwe, H. (2007). FEBS Lett.581, 1297–1301. [DOI] [PubMed]
- Kume, A., Koyata, H., Sakakibara, T., Ishiguro, Y., Kure, S. & Hiraga, K. (1991). J. Biol. Chem.266, 3323–3329. [PubMed]
- Kume, A., Kure, S., Tada, K. & Hiraga, K. (1988). Biochem. Biophys. Res. Commun.154, 292–297. [DOI] [PubMed]
- Kure, S., Mandel, H., Rolland, M. O., Sakata, Y., Shinka, T., Drugan, A., Boneh, A., Tada, K., Matsubara, Y. & Narisawa, K. (1998). Hum. Genet.102, 430–434. [DOI] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M. & Penny, D. (2002). Proc. Natl Acad. Sci. USA, 99, 12246–12251. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Nakai, T., Nakagawa, N., Maoka, N., Masui, R., Kuramitsu, S. & Kamiya, N. (2005). EMBO J.24, 1523–1536. [DOI] [PMC free article] [PubMed]
- Okamura-Ikeda, K., Ohmura, Y., Fujiwara, K. & Motokawa, Y. (1993). FEBS J.216, 539–548. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Page, R. D. M. (1996). Comput. Appl. Biosci.12, 357–358. [DOI] [PubMed]
- Turner, S. R., Ireland, R. & Rawsthorne, S. (1992). J. Biol. Chem.267, 5355–5360. [PubMed]