Full text
PDF

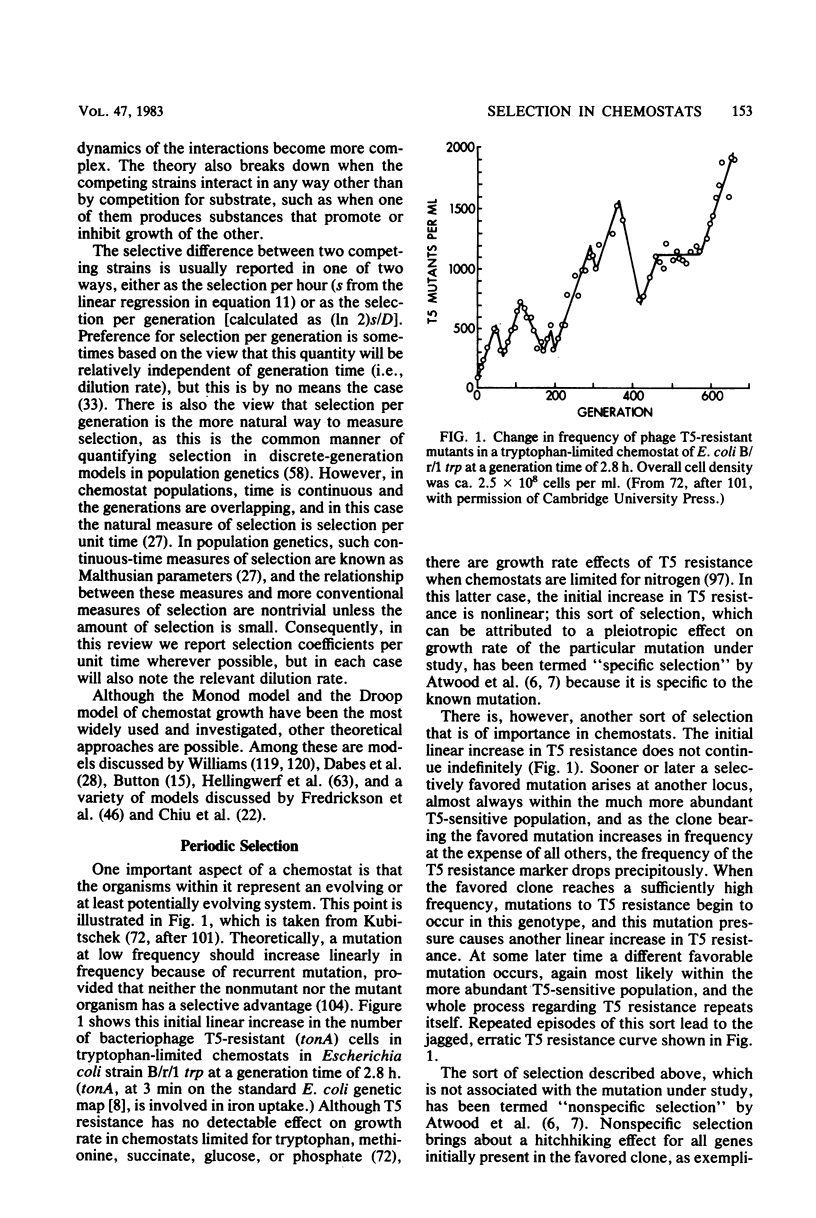


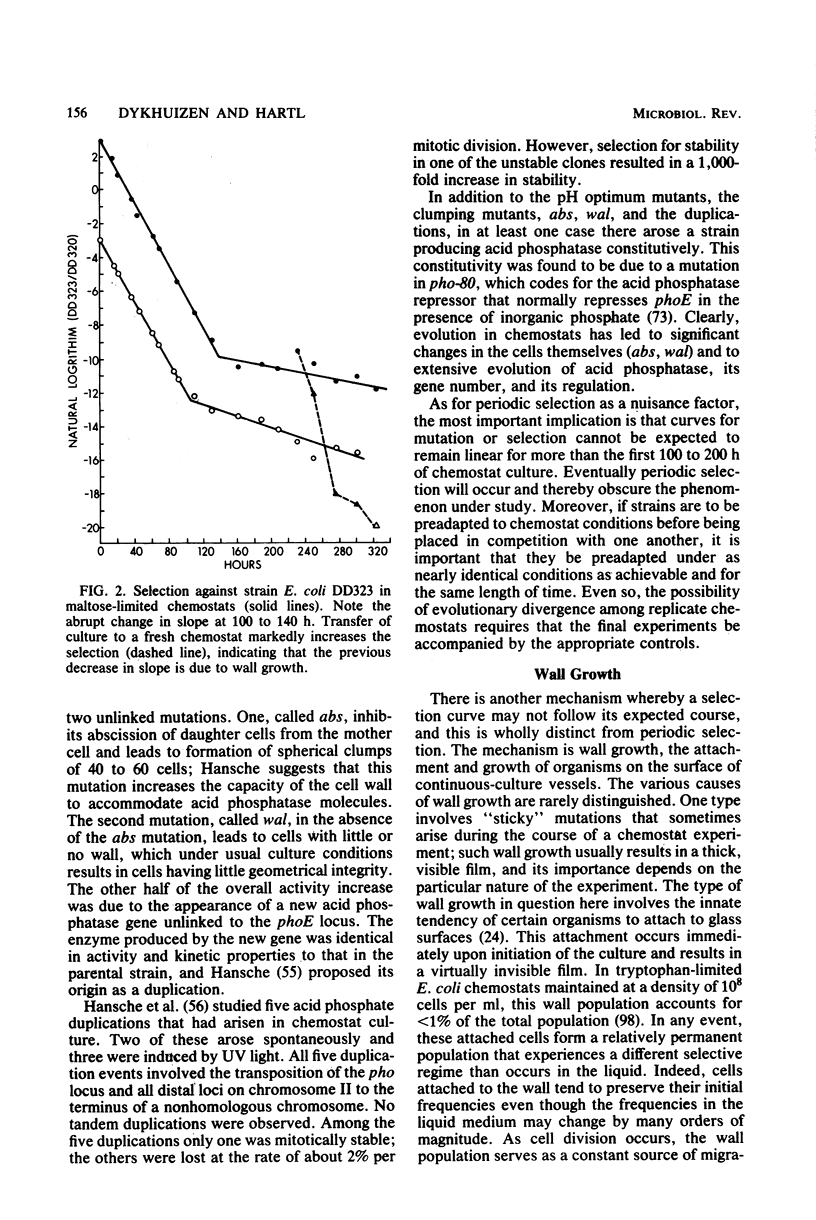












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ATWOOD K. C., SCHNEIDER L. K., RYAN F. J. Periodic selection in Escherichia coli. Proc Natl Acad Sci U S A. 1951 Mar;37(3):146–155. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ATWOOD K. C., SCHNEIDER L. K., RYAN F. J. Selective mechanisms in bacteria. Cold Spring Harb Symp Quant Biol. 1951;16:345–355. doi: 10.1101/sqb.1951.016.01.026. [DOI] [PubMed] [Google Scholar]
- Adams J., Hansche P. E. Population studies in microorganisms. I. Evolution of diploidy in Saccharomyces cerevisiae. Genetics. 1974 Feb;76(2):327–338. doi: 10.1093/genetics/76.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson T. F., Lustbader E. Inheritability of plasmids and population dynamics of cultured cells. Proc Natl Acad Sci U S A. 1975 Oct;72(10):4085–4089. doi: 10.1073/pnas.72.10.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel S. W., Hartl D. L. Evolution of transposons: natural selection for Tn5 in Escherichia coli K12. Genetics. 1983 Apr;103(4):581–592. doi: 10.1093/genetics/103.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom F. R., McFall E. Isolation and characterization of D-serine deaminase constitutive mutants by utilization of D-serine as sole carbon or nitrogen source. J Bacteriol. 1975 Mar;121(3):1078–1084. doi: 10.1128/jb.121.3.1078-1084.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONTOIS D. E. Kinetics of bacterial growth: relationship between population density and specific growth rate of continuous cultures. J Gen Microbiol. 1959 Aug;21:40–50. doi: 10.1099/00221287-21-1-40. [DOI] [PubMed] [Google Scholar]
- Campbell A. Evolutionary significance of accessory DNA elements in bacteria. Annu Rev Microbiol. 1981;35:55–83. doi: 10.1146/annurev.mi.35.100181.000415. [DOI] [PubMed] [Google Scholar]
- Chao L., Levin B. R. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. H., Jarvis A. W., Lindsay R. J., Hamilton W. A. Proton movements coupled to lactate and alanine transport in Escherichia coli: isolation of mutants with altered stoichiometry in alanine transport. J Bacteriol. 1976 Jun;126(3):1232–1244. doi: 10.1128/jb.126.3.1232-1244.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney C. L., Wang D. I. Transient response of Enterobacter aerogenes under a dual nutrient limitation in a chemostat. Biotechnol Bioeng. 1976 Feb;18(2):189–198. doi: 10.1002/bit.260180205. [DOI] [PubMed] [Google Scholar]
- Cox E. C., Gibson T. C. Selection for high mutation rates in chemostats. Genetics. 1974 Jun;77(2):169–184. doi: 10.1093/genetics/77.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabes J. N., Finn R. K., Welke C. R. Equations of substrate-limited growth: the case for blackman kinetics. Biotechnol Bioeng. 1973 Nov;15(6):1159–1177. doi: 10.1002/bit.260150613. [DOI] [PubMed] [Google Scholar]
- Dykhuizen D., Campbell J. H., Rolfe B. G. The influences of a lambda prophage on the growth rate of Escherichia coli. Microbios. 1978;23(92):99–113. [PubMed] [Google Scholar]
- Edlin G., Lin L., Kudrna R. Lambda lysogens of E. coli reproduce more rapidly than non-lysogens. Nature. 1975 Jun 26;255(5511):735–737. doi: 10.1038/255735a0. [DOI] [PubMed] [Google Scholar]
- Francis J. C., Hansche P. E. Directed Evolution of Metabolic Pathways in Microbial Populations II. a Repeatable Adaptation in SACCHAROMYCES CEREVISIAE. Genetics. 1973 Jun;74(2):259–265. doi: 10.1093/genetics/74.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R., Brickner H., Fekete J., Vickerman M. M., Carey K. E. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun. 1983 Feb;39(2):686–703. doi: 10.1128/iai.39.2.686-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R., Freter R. R., Brickner H. Experimental and mathematical models of Escherichia coli plasmid transfer in vitro and in vivo. Infect Immun. 1983 Jan;39(1):60–84. doi: 10.1128/iai.39.1.60-84.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R., Stauffer E., Cleven D., Holdeman L. V., Moore W. E. Continuous-flow cultures as in vitro models of the ecology of large intestinal flora. Infect Immun. 1983 Feb;39(2):666–675. doi: 10.1128/iai.39.2.666-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T. C., Scheppe M. L., Cox E. C. Fitness of an Escherichia coli mutator gene. Science. 1970 Aug 14;169(3946):686–688. doi: 10.1126/science.169.3946.686. [DOI] [PubMed] [Google Scholar]
- Godwin D., Slater J. H. The influence of the growth environment on the stability of a drug resistance plasmid in Escherichia coli K12. J Gen Microbiol. 1979 Mar;111(1):201–210. doi: 10.1099/00221287-111-1-201. [DOI] [PubMed] [Google Scholar]
- HERBERT D., ELSWORTH R., TELLING R. C. The continuous culture of bacteria; a theoretical and experimental study. J Gen Microbiol. 1956 Jul;14(3):601–622. doi: 10.1099/00221287-14-3-601. [DOI] [PubMed] [Google Scholar]
- HORIUCHI T., TOMIZAWA J. I., NOVICK A. Isolation and properties of bacteria capable of high rates of beta-galactosidase synthesis. Biochim Biophys Acta. 1962 Jan 22;55:152–163. doi: 10.1016/0006-3002(62)90941-1. [DOI] [PubMed] [Google Scholar]
- Hansche P. E., Beres V., Lange P. Gene duplication in Saccharomyces cerevisiae. Genetics. 1978 Apr;88(4 Pt 1):673–687. [PMC free article] [PubMed] [Google Scholar]
- Hansche P. E. Gene duplication as a mechanism of genetic adaptation in Saccharomyces cerevisiae. Genetics. 1975 Apr;79(4):661–674. doi: 10.1093/genetics/79.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder W., Kuenen J. G. A review. Microbial selection in continuous culture. J Appl Bacteriol. 1977 Aug;43(1):1–24. doi: 10.1111/j.1365-2672.1977.tb00717.x. [DOI] [PubMed] [Google Scholar]
- Hartl D. L., Dykhuizen D. E. Potential for selection among nearly neutral allozymes of 6-phosphogluconate dehydrogenase in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6344–6348. doi: 10.1073/pnas.78.10.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D., Dykhuizen D. A selectively driven molecular clock. Nature. 1979 Sep 20;281(5728):230–231. doi: 10.1038/281230a0. [DOI] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. 3. Isolation and properties of constitutive mutants. J Bacteriol. 1966 Mar;91(3):1161–1167. doi: 10.1128/jb.91.3.1161-1167.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Kinney T., Adams J. The maintenance of Plasmid-containing organisms in populations of Escherichia coli. J Gen Microbiol. 1981 Mar;123(1):129–141. doi: 10.1099/00221287-123-1-129. [DOI] [PubMed] [Google Scholar]
- Jones I. M., Primrose S. B., Robinson A., Ellwood D. C. Maintenance of some ColE1-type plasmids in chemostat culture. Mol Gen Genet. 1980;180(3):579–584. doi: 10.1007/BF00268063. [DOI] [PubMed] [Google Scholar]
- Koch A. L. The pertinence of the periodic selection phenomenon to prokaryote evolution. Genetics. 1974 May;77(1):127–142. doi: 10.1093/genetics/77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LARSEN D. H., DIMMICK R. L. ATTACHMENT AND GROWTH OF BACTERIA ON SURFACES OF CONTINUOUS-CULTURE VESSELS. J Bacteriol. 1964 Nov;88:1380–1387. doi: 10.1128/jb.88.5.1380-1387.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange P., Hansche P. E. Mapping of a centromere-linked gene responsible for constitutive acid phosphatase synthesis in yeast. Mol Gen Genet. 1980;180(3):605–607. doi: 10.1007/BF00268067. [DOI] [PubMed] [Google Scholar]
- Levin B. R. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics. 1981 Sep;99(1):1–23. doi: 10.1093/genetics/99.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. R., Rice V. A. The kinetics of transfer of nonconjugative plasmids by mobilizing conjugative factors. Genet Res. 1980 Jun;35(3):241–259. doi: 10.1017/s0016672300014117. [DOI] [PubMed] [Google Scholar]
- Levin B. R., Stewart F. M., Rice V. A. The kinetics of conjugative plasmid transmission: fit of a simple mass action model. Plasmid. 1979 Apr;2(2):247–260. doi: 10.1016/0147-619x(79)90043-x. [DOI] [PubMed] [Google Scholar]
- Lin L., Bitner R., Edlin G. Increased reproductive fitness of Escherichia coli lambda lysogens. J Virol. 1977 Feb;21(2):554–559. doi: 10.1128/jvi.21.2.554-559.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOSER H. Structure and dynamics of bacterial populations maintained in the chemostat. Cold Spring Harb Symp Quant Biol. 1957;22:121–137. doi: 10.1101/sqb.1957.022.01.015. [DOI] [PubMed] [Google Scholar]
- MUNSON R. J., BRIDGES B. A. 'TAKE-OVER'--AN UNUSUAL SELECTION PROCESS IN STEADY-STATE CULTURES OF ESCHERICHIA COLI. J Gen Microbiol. 1964 Dec;37:411–418. doi: 10.1099/00221287-37-3-411. [DOI] [PubMed] [Google Scholar]
- Martin G. A., Hempfling W. P. A method for the regulation of microbial population density during continuous culture at high growth rates. Arch Microbiol. 1976 Feb;107(1):41–47. doi: 10.1007/BF00427865. [DOI] [PubMed] [Google Scholar]
- Maruyama T., Kimura M. Genetic variability and effective population size when local extinction and recolonization of subpopulations are frequent. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6710–6714. doi: 10.1073/pnas.77.11.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason T. G., Richardson G. Escherichia coli and the human gut: some ecological considerations. J Appl Bacteriol. 1981 Aug;51(1):1–16. doi: 10.1111/j.1365-2672.1981.tb00903.x. [DOI] [PubMed] [Google Scholar]
- Mason T. G., Richardson G. Observations on the in vivo and in vitro competition between strains of Escherichia coli isolated from the human gut. J Appl Bacteriol. 1982 Aug;53(1):19–27. doi: 10.1111/j.1365-2672.1982.tb04730.x. [DOI] [PubMed] [Google Scholar]
- Mason T. G., Slater J. H. Competition between an Escherichia coli tyrosine auxotroph and a prototrophic revertant in glucose- and tyrosine-limited chemostats. Antonie Van Leeuwenhoek. 1979;45(2):253–263. doi: 10.1007/BF00418588. [DOI] [PubMed] [Google Scholar]
- Matz K., Schmandt M., Gussin G. N. The rex gene of bacteriophage lambda is really two genes. Genetics. 1982 Nov;102(3):319–327. doi: 10.1093/genetics/102.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D J. Segregation of the Selective Advantage Obtained through Orthoselection in Escherichia Coli. Genetics. 1955 Nov;40(6):937–950. doi: 10.1093/genetics/40.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Milkman R. Electrophoretic variation in Escherichia coli from natural sources. Science. 1973 Dec 7;182(4116):1024–1026. doi: 10.1126/science.182.4116.1024. [DOI] [PubMed] [Google Scholar]
- NOVICK A., HORIUCHI T. Hyper-production of beta-galactosidase by Escherichia coli bacteria. Cold Spring Harb Symp Quant Biol. 1961;26:239–245. doi: 10.1101/sqb.1961.026.01.029. [DOI] [PubMed] [Google Scholar]
- NOVICK A., SZILARD L. Experiments with the Chemostat on spontaneous mutations of bacteria. Proc Natl Acad Sci U S A. 1950 Dec;36(12):708–719. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestmann E. R., Hill R. F. Population changes in continuously growing mutator cultures of Escherichia coli. Genetics. 1973 Apr;73(Suppl):41–44. [PubMed] [Google Scholar]
- Paynter M. J., Bungay H. R., 3rd Responses in continuous cultures of lysogenic Escherichia coli following induction. Biotechnol Bioeng. 1970 May;12(3):347–351. doi: 10.1002/bit.260120304. [DOI] [PubMed] [Google Scholar]
- Roth M., Noack D. Genetic stability of differentiated functions in Streptomyces hygroscopicus in relation to conditions of continuous culture. J Gen Microbiol. 1982 Jan;128(1):107–114. doi: 10.1099/00221287-128-1-107. [DOI] [PubMed] [Google Scholar]
- Weiss R. L., Kukora J. R., Adams J. The relationship between enzyme activity, cell geometry, and fitness in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1975 Mar;72(3):794–798. doi: 10.1073/pnas.72.3.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters J. T., van Andel J. G. R-plasmid persistence in Escherichia coli grown in chemostat cultures [proceedings]. Antonie Van Leeuwenhoek. 1979;45(2):317–318. doi: 10.1007/BF00418597. [DOI] [PubMed] [Google Scholar]
- Zünd P., Lebek G. Generation time-prolonging R plasmids: correlation between increases in the generation time of Escherichia coli caused by R plasmids and their molecular size. Plasmid. 1980 Jan;3(1):65–69. doi: 10.1016/s0147-619x(80)90034-7. [DOI] [PubMed] [Google Scholar]



