The production and crystallization of the human DEAD box polypeptide 5 are reported. A 2.7 Å native diffraction data set has been obtained.
Keywords: DEAD-box helicases, DDX5
Abstract
The DEAD-box RNA helicase DDX5 is involved in many aspects of RNA processing and has been implicated in a number of cellular processes involving alteration of RNA secondary structure. The N-terminal region of DDX5, which contains the conserved domain 1 of the DEAD-box helicases, has been cloned and expressed in Escherichia coli and purified. Here, the crystallization and preliminary diffraction analysis of this region is reported. X-ray diffraction data were processed to a resolution of 2.7 Å. The crystals belonged to space group I222, with unit-cell parameters a = 66.18, b = 73.80, c = 104.00 Å, α = β = γ = 90°.
1. Introduction
RNA helicases are involved in all aspects of RNA metabolism. They are ATPases with RNA-binding and RNA-unwinding activities and are involved in transcription, pre-mRNA splicing, ribosome biogenesis, mRNA nuclear export, translation initiation and degradation of mRNA (Linder, 2006 ▶; Cordin et al., 2006 ▶).
DDX5 (also known as p68) was first discovered owing to its cross-reactivity with a monoclonal antibody of the large T antigen of simian virus 40 (Lane & Hoeffler, 1980 ▶). Subsequent analysis of the DNA sequence of DDX5 revealed extensive homology to the translation initiation factor eIF-4A. The molecular similarity of DDX5 to both the large T antigen and eIF-4A, which are an ATP-dependent DNA helicase and an ATP-dependent RNA helicase, respectively, implied that DDX5 may function as an RNA or DNA helicase (Ford et al., 1988 ▶). DDX5 belongs to the DEAD-box family of proteins, which are named after their conserved amino-acid sequence Asp-Glu-Ala-Asp (D-E-A-D; Linder et al., 1989 ▶). Based on their conserved sequence motifs, they are classified into superfamily 2 (SF2) of the helicases (Gorbalenya & Koonin, 1993 ▶). The DEAD-box family of helicases is characterized by the unique Q motif as well as eight other conserved motifs (Tanner et al., 2003 ▶). The motifs Q, I (Walker A, phosphate-binding P-loop), Ia, Ib, II (Walker B, DExD box) and III form domain 1, while motifs IV, V and VI form domain 2 (Cordin et al., 2006 ▶; Tanner et al., 2003 ▶). Over the past decade, an increasing number of DEAD-box RNA helicase structures have been determined (Hogbom et al., 2007 ▶; Sengoku et al., 2006 ▶; Shi et al., 2004 ▶; Carmel & Matthews, 2004 ▶; Caruthers et al., 2000 ▶; Benz et al., 1999 ▶).
DDX5 has been implicated in a wide range of biological processes, with early studies reporting its possible involvement in the regulation of differentiation/maturation of various animal and human cells (Abdelhaleem, 2005 ▶). Subsequently, it was shown to bind, unwind and rearrange RNA secondary structures (Rossler et al., 2001 ▶) and was also shown to be crucial in the processing, alternate splicing and degradation of mRNA (Fuller-Pace, 2006 ▶; Abdelhaleem, 2005 ▶). DDX5 also acts as a transcriptional co-regulator of hormone receptors and transcription factors of differentiation and transcriptional initiation and mRNA decay (Fuller-Pace & Ali, 2008 ▶; Abdelhaleem, 2005 ▶). In this paper, we report the cloning, expression, purification and preliminary X-ray diffraction analysis of the N-terminal region of DDX5 consisting of amino acids 1–305 and spanning domain 1 of the DEAD-box helicase.
2. Experimental procedures
2.1. Cloning, expression and purification
The open reading frame encoding human DDX5 was amplified from a spleen cDNA expression library (Goh et al., 2004 ▶). The 5′ primer 5′-CGGGATCCATGTCGGGTTATTCGAGTGACCGAGAC-3′ and 3′ primer 5′-CCGCTCGAGTTAAAGTGCACCAATGTTTATATGAA 3′ were used to clone the N-terminal region of DDX5 (residues 1–305) in an expression plasmid with an amino-terminal glutathione S-transferease (GST) fusion partner (pGEX6p1, GE Healthcare). The sequence identity was confirmed by DNA sequencing (DNA core facility, IMCB, Singapore).
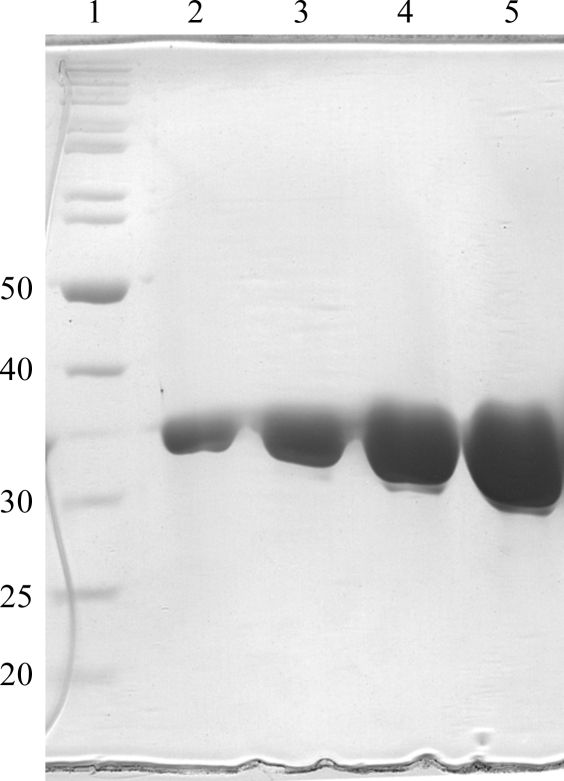
The fusion protein was expressed in Escherichia coli BL21-CodonPlus-RIL (Stratagene). Cultures were first grown overnight at 310 K in Luria–Bertani (LB) medium supplemented with 100 µg ml−1 ampicillin and 20 µg ml−1 chloramphenicol before inoculation into Terrific Broth (TB) supplemented with the same amounts of antibiotics. On reaching an OD600 of 0.8, the cultures were cooled to 291 K and induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 0.2 mM. After an incubation period of 24 h, the cells were harvested at 4000g for 10 min. Bacterial pellets were resuspended in lysis buffer (50 mM Tris–HCl pH 7.4, 300 mM NaCl, 10% glycerol and 2 mM DTT) supplemented with Complete Protease Inhibitor (Roche). For purification, the cells were subjected to sonication. The lysate was cleared by centrifugation (1 h, 15 000g) and the supernatant was mixed with 3 ml glutathione Sepharose resin (GE Healthcare) pre-washed with lysis buffer. Following 3 h of batch binding at 277 K with constant-rotation mixing, four washes with lysis buffer were performed to remove unbound proteins. Removal of the GST tag from the N-terminus of DDX5 (1–305) was achieved by proteolytic cleavage using recombinant 3C protease (GE Healthcare). Approximately 100 µg 3C protease was used per milligram of GST-DDX5 (1–305). Briefly, the resin-bound fusion protein was resuspended in 30 ml lysis buffer containing 3C protease. Cleavage took place overnight at 277 K with constant rotation. The supernatant containing cleaved DDX5 (1–305) was separated by pouring the resin into an empty Econo-Pac column (Bio-Rad). The elutant was concentrated using an Amicon Ultra-15 Centrifugal Filter Device (Millipore) and further purified by size-exclusion chromatography through a Superdex S200 column (GE Healthcare) pre-equilibrated with 25 mM Tris–HCl pH 7.4, 300 mM NaCl, 10% glycerol and 2 mM DTT. Peak fractions were analyzed by SDS–PAGE to assess their purity (Fig. 1 ▶). The protein concentration was determined by spectrometry using the molar extinction coefficient of DDX5 (1–305) at 280 nm (35 785 M −1 cm−1). The molar extinction coefficient was determined from the protein sequence of DDX5 (1–305) using the ProtParam tool (Gasteiger et al., 2005 ▶). Pure DDX5 (1–305) was concentrated to 11 mg ml−1. As a consequence of cleavage, five amino acids (GPLGS) remained fused to the N-terminus of DDX5 (1–305).
Figure 1.
SDS–PAGE analysis of the purification stages of DDX5 (1–305). Lane 1, protein molecular-weight standards (kDa); lanes 2–5, fractions collected from the gel-filtration column.
2.2. Crystallization
DDX5 (1–305) was crystallized via the sitting-drop vapour-diffusion method at 288 K using 24-well plates. 1 µl protein solution was mixed with 1 µl reservoir solution and equilibrated against 0.5 ml reservoir solution (Crystal Screens I and II and PEG/Ion Screens I and II; Hampton Research). X-ray diffraction-quality crystals were obtained within two months from drops containing 1 µl protein solution [DDX5 (1–305) at 11 mg ml−1 in 25 mM Tris–HCl pH 7.4, 300 mM NaCl, 10% glycerol and 2 mM DTT] and 1 µl 2%(v/v) Tacsimate pH 4.0, 0.1 M bis-tris pH 6.5 and 20%(w/v) polyethylene glycol (PEG) 3350.
2.3. X-ray diffraction analysis
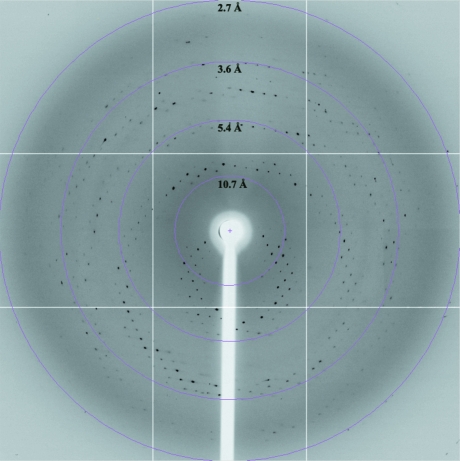
For cryoprotection, the crystal was soaked in cryosolution [2%(v/v) Tacsimate pH 4.0, 0.1 M bis-tris pH 6.5, 20%(w/v) PEG 3350 and 25% glycerol] and flash-cooled in liquid nitrogen. Native data sets were collected using a Quantum CCD image plate on beamline 13B1 at the National Synchrotron Radiation Research Center (NSRRC), Taiwan. 150 images were collected as 1° oscillations with 1 s exposure. The crystal-to-detector distance was set to 400 mm. The X-ray diffraction pattern of a DDX5 (1–305) crystal is shown in Fig. 2 ▶. The raw data were integrated and scaled using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
Figure 2.
Diffraction pattern of DDX5 (1–305) together with resolution rings. The crystal-to-detector distance was 400 mm and the oscillation angle was 1°.
3. Results and discussion
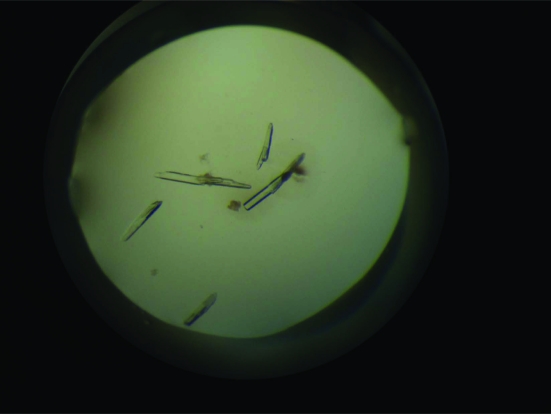
The N-terminal region of DDX5 (1–305) was cloned and expressed in E. coli BL21 (DE3). The typical yield of pure DDX5 (1–305) was 2 mg per litre of bacterial culture. Crystals first appeared within two weeks of incubation. Crystals grew to maximum dimensions of 0.15 × 0.05 × 0.05 mm (Fig. 3 ▶). Native data were collected from a single crystal of DDX5 (1–305) grown in 2%(v/v) Tacsimate pH 4.0, 0.1 M bis-tris pH 6.5 and 20%(w/v) PEG 3350 (Fig. 2 ▶). The space group was determined to be I222, with unit-cell parameters a = 66.18, b = 73.80, c = 104.00 Å, α = β = γ = 90°. The asymmetric unit contained one copy of DDX5 (1–305); the crystal volume per unit molecular weight (V M) was calculated to be 2.32 Å3 Da−1, corresponding to a solvent content of 47% (Matthews, 1968 ▶). Crystallographic statistics of the native data are summarized in Table 1 ▶. Crystallization of full-length DDX5 is actively being pursued, as is a molecular-replacement solution for DDX5 (1–305) using the structure of the conserved domain 1 of DDX3X (Hogbom et al., 2007 ▶) as a search model. The percentage sequence identity between the first 305 residues of DDX5 and DDX3X is ∼41%.
Figure 3.
Crystals of DDX5 (1–305). Typically, crystals grew to maximum dimensions of 0.15 × 0.05 × 0.05 mm.
Table 1. Statistics of preliminary data analysis.
Values in parentheses are for the highest resolution shell.
| Space group | I222 |
| Unit-cell parameters (Å, °) | a = 66.18, b = 73.80, c = 104.00, α = β = γ = 90 |
| No. of molecules per ASU | 1 |
| Resolution (Å) | 25–2.7 (2.8–2.7) |
| Wavelength (Å) | 0.99 |
| Observed reflections | 42651 (4013) |
| Unique reflections | 7229 (704) |
| Redundancy | 5.9 (5.7) |
| Completeness (%) | 99.8 (99.9) |
| Rmerge (%) | 8.5 (48.3) |
| 〈I/σ(I)〉 | 19.6 (2.7) |
Acknowledgments
This work was supported by grants from the Agency for Science, Technology and Research (A*STAR), Singapore. The authors wish to thank the personnel at NSRRC, Taiwan for their kind help during data collection.
References
- Abdelhaleem, M. (2005). Clin. Biochem.38, 499–503. [DOI] [PubMed]
- Benz, J., Trachsel, H. & Baumann, U. (1999). Structure, 7, 671–679. [DOI] [PubMed]
- Carmel, A. B. & Matthews, B. W. (2004). RNA, 10, 66–74. [DOI] [PMC free article] [PubMed]
- Caruthers, J. M., Johnson, E. R. & McKay, D. B. (2000). Proc. Natl Acad. Sci. USA, 97, 13080–13085. [DOI] [PMC free article] [PubMed]
- Cordin, O., Banroques, J., Tanner, N. K. & Linder, P. (2006). Gene, 367, 17–37. [DOI] [PubMed]
- Ford, M. J., Anton, I. A. & Lane, D. P. (1988). Nature (London), 332, 736–738. [DOI] [PubMed]
- Fuller-Pace, F. V. (2006). Nucleic Acids Res.34, 4206–4215. [DOI] [PMC free article] [PubMed]
- Fuller-Pace, F. V. & Ali, S. (2008). Biochem. Soc. Trans.36, 609–612. [DOI] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Goh, P. Y., Tan, Y. J., Lim, S. P., Tan, Y. H., Lim, S. G., Fuller-Pace, F. & Hong, W. (2004). J. Virol.78, 5288–5298. [DOI] [PMC free article] [PubMed]
- Gorbalenya, A. E. & Koonin, E. V. (1993). Curr. Opin. Struct. Biol.3, 419–429.
- Hogbom, M., Collins, R., van den Berg, S., Jenvert, R. M., Karlberg, T., Kotenyova, T., Flores, A., Karlsson Hedestam, G. B. & Schiavone, L. H. (2007). J. Mol. Biol.372, 150–159. [DOI] [PubMed]
- Lane, D. P. & Hoeffler, W. K. (1980). Nature (London), 288, 167–170. [DOI] [PubMed]
- Linder, P. (2006). Nucleic Acids Res.34, 4168–4180. [DOI] [PMC free article] [PubMed]
- Linder, P., Lasko, P. F., Ashburner, M., Leroy, P., Nielsen, P. J., Nishi, K., Schnier, J. & Slonimski, P. P. (1989). Nature (London), 337, 121–122. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rossler, O. G., Straka, A. & Stahl, H. (2001). Nucleic Acids Res.29, 2088–2096. [DOI] [PMC free article] [PubMed]
- Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S. & Yokoyama, S. (2006). Cell, 125, 287–300. [DOI] [PubMed]
- Shi, H., Cordin, O., Minder, C. M., Linder, P. & Xu, R. M. (2004). Proc. Natl Acad. Sci. USA, 101, 17628–17633. [DOI] [PMC free article] [PubMed]
- Tanner, N. K., Cordin, O., Banroques, J., Doere, M. & Linder, P. (2003). Mol. Cell, 11, 127–138. [DOI] [PubMed]