Full text
PDF























Selected References
These references are in PubMed. This may not be the complete list of references from this article.
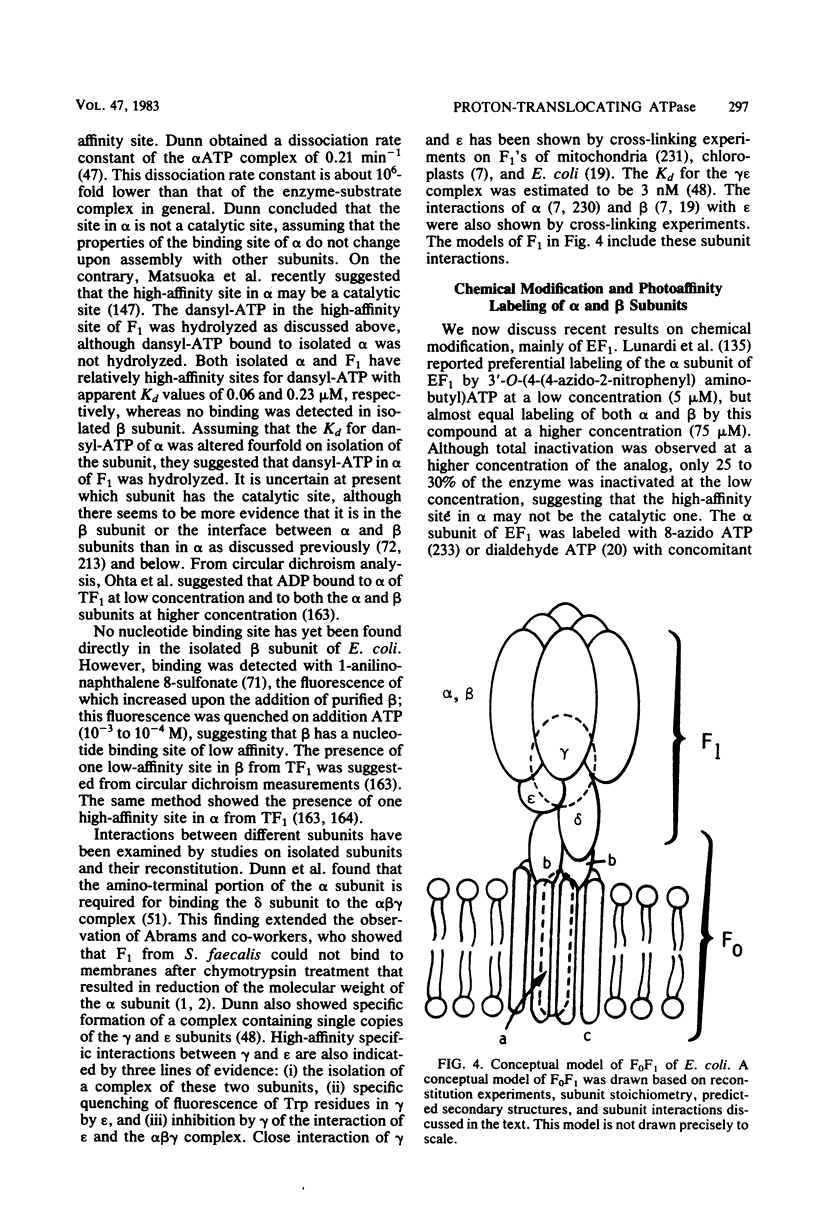
- Abrams A., Morris D., Jensen C. Chymotryptic conversion of bacterial membrane ATPase to an active form with modified alpha chains and defective membrane binding properties. Biochemistry. 1976 Dec 14;15(25):5560–5566. doi: 10.1021/bi00670a021. [DOI] [PubMed] [Google Scholar]
- Alfonzo M., Racker E. Components and mechanism of action of ATP-driven proton pumps. Can J Biochem. 1979 Dec;57(12):1351–1358. doi: 10.1139/o79-180. [DOI] [PubMed] [Google Scholar]
- Amzel L. M., McKinney M., Narayanan P., Pedersen P. L. Structure of the mitochondrial F1 ATPase at 9-A resolution. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5852–5856. doi: 10.1073/pnas.79.19.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amzel L. M., Pedersen P. L. Adenosine triphosphatase from rat liver mitochondria. Crystallization and x-ray diffraction studies of the F1-component of the enzyme. J Biol Chem. 1978 Apr 10;253(7):2067–2069. [PubMed] [Google Scholar]
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Baird B. A., Hammes G. G. Chemical cross-linking studies of chloroplast coupling factor 1. J Biol Chem. 1976 Nov 25;251(22):6953–6962. [PubMed] [Google Scholar]
- Baird B. A., Hammes G. G. Structure of oxidative- and photo-phosphorylation coupling factor complexes. Biochim Biophys Acta. 1979 Jul 3;549(1):31–53. doi: 10.1016/0304-4173(79)90017-x. [DOI] [PubMed] [Google Scholar]
- Bengis-Garber C., Gromet-Elhanan Z. Purification of the energy-transducing adenosine triphosphatase complex from Rhodospirillum rubrum. Biochemistry. 1979 Aug 7;18(16):3577–3581. doi: 10.1021/bi00583a022. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Boyer P. D., Kohlbrenner W. E., McIntosh D. B., Smith L. T., O'Neal C. C. ATP and ADP modulations of catalysis by F1 and Ca2+, Mg2+-ATPases. Ann N Y Acad Sci. 1982;402:65–83. doi: 10.1111/j.1749-6632.1982.tb25732.x. [DOI] [PubMed] [Google Scholar]
- Bragg P. D., Hou C. A cross-linking study of the Ca2+, Mg2+-activated adenosine triphosphatase of Escherichia coli. Eur J Biochem. 1980 May;106(2):495–503. doi: 10.1111/j.1432-1033.1980.tb04596.x. [DOI] [PubMed] [Google Scholar]
- Bragg P. D., Hou C. Purification and characterization of the inactive Ca2+, Mg2+-activated adenosine triphosphatase of the unc A- mutant Escherichia coli AN120. Arch Biochem Biophys. 1977 Jan 30;178(2):486–494. doi: 10.1016/0003-9861(77)90219-3. [DOI] [PubMed] [Google Scholar]
- Bragg P. D., Hou C. Solubilization of a phospholipid-stimulated adenosine triphosphatase complex from membranes of Escherichia coli. Arch Biochem Biophys. 1976 Jun;174(2):553–561. doi: 10.1016/0003-9861(76)90383-0. [DOI] [PubMed] [Google Scholar]
- Bragg P. D., Hou C. Subunit composition, function, and spatial arrangement in the Ca2+-and Mg2+-activated adenosine triphosphatases of Escherichia coli and Salmonella typhimurium. Arch Biochem Biophys. 1975 Mar;167(1):311–321. doi: 10.1016/0003-9861(75)90467-1. [DOI] [PubMed] [Google Scholar]
- Bragg P. D., Stan-Lotter H., Hou C. Adenine nucleotide binding sites in normal and mutant adenosine triphosphatases of Escherichia coli. Arch Biochem Biophys. 1982 Feb;213(2):669–679. doi: 10.1016/0003-9861(82)90597-5. [DOI] [PubMed] [Google Scholar]
- Bragg P. D., Stan-Lotter H., Hou C. Affinity labeling of purified Ca2+,Mg2+-activated ATPase of Escherichia coli by the 2',3'-dialdehydes of adenosine 5'-di- and triphosphates. Arch Biochem Biophys. 1981 Apr 1;207(2):290–299. doi: 10.1016/0003-9861(81)90036-9. [DOI] [PubMed] [Google Scholar]
- Bruist M. F., Hammes G. G. Further characterization of nucleotide binding sites on chloroplast coupling factor one. Biochemistry. 1981 Oct 27;20(22):6298–6305. doi: 10.1021/bi00525a003. [DOI] [PubMed] [Google Scholar]
- Brusilow W. S., Gunsalus R. P., Hardeman E. C., Decker K. P., Simoni R. D. In vitro synthesis of the F0 and F1 components of the proton translocating ATPase of Escherichia coli. J Biol Chem. 1981 Apr 10;256(7):3141–3144. [PubMed] [Google Scholar]
- Brusilow W. S., Klionsky D. J., Simoni R. D. Differential polypeptide synthesis of the proton-translocating ATPase of Escherichia coli. J Bacteriol. 1982 Sep;151(3):1363–1371. doi: 10.1128/jb.151.3.1363-1371.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
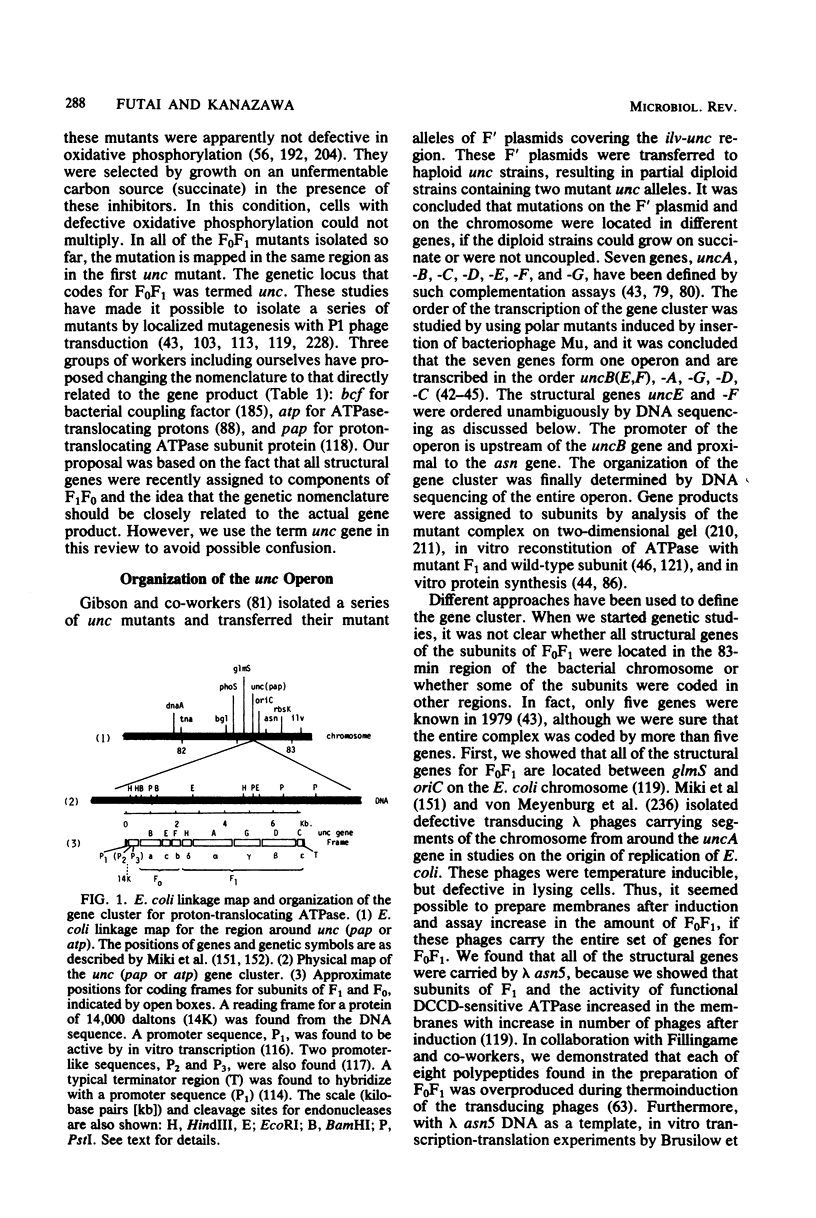
- Butlin J. D., Cox G. B., Gibson F. Oxidative phosphorylation in Escherichia coli K-12: the genetic and biochemical characterisations of a strain carrying a mutation in the uncB gene. Biochim Biophys Acta. 1973 Feb 22;292(2):366–375. doi: 10.1016/0005-2728(73)90043-1. [DOI] [PubMed] [Google Scholar]
- Butlin J. D., Cox G. B., Gibson F. Oxidative phosphorylation in Escherichia coli K12. Mutations affecting magnesium ion- or calcium ion-stimulated adenosine triphosphatase. Biochem J. 1971 Aug;124(1):75–81. doi: 10.1042/bj1240075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley L. C., Jr, Hammes G. G. Characterization of nucleotide binding sites on chloroplast coupling factor 1. Biochemistry. 1975 Jul;14(13):2968–2975. doi: 10.1021/bi00684a027. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choate G. L., Hutton R. L., Boyer P. D. Occurrence and significance of oxygen exchange reactions catalyzed by mitochondrial adenosine triphosphatase preparations. J Biol Chem. 1979 Jan 25;254(2):286–290. [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Clarke D. J., Morris J. G. Partial purification of a dicyclohexylcarbodi-imide-sensitive membrane adenosine triphosphatase complex from the obligately anaerobic bacterium Clostridium Pasteurianum. Biochem J. 1976 Mar 15;154(3):725–729. doi: 10.1042/bj1540725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. B., Crane F. L., Downie J. A., Radik J. Different effects of inhibitors on two mutants of Escherichia coli K12 affected in the Fo portion of the adenosine triphosphatase complex. Biochim Biophys Acta. 1977 Oct 12;462(1):113–120. doi: 10.1016/0005-2728(77)90193-1. [DOI] [PubMed] [Google Scholar]
- Cox G. B., Downie J. A., Langman L., Senior A. E., Ash G., Fayle D. R., Gibson F. Assembly of the adenosine triphosphatase complex in Escherichia coli: assembly of F0 is dependent on the formation of specific F1 subunits. J Bacteriol. 1981 Oct;148(1):30–42. doi: 10.1128/jb.148.1.30-42.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. B., Gibson F., McCann L. Reconstitution of oxidative phosphorylation and the adenosine triphosphate-dependent transhydrogenase activity by a combination of membrane fractions from unCA- and uncB- mutant strains of Escherichia coli K12. Biochem J. 1973 Aug;134(4):1015–1021. doi: 10.1042/bj1341015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criddle R. S., Packer L., Shieh P. Oligomycin-dependent ionophoric protein subunit of mitochondrial adenosinetriphosphatase. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4306–4310. doi: 10.1073/pnas.74.10.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R. L., Grubmeyer C., Penefsky H. S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase. Rate enhancements resulting from cooperative interactions between multiple catalytic sites. J Biol Chem. 1982 Oct 25;257(20):12101–12105. [PubMed] [Google Scholar]
- Cross R. L., Nalin C. M. Adenine nucleotide binding sites on beef heart F1-ATPase. Evidence for three exchangeable sites that are distinct from three noncatalytic sites. J Biol Chem. 1982 Mar 25;257(6):2874–2881. [PubMed] [Google Scholar]
- Cross R. L. The mechanism and regulation of ATP synthesis by F1-ATPases. Annu Rev Biochem. 1981;50:681–714. doi: 10.1146/annurev.bi.50.070181.003341. [DOI] [PubMed] [Google Scholar]
- Decker K. P., Brusilow W. S., Gunsalus R. P., Simoni R. D. In vitro membrane association of the F0 polypeptides of the Escherichia coli proton translocating ATPase. J Bacteriol. 1982 Nov;152(2):815–821. doi: 10.1128/jb.152.2.815-821.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A., Cox G. B., Langman L., Ash G., Becker M., Gibson F. Three genes coding for subunits of the membrane sector (F0) of the Escherichia coli adenosine triphosphatase complex. J Bacteriol. 1981 Jan;145(1):200–210. doi: 10.1128/jb.145.1.200-210.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A., Gibson F., Cox G. B. Membrane adenosine triphosphatases of prokaryotic cells. Annu Rev Biochem. 1979;48:103–131. doi: 10.1146/annurev.bi.48.070179.000535. [DOI] [PubMed] [Google Scholar]
- Downie J. A., Langman L., Cox G. B., Yanofsky C., Gibson F. Subunits of the adenosine triphosphatase complex translated in vitro from the Escherichia coli unc operon. J Bacteriol. 1980 Jul;143(1):8–17. doi: 10.1128/jb.143.1.8-17.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A., Senior A. E., Gibson F., Cox G. B. A fifth gene (uncE) in the operon concerned with oxidative phosphorylation in Escherichia coli. J Bacteriol. 1979 Feb;137(2):711–718. doi: 10.1128/jb.137.2.711-718.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S. D. ATP causes a large change in the conformation of the isolated alpha subunit of Escherichia coli F1 ATPase. J Biol Chem. 1980 Dec 25;255(24):11857–11860. [PubMed] [Google Scholar]
- Dunn S. D., Futai M. Reconstitution of a functional coupling factor from the isolated subunits of Escherichia coli F1 ATPase. J Biol Chem. 1980 Jan 10;255(1):113–118. [PubMed] [Google Scholar]
- Dunn S. D., Heppel L. A., Fullmer C. S. The NH2-terminal portion of the alpha subunit of Escherichia coli F1 ATPase is required for binding the delta subunit. J Biol Chem. 1980 Jul 25;255(14):6891–6896. [PubMed] [Google Scholar]
- Dunn S. D., Heppel L. A. Properties and functions of the subunits of the Escherichia coli coupling factor ATPase. Arch Biochem Biophys. 1981 Sep;210(2):421–436. doi: 10.1016/0003-9861(81)90206-x. [DOI] [PubMed] [Google Scholar]
- Dunn S. D. Identification of the altered subunit in the inactive F1ATPase of an Escherichia coli uncA mutant. Biochem Biophys Res Commun. 1978 May 30;82(2):596–602. doi: 10.1016/0006-291x(78)90916-6. [DOI] [PubMed] [Google Scholar]
- Dunn S. D. The isolated gamma subunit of Escherichia coli F1 ATPase binds the epsilon subunit. J Biol Chem. 1982 Jul 10;257(13):7354–7359. [PubMed] [Google Scholar]
- Esch F. S., Allison W. S. Identification of a tyrosine residue at a nucleotide binding site in the beta subunit of the mitochondrial ATPase with p-fluorosulfonyl[14C]-benzoyl-5'-adenosine. J Biol Chem. 1978 Sep 10;253(17):6100–6106. [PubMed] [Google Scholar]
- Esch F. S., Allison W. S. On the subunit stoichiometry of the F1-ATPase and the sites in it that react specifically with p-fluorosulfonylbenzoyl-5'-adenosine. J Biol Chem. 1979 Nov 10;254(21):10740–10746. [PubMed] [Google Scholar]
- Esch F. S., Böhlen P., Otsuka A. S., Yoshida M., Allison W. S. Inactivation of the bovine mitochondrial F1-ATPase with dicyclohexyl[14C]carbodiimide leads to the modification of a specific glutamic acid residue in the beta subunit. J Biol Chem. 1981 Sep 10;256(17):9084–9089. [PubMed] [Google Scholar]
- Feldman R. I., Sigman D. S. The synthesis of enzyme-bound ATP by soluble chloroplast coupling factor 1. J Biol Chem. 1982 Feb 25;257(4):1676–1683. [PubMed] [Google Scholar]
- Fillingame R. H. Identification of the dicyclohexylcarbodiimide-reactive protein component of the adenosine 5'-triphosphate energy-transducing system of Escherichia coli. J Bacteriol. 1975 Nov;124(2):870–883. doi: 10.1128/jb.124.2.870-883.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingame R. H., Mosher M. E., Negrin R. S., Peters L. K. H+-ATPase of Escherichia coli uncB402 mutation leads to loss of chi subunit of subunit of F0 sector. J Biol Chem. 1983 Jan 10;258(1):604–609. [PubMed] [Google Scholar]
- Fillingame R. H. The proton-translocating pumps of oxidative phosphorylation. Annu Rev Biochem. 1980;49:1079–1113. doi: 10.1146/annurev.bi.49.070180.005243. [DOI] [PubMed] [Google Scholar]
- Foster D. L., Boublik M., Kaback H. R. Structure of the lac carrier protein of Escherichia coli. J Biol Chem. 1983 Jan 10;258(1):31–34. [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Energy-transducing H+-ATPase of Escherichia coli. Purification, reconstitution, and subunit composition. J Biol Chem. 1979 Sep 10;254(17):8230–8236. [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982 Feb 25;257(4):2009–2015. [PubMed] [Google Scholar]
- Foster D. L., Mosher M. E., Futai M., Fillingame R. H. Subunits of the H+-ATPase of Escherichia coli. Overproduction of an eight-subunit F1F0-ATPase following induction of a lambda-transducing phage carrying the unc operon. J Biol Chem. 1980 Dec 25;255(24):12037–12041. [PubMed] [Google Scholar]
- Frangione B., Rosenwasser E., Penefsky H. S., Pullman M. E. Amino acid sequence of the protein inhibitor of mitochondrial adenosine triphosphatase. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7403–7407. doi: 10.1073/pnas.78.12.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Bienhaus G., Hoppe J., Schairer H. U. The dicyclohexylcarbodiimide-binding protein c of ATP synthase from Escherichia coli is not sufficient to express an efficient H+ conduction. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6643–6646. doi: 10.1073/pnas.78.11.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Friedl C., Schairer H. U. F0 of Escherichia coli ATP-synthase containing mutant and wild-type carbodiimide-binging proteins is impaired in H+ -conduction. FEBS Lett. 1980 Oct 6;119(2):254–256. doi: 10.1016/0014-5793(80)80265-1. [DOI] [PubMed] [Google Scholar]
- Friedl P., Friedl C., Schairer H. U. The ATP synthetase of Escherichia coli K12: purification of the enzyme and reconstitution of energy-transducing activities. Eur J Biochem. 1979 Oct;100(1):175–180. doi: 10.1111/j.1432-1033.1979.tb02046.x. [DOI] [PubMed] [Google Scholar]
- Friedl P., Hoppe J., Gunsalus R. P., Michelsen O., von Meyenburg K., Schairer H. U. Membrane integration and function of the three F0 subunits of the ATP synthase of Escherichia coli K12. EMBO J. 1983;2(1):99–103. doi: 10.1002/j.1460-2075.1983.tb01388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., Schairer H. U. The isolated F0 of Escherichia coli aTP-synthase is reconstitutively active in H+-conduction and ATP-dependent energy-transduction. FEBS Lett. 1981 Jun 15;128(2):261–264. doi: 10.1016/0014-5793(81)80094-4. [DOI] [PubMed] [Google Scholar]
- Futai M., Kanazawa H., Takeda K., Kagawa Y. Reconstitution of ATPase from the isolated subunits of coupling factor F1's of Escherichia coli and thermophilic bacterium PS3. Biochem Biophys Res Commun. 1980 Sep 16;96(1):227–234. doi: 10.1016/0006-291x(80)91204-8. [DOI] [PubMed] [Google Scholar]
- Futai M. Reconstitution of ATPase activity from the isolated alpha, beta, and gamma subunits of the coupling factor, F1, of Escherichia coli. Biochem Biophys Res Commun. 1977 Dec 21;79(4):1231–1237. doi: 10.1016/0006-291x(77)91138-x. [DOI] [PubMed] [Google Scholar]
- Gay N. J., Walker J. E. Homology between human bladder carcinoma oncogene product and mitochondrial ATP-synthase. Nature. 1983 Jan 20;301(5897):262–264. doi: 10.1038/301262a0. [DOI] [PubMed] [Google Scholar]
- Gay N. J., Walker J. E. The atp operon: nucleotide sequence of the promoter and the genes for the membrane proteins, and the delta subunit of Escherichia coli ATP-synthase. Nucleic Acids Res. 1981 Aug 25;9(16):3919–3926. doi: 10.1093/nar/9.16.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay N. J., Walker J. E. The atp operon: nucleotide sequence of the region encoding the alpha-subunit of Escherichia coli ATP-synthase. Nucleic Acids Res. 1981 May 11;9(9):2187–2194. doi: 10.1093/nar/9.9.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson F., Cox G. B., Downie J. A., Radik J. A mutation affecting a second component of the F0 portion of the magnesium ion-stimulated adenosine triphosphatase of Escherichia coli K12. The uncC424 allele. Biochem J. 1977 Apr 15;164(1):193–198. doi: 10.1042/bj1640193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresser M. J., Myers J. A., Boyer P. D. Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model. J Biol Chem. 1982 Oct 25;257(20):12030–12038. [PubMed] [Google Scholar]
- Grubmeyer C., Cross R. L., Penefsky H. S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase. Rate constants for elementary steps in catalysis at a single site. J Biol Chem. 1982 Oct 25;257(20):12092–12100. [PubMed] [Google Scholar]
- Grubmeyer C., Penefsky H. S. Cooperatively between catalytic sites in the mechanism of action of beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1981 Apr 25;256(8):3728–3734. [PubMed] [Google Scholar]
- Grubmeyer C., Penefsky H. S. The presence of two hydrolytic sites on beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1981 Apr 25;256(8):3718–3727. [PubMed] [Google Scholar]
- Gunsalus R. P., Brusilow W. S., Simoni R. D. Gene order and gene-polypeptide relationships of the proton-translocating ATPase operon (unc) of Escherichia coli. Proc Natl Acad Sci U S A. 1982 Jan;79(2):320–324. doi: 10.1073/pnas.79.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes G. G. Unifying concept for the coupling between ion pumping and ATP hydrolysis or synthesis. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6881–6884. doi: 10.1073/pnas.79.22.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen F. G., Nielsen J., Riise E., von Meyenburg K. The genes for the eight subunits of the membrane bound ATP synthase of Escherichia coli. Mol Gen Genet. 1981;183(3):463–472. doi: 10.1007/BF00268766. [DOI] [PubMed] [Google Scholar]
- Hasan S. M., Tsuchiya T., Rosen B. P. Energy transduction in Escherichia coli: physiological and biochemical effects of mutation in the uncB locus. J Bacteriol. 1978 Jan;133(1):108–113. doi: 10.1128/jb.133.1.108-113.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensgens L. A., Grivell L. A., Borst P., Bos J. L. Nucleotide sequence of the mitochondrial structural gene for subunit 9 of yeast ATPase complex. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1663–1667. doi: 10.1073/pnas.76.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe J., Friedl P., Schairer H. U., Sebald W., von Meyenburg K., Jørgensen B. B. The topology of the proton translocating F0 component of the ATP synthase from E. coli K12: studies with proteases. EMBO J. 1983;2(1):105–110. doi: 10.1002/j.1460-2075.1983.tb01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe J., Montecucco C., Friedl P. Labeling of subunit b of the ATP synthase from Escherichia coli with a photoreactive phospholipid analogue. J Biol Chem. 1983 Mar 10;258(5):2882–2885. [PubMed] [Google Scholar]
- Hoppe J., Schairer H. U., Friedl P., Sebald W. An Asp-Asn substitution in the proteolipid subunit of the ATP-synthase from Escherichia coli leads to a non-functional proton channel. FEBS Lett. 1982 Aug 16;145(1):21–29. doi: 10.1016/0014-5793(82)81198-8. [DOI] [PubMed] [Google Scholar]
- Hoppe J., Schairer H. U., Sebald W. Identification of amino-acid substitutions in the proteolipid subunit of the ATP synthase from dicyclohexylcarbodiimide-resistant mutants of Escherichia coli. Eur J Biochem. 1980 Nov;112(1):17–24. doi: 10.1111/j.1432-1033.1980.tb04981.x. [DOI] [PubMed] [Google Scholar]
- Hoppe J., Schairer H. U., Sebald W. The proteolipid of a mutant ATPase from Escherichia coli defective in H+-conduction contains a glycine instead of the carbodiimide-reactive aspartyl residue. FEBS Lett. 1980 Jan 1;109(1):107–111. doi: 10.1016/0014-5793(80)81321-4. [DOI] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Ogawa H. Organization of the recA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):313–317. doi: 10.1073/pnas.77.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe C. J., Auffret A. D., Doherty A., Bowman C. M., Dyer T. A., Gray J. C. Location and nucleotide sequence of the gene for the proton-translocating subunit of wheat chloroplast ATP synthase. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6903–6907. doi: 10.1073/pnas.79.22.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. B., Joshi S., Sanadi D. R. On the role of factor B and oligomycin on generation and discharge of the proton gradient. J Biol Chem. 1982 Jun 25;257(12):6697–6701. [PubMed] [Google Scholar]
- Hulla F. W., Höckel M., Rack M., Risi S., Dose K. Characterization and affinity labeling of nucleotide binding sites of bacterial plasma membrane adenosine triphosphatase (F1). Biochemistry. 1978 Mar 7;17(5):823–828. doi: 10.1021/bi00598a012. [DOI] [PubMed] [Google Scholar]
- Humbert R., Brusilow W. S., Gunsalus R. P., Klionsky D. J., Simoni R. D. Escherichia coli mutants defective in the uncH gene. J Bacteriol. 1983 Jan;153(1):416–422. doi: 10.1128/jb.153.1.416-422.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton R. L., Boyer P. D. Subunit interaction during catalysis. Alternating site cooperativity of mitochondrial adenosine triphosphatase. J Biol Chem. 1979 Oct 25;254(20):9990–9993. [PubMed] [Google Scholar]
- Höckel M., Hulla F. W., Risi S., Dose K. Kinetic studies on bacterial plasma membrane ATPase (F1). Nucleotide-induced long term inactivation of ATP hydrolyzing activity is linked to the formation of multiple "tight" enzyme nucleotide complexes. J Biol Chem. 1978 Jun 25;253(12):4292–4296. [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Jagendorf A. T., Uribe E. ATP formation caused by acid-base transition of spinach chloroplasts. Proc Natl Acad Sci U S A. 1966 Jan;55(1):170–177. doi: 10.1073/pnas.55.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa Y., Nukiwa N. Conversion of stable ATPase to labile ATPase by acetylation, and the alpha beta and alpha gamma subunit complexes during its reconstitution. Biochem Biophys Res Commun. 1981 Jun 16;100(3):1370–1376. doi: 10.1016/0006-291x(81)91975-6. [DOI] [PubMed] [Google Scholar]
- Kagawa Y. Reconstitution of the energy transformer, gate and channel subunit reassembly, crystalline ATPase and ATP synthesis. Biochim Biophys Acta. 1978 Sep 21;505(1):45–93. doi: 10.1016/0304-4173(78)90008-3. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Futai M. Release of the alpha subunit of coupling factor F1 ATPase from membranes of an uncoupled mutant of Escherichia coli. FEBS Lett. 1980 Jan 1;109(1):104–106. doi: 10.1016/0014-5793(80)81320-2. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Futai M. Structure and function of H+-ATPase: what we have learned from Escherichia coli H+-ATPase. Ann N Y Acad Sci. 1982;402:45–64. doi: 10.1111/j.1749-6632.1982.tb25731.x. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Horiuchi Y., Takagi M., Ishino Y., Futai M. Coupling factor F1 ATPase with defective beta subunit from a mutant of Escherichia coli. J Biochem. 1980 Sep;88(3):695–703. doi: 10.1093/oxfordjournals.jbchem.a133022. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Kayano T., Kiyasu T., Futai M. Nucleotide sequence of the genes for beta and epsilon subunits of proton-translocating ATPase from Escherichia coli. Biochem Biophys Res Commun. 1982 Apr 29;105(4):1257–1264. doi: 10.1016/0006-291x(82)90922-6. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Mabuchi K., Futai M. Nucleotide sequence of the promoter region of the gene cluster for proton-translocating ATPase from Escherichia coli and identification of the active promotor. Biochem Biophys Res Commun. 1982 Jul 30;107(2):568–575. doi: 10.1016/0006-291x(82)91529-7. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Mabuchi K., Kayano T., Noumi T., Sekiya T., Futai M. Nucleotide sequence of the genes for F0 components of the proton-translocating ATPase from Escherichia coli: prediction of the primary structure of F0 subunits. Biochem Biophys Res Commun. 1981 Nov 30;103(2):613–620. doi: 10.1016/0006-291x(81)90495-2. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Mabuchi K., Kayano T., Tamura F., Futai M. Nucleotide sequence of genes coding for dicyclohexylcarbodiimide-binding protein and the alpha subunit of proton-translocating ATPase of Escherichia coli. Biochem Biophys Res Commun. 1981 May 15;100(1):219–225. doi: 10.1016/s0006-291x(81)80085-x. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Miki T., Tamura F., Yura T., Futai M. Specialized transducing phage lambda carrying the genes for coupling factor of oxidative phosphorylation of Escherichia coli: increased synthesis of coupling factor on induction of prophage lambda asn. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1126–1130. doi: 10.1073/pnas.76.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa H., Noumi T., Futai M., Nitta T. Escherichia coli mutants defective in the gamma subunit of proton-translocating ATPase: intracistronic mapping of the defective site and the biochemical properties of the mutants. Arch Biochem Biophys. 1983 Jun;223(2):521–532. doi: 10.1016/0003-9861(83)90617-3. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Saito S., Futai M. Coupling factor ATPase from Escherichia coli. An uncA mutant (uncA401) with defective alpha subunit. J Biochem. 1978 Dec;84(6):1513–1517. doi: 10.1093/oxfordjournals.jbchem.a132276. [DOI] [PubMed] [Google Scholar]
- Kanazawa H., Tamura F., Mabuchi K., Miki T., Futai M. Organization of unc gene cluster of Escherichia coli coding for proton-translocating ATPase of oxidative phosphorylation. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7005–7009. doi: 10.1073/pnas.77.12.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanner B. I., Gutnick D. L. Use of neomycin in the isolation of mutants blocked in energy conservation in Escherichia coli. J Bacteriol. 1972 Jul;111(1):287–289. doi: 10.1128/jb.111.1.287-289.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashket E. R. Stoichiometry of the H+-ATPase of growing and resting, aerobic Escherichia coli. Biochemistry. 1982 Oct 26;21(22):5534–5538. doi: 10.1021/bi00265a024. [DOI] [PubMed] [Google Scholar]
- Khananshvili D., Gromet-Elhanan Z. Chemical modification of the beta-subunit isolated from a membrane-bound Fo-F1-ATP synthase: modification by 4-chloro-7-nitrobenzofurazan does not inhibit restoration of ATP synthesis or hydrolysis. Biochem Biophys Res Commun. 1982 Sep 30;108(2):881–887. doi: 10.1016/0006-291x(82)90913-5. [DOI] [PubMed] [Google Scholar]
- Khananshvili D., Gromet-Elhanan Z. Isolation and purification of an active gamma-subunit of the F0.F1-ATP synthase from chromatophore membranes of Rhodospirillum rubrum. The role of gamma in ATP synthesis and hydrolysis as compared to proton translocation. J Biol Chem. 1982 Oct 10;257(19):11377–11383. [PubMed] [Google Scholar]
- Khananshvili D., Gromet-Elhanan Z. The interaction of carboxyl group reagents with the Rhodospirillum rubrum F1-ATPase and its isolated beta-subunit. J Biol Chem. 1983 Mar 25;258(6):3720–3725. [PubMed] [Google Scholar]
- Kobayashi H., Murakami N., Unemoto T. Regulation of the cytoplasmic pH in Streptococcus faecalis. J Biol Chem. 1982 Nov 25;257(22):13246–13252. [PubMed] [Google Scholar]
- Krebbers E. T., Larrinua I. M., McIntosh L., Bogorad L. The maize chloroplast genes for the beta and epsilon subunits of the photosynthetic coupling factor CF1 are fused. Nucleic Acids Res. 1982 Aug 25;10(16):4985–5002. doi: 10.1093/nar/10.16.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauquin G., Pougeois R., Vignais P. V. 4-Azido-2-nitrophenyl phosphate, a new photoaffinity derivative of inorganic phosphate. Study of its interaction with the inorganic phosphate binding site of beef heart mitochondrial adenosine triphosphatase. Biochemistry. 1980 Sep 30;19(20):4620–4626. doi: 10.1021/bi00561a013. [DOI] [PubMed] [Google Scholar]
- Leimgruber R. M., Jensen C., Abrams A. Purification and characterization of the membrane adenosine triphosphatase complex from the wild-type and N,N'-dicyclohexylcarbodiimide-resistant strains of Streptococcus faecalis. J Bacteriol. 1981 Aug;147(2):363–372. doi: 10.1128/jb.147.2.363-372.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser M., Gromet-Elhanan Z. Demonstration of acid-base phosphorylation in chromatophores in the presence of a K+ diffusion potential. FEBS Lett. 1974 Aug 1;43(3):267–270. doi: 10.1016/0014-5793(74)80658-7. [DOI] [PubMed] [Google Scholar]
- Loo T. W., Bragg P. D. The DCCD-binding polypeptide is close to the F1 ATPase-binding site on the cytoplasmic surface of the cell membrane of Escherichia coli. Biochem Biophys Res Commun. 1982 May 31;106(2):400–406. doi: 10.1016/0006-291x(82)91124-x. [DOI] [PubMed] [Google Scholar]
- Lunardi J., Satre M., Vignais P. V. Exploration of adenosine 5'-diphosphate-adenosine 5'-triphosphate binding sites of Escherichia coli adenosine 5'-triphosphatase with arylazido adenine nucleotides. Biochemistry. 1981 Feb 3;20(3):473–480. doi: 10.1021/bi00506a005. [DOI] [PubMed] [Google Scholar]
- Lunardi J., Vignais P. V. Adenine nucleotide binding sites in chemically modified F1-ATPase: inhibitory effect of 4-chloro-7-nitrobenzofurazan on photolabeling by arylazido nucleotides. FEBS Lett. 1979 Jun 1;102(1):23–28. doi: 10.1016/0014-5793(79)80920-5. [DOI] [PubMed] [Google Scholar]
- Lunardi J., Vignais P. V. Studies of the nucleotide-binding sites on the mitochondrial F1-ATPase through the use of a photoactivable derivative of adenylyl imidodiphosphate. Biochim Biophys Acta. 1982 Oct 18;682(1):124–134. doi: 10.1016/0005-2728(82)90126-8. [DOI] [PubMed] [Google Scholar]
- Mabuchi K., Kanazawa H., Kayano T., Futai M. Nucleotide sequence of the gene coding for the delta subunit of proton translocating ATPase of Escherichia coli. Biochem Biophys Res Commun. 1981 Sep 16;102(1):172–179. doi: 10.1016/0006-291x(81)91504-7. [DOI] [PubMed] [Google Scholar]
- Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system. The DNA sequence of a mitochondrial ATPase gene in Saccharomyces cerevisiae. J Biol Chem. 1979 Jun 10;254(11):4617–4623. [PubMed] [Google Scholar]
- Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system: sequence analysis of a yeast mitochondrial ATPase gene containing the oli-2 and oli-4 loci. Cell. 1980 Jun;20(2):507–517. doi: 10.1016/0092-8674(80)90637-6. [DOI] [PubMed] [Google Scholar]
- Maeda M., Futai M., Anraku Y. Biochemical characterization of the uncA phenotype of Escherichia coli. Biochem Biophys Res Commun. 1976 May 23;76(2):331–338. doi: 10.1016/0006-291x(77)90729-x. [DOI] [PubMed] [Google Scholar]
- Maeda M., Kobayashi H., Futai M., Anraku Y. Non-covalently bound adenine nucleotides in adenosine triphosphatase of Escherichia coli. Biochem Biophys Res Commun. 1976 May 3;70(1):228–234. doi: 10.1016/0006-291x(76)91132-3. [DOI] [PubMed] [Google Scholar]
- Maeda M., Kobayashi H., Futai M., Anraku Y. Studies on the turnovers in vivo of adenosine di- and triphosphates in a coupling factor of Escherichia coli. J Biochem. 1977 Jul;82(1):311–314. doi: 10.1093/oxfordjournals.jbchem.a131687. [DOI] [PubMed] [Google Scholar]
- Maloney P. C. Energy coupling to ATP synthesis by the proton-translocating ATPase. J Membr Biol. 1982;67(1):1–12. doi: 10.1007/BF01868643. [DOI] [PubMed] [Google Scholar]
- Maloney P. C., Schattschneider S. Voltage sensitivity of the proton-translocating adenosine 5'-triphosphatase in Streptococcus lactis. FEBS Lett. 1980 Feb 11;110(2):337–340. doi: 10.1016/0014-5793(80)80106-2. [DOI] [PubMed] [Google Scholar]
- Matsubara H., Hase T., Hashimoto T., Tagawa K. Amino acid sequence of an intrinsic inhibitor of mitochondrial ATPase from yeast. J Biochem. 1981 Oct;90(4):1159–1165. doi: 10.1093/oxfordjournals.jbchem.a133568. [DOI] [PubMed] [Google Scholar]
- Matsuoka I., Takeda K., Futai M., Tonomura Y. Reactions of a fluorescent ATP analog, 2'-(5-dimethyl-aminonaphthalene-1-sulfonyl) amino-2'-deoxyATP, with E. coli F1-ATPase and its subunits: the roles of the high affinity binding site in the alpha subunit and the low affinity binding site in the beta subunit. J Biochem. 1982 Nov;92(5):1383–1398. doi: 10.1093/oxfordjournals.jbchem.a134062. [DOI] [PubMed] [Google Scholar]
- Miki T., Hiraga S., Nagata T., Yura T. Bacteriophage lambda carrying the Escherichia coli chromosomal region of the replication origin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):5099–5103. doi: 10.1073/pnas.75.10.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Kimura M., Hiraga S., Nagata T., Yura T. Cloning and physical mapping of the dnaA region of the Escherichia coli chromosome. J Bacteriol. 1979 Dec;140(3):817–824. doi: 10.1128/jb.140.3.817-824.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. Keilin's respiratory chain concept and its chemiosmotic consequences. Science. 1979 Dec 7;206(4423):1148–1159. doi: 10.1126/science.388618. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Koppenol W. H. Chemiosmotic ATPase mechanisms. Ann N Y Acad Sci. 1982;402:584–601. doi: 10.1111/j.1749-6632.1982.tb25785.x. [DOI] [PubMed] [Google Scholar]
- Muñoz E. Polymorphism and conformational dynamics of F1-ATPases from bacterial membranes. A model for the regulation of these enzymes on the basis of molecular plasticity. Biochim Biophys Acta. 1982 May 12;650(4):233–265. doi: 10.1016/0304-4157(82)90018-1. [DOI] [PubMed] [Google Scholar]
- Negrin R. S., Foster D. L., Fillingame R. H. Energy-transducing H+-ATPase of Escherichia coli. Reconstitution of proton translocation activity of the intrinsic membrane sector. J Biol Chem. 1980 Jun 25;255(12):5643–5648. [PubMed] [Google Scholar]
- Nelson N., Eytan E., Notsani B. E., Sigrist H., Sigrist-Nelson K., Gitler C. Isolation of a chloroplast N,N'-dicyclohexylcarbodiimide-binding proteolipid, active in proton translocation. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2375–2378. doi: 10.1073/pnas.74.6.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N., Kanner B. I., Gutnick D. L. Purification and properties of Mg2+-Ca2+ adenosinetriphosphatase from Escherichia coli. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2720–2724. doi: 10.1073/pnas.71.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N., Nelson H., Schatz G. Biosynthesis and assembly of the proton-translocating adenosine triphosphatase complex from chloroplasts. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1361–1364. doi: 10.1073/pnas.77.3.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J., Hansen F. G., Hoppe J., Friedl P., von Meyenburg K. The nucleotide sequence of the atp genes coding for the F0 subunits a, b, c and the F1 subunit delta of the membrane bound ATP synthase of Escherichia coli. Mol Gen Genet. 1981;184(1):33–39. doi: 10.1007/BF00271191. [DOI] [PubMed] [Google Scholar]
- Noumi T., Kanazawa H. Mutants of Escherichia coli H+-ATPase defective in the delta subunit of F1 and the b subunit of F0. Biochem Biophys Res Commun. 1983 Feb 28;111(1):143–149. doi: 10.1016/s0006-291x(83)80128-4. [DOI] [PubMed] [Google Scholar]
- Ohta S., Tsubo M., Oshima T., Yoshida M., Kagawa Y. Nucleotide binding to isolated alpha and beta subunits of proton translocating adenosine triphosphatase studied with circular dichroism. J Biochem. 1980 Jun;87(6):1609–1617. doi: 10.1093/oxfordjournals.jbchem.a132904. [DOI] [PubMed] [Google Scholar]
- Ohta S., Tsuboi M., Yoshida M., Kagawa Y. Intersubunit interactions in proton-translocating adenosine triphosphatase as revealed by hydrogen-exchange kinetics. Biochemistry. 1980 May 13;19(10):2160–2165. doi: 10.1021/bi00551a025. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Sone N., Hirata H., Yoshida M., Kagawa Y. Purified proton conductor in proton translocating adenosine triphosphatase of a thermophilic bacterium. J Biol Chem. 1977 Sep 10;252(17):6125–6131. [PubMed] [Google Scholar]
- Oren R., Weiss S., Garty H., Caplan S. R., Gromet-Elhanan Z. ATP synthesis catalyzed by the ATPase complex from Rhodospirillum rubrum reconstituted into phospholipid vesicles together with bacteriorhodopsin. Arch Biochem Biophys. 1980 Dec;205(2):503–509. doi: 10.1016/0003-9861(80)90133-2. [DOI] [PubMed] [Google Scholar]
- Paradies H. H. Crystallographic study of single crystals of mitochondrial coupling factor (BF1) from beef heart. Biochem Biophys Res Commun. 1980 Feb 27;92(4):1076–1082. doi: 10.1016/0006-291x(80)90396-4. [DOI] [PubMed] [Google Scholar]
- Paradies H. H. Effect of ATP on the translational diffusion coefficient of the alpha-subunit of Escherichia coli F1-ATPase. FEBS Lett. 1980 Nov 3;120(2):289–292. doi: 10.1016/0014-5793(80)80319-x. [DOI] [PubMed] [Google Scholar]
- Paradies H. H., Kagawa Y. Stability and flexibility of the alpha-subunit of F1-ATPase from the thermophilic bacterium PS3. FEBS Lett. 1981 Dec 21;136(1):3–7. doi: 10.1016/0014-5793(81)81203-3. [DOI] [PubMed] [Google Scholar]
- Paradies H. H., Mertens G., Schmid R., Schneider E., Altendorf K. Molecular properties of the ATP synthetase from Escherichia coli. Biochem Biophys Res Commun. 1981 Feb 12;98(3):595–606. doi: 10.1016/0006-291x(81)91156-6. [DOI] [PubMed] [Google Scholar]
- Paradies H. H., Schmidt U. D. Size and molecular parameters of adenosine triphosphatase from Escherichia coli. J Biol Chem. 1979 Jun 25;254(12):5257–5263. [PubMed] [Google Scholar]
- Paradies H. H. Size and shape of the alpha subunit of the (Ca,Mg)-dependent adenosinetriphosphate from Escherichia coli in solution in the presence and absence of ATP. Eur J Biochem. 1981 Aug;118(1):187–194. doi: 10.1111/j.1432-1033.1981.tb05504.x. [DOI] [PubMed] [Google Scholar]
- Pedersen P. L., Hullihen J., Wehrle J. P. Proton adenosine triphosphatase complex of rat liver. The effect of trypsin on the F1 and F0 moieties of the enzyme. J Biol Chem. 1981 Feb 10;256(3):1362–1369. [PubMed] [Google Scholar]
- Penefsky H. S. Mitochondrial ATPase. Adv Enzymol Relat Areas Mol Biol. 1979;49:223–280. doi: 10.1002/9780470122945.ch6. [DOI] [PubMed] [Google Scholar]
- Philosoph S., Binder A., Gromet-Elhanan Z. Coupling factor ATPase complex of Rhodospirillum rubrum. Purification and properties of a reconstitutively active single subunit. J Biol Chem. 1977 Dec 10;252(23):8747–8752. [PubMed] [Google Scholar]
- Philosoph S., Gromet-Elhanan Z. Antibodies to the F1-ATPase of Rhodospirillum rubrum and its purified native beta-subunit: inhibition of ATP-linked activities in R. rubrum and in lettuce. Eur J Biochem. 1981 Sep;119(1):107–113. doi: 10.1111/j.1432-1033.1981.tb05583.x. [DOI] [PubMed] [Google Scholar]
- Pick U., Racker E. Purification and reconstitution of the N,N'-dicyclohexylcarbodiimide-sensitive ATPase complex from spinach chloroplasts. J Biol Chem. 1979 Apr 25;254(8):2793–2799. [PubMed] [Google Scholar]
- Pougeois R., Satre M., Vignais P. V. Characterization of dicyclohexylcarbodiimide binding site on coupling factor 1 of mitochondrial and bacterial membrane-bound ATPases. FEBS Lett. 1980 Aug 11;117(1):344–348. doi: 10.1016/0014-5793(80)80977-x. [DOI] [PubMed] [Google Scholar]
- Rosen B. P., Brey R. N., Hasan S. M. Energy transduction in Escherichia coli: new mutation affecting the Fo portion of the ATP synthetase complex. J Bacteriol. 1978 Jun;134(3):1030–1038. doi: 10.1128/jb.134.3.1030-1038.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen B. P., Hasan S. M. Purification of an N,N'-dicyclohexylcarbodiimide-sensitive ATPase from Escherichia coli. FEBS Lett. 1979 Aug 15;104(2):339–342. doi: 10.1016/0014-5793(79)80847-9. [DOI] [PubMed] [Google Scholar]
- Rosen B. P. Restoration of active transport in an Mg2+-adenosine triphosphatase-deficient mutant of Escherichia coli. J Bacteriol. 1973 Dec;116(3):1124–1129. doi: 10.1128/jb.116.3.1124-1129.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G., Argos P. Protein folding. Annu Rev Biochem. 1981;50:497–532. doi: 10.1146/annurev.bi.50.070181.002433. [DOI] [PubMed] [Google Scholar]
- Rott R., Nelson N. Purification and immunological properties of proton-ATPase complexes from yeast and rat liver mitochondria. J Biol Chem. 1981 Sep 10;256(17):9224–9228. [PubMed] [Google Scholar]
- Ryrie I. J. Reconstitution of ATP-32-Pi exchange by phospholipid addition to the purified oligomycin-sensitive ATPase from yeast mitochondria. Arch Biochem Biophys. 1975 Jun;168(2):704–711. doi: 10.1016/0003-9861(75)90304-5. [DOI] [PubMed] [Google Scholar]
- Rögner M., Ohno K., Hamamoto T., Sone N., Kagawa Y. Net ATP synthesis in H+ -atpase macroliposomes by an external electric field. Biochem Biophys Res Commun. 1979 Nov 14;91(1):362–367. doi: 10.1016/0006-291x(79)90627-2. [DOI] [PubMed] [Google Scholar]
- Sakamoto J., Tonomura Y. Synthesis of enzyme-bound ATP by mitochondrial soluble F1-ATPase in the presence of dimethylsulfoxide. J Biochem. 1983 Jun;93(6):1601–1614. doi: 10.1093/oxfordjournals.jbchem.a134299. [DOI] [PubMed] [Google Scholar]
- Saraste M., Gay N. J., Eberle A., Runswick M. J., Walker J. E. The atp operon: nucleotide sequence of the genes for the gamma, beta, and epsilon subunits of Escherichia coli ATP synthase. Nucleic Acids Res. 1981 Oct 24;9(20):5287–5296. doi: 10.1093/nar/9.20.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satre M., Bof M., Issartel J. P., Vignais P. V. Chemical modification of F1-ATPase by dicyclohexylcarbodiimide: application to analysis of the stoichiometry of subunits in Escherichia coli F1. Biochemistry. 1982 Sep 14;21(19):4772–4776. doi: 10.1021/bi00262a038. [DOI] [PubMed] [Google Scholar]
- Satre M., Klein G., Vignais P. V. Isolation of Escherichia coli mutants with an adenosine triphosphatase insensitive to aurovertin. J Bacteriol. 1978 Apr;134(1):17–23. doi: 10.1128/jb.134.1.17-23.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satre M., Lunardi J., Pougeois R., Vignais P. V. Inactivation of Escherichia coli BF1-ATPase by dicyclohexylcarbodiimide. Chemical modification of the beta subunit. Biochemistry. 1979 Jul 10;18(14):3134–3140. doi: 10.1021/bi00581a034. [DOI] [PubMed] [Google Scholar]
- Satre M., Zaccaï G. Small angle neutron scattering of Escherichia coli BF1-ATPase. FEBS Lett. 1979 Jun 15;102(2):244–248. doi: 10.1016/0014-5793(79)80010-1. [DOI] [PubMed] [Google Scholar]
- Schairer H. U., Hoppe J., Sebald W., Friedl P. Topological and functional aspects of the proton conductor, F0, of the Escherichia coli ATP-synthase. Biosci Rep. 1982 Aug;2(8):631–639. doi: 10.1007/BF01314228. [DOI] [PubMed] [Google Scholar]
- Scheurich P., Schäfer H. J., Dose K. 8-Azido-adenosine 5'-triphosphate as a photoaffinity label for bacterial F1 ATPase. Eur J Biochem. 1978 Jul 17;88(1):253–257. doi: 10.1111/j.1432-1033.1978.tb12445.x. [DOI] [PubMed] [Google Scholar]
- Schindler H., Nelson N. Proteolipid of adenosinetriphosphatase from yeast mitochondria forms proton-selective channels in planar lipid bilayers. Biochemistry. 1982 Nov 9;21(23):5787–5794. doi: 10.1021/bi00266a010. [DOI] [PubMed] [Google Scholar]
- Schmitt M., Rittinghaus K., Scheurich P., Schwulera U., Dose K. Immunological properties of membrane-bound adenosine triphosphatase: immunological identification of rutamycin-sensitive F0.F1ATPase from Micrococcus luteus ATCC 4698 established by crossed immunoelectrophoresis. Biochim Biophys Acta. 1978 Jun 2;509(3):410–418. doi: 10.1016/0005-2736(78)90235-3. [DOI] [PubMed] [Google Scholar]
- Schneider E., Altendorf K. ATP synthetase (F1F0) of Escherichia coli K-12. High-yield preparation of functional F0 by hydrophobic affinity chromatography. Eur J Biochem. 1982 Aug;126(1):149–153. doi: 10.1111/j.1432-1033.1982.tb06759.x. [DOI] [PubMed] [Google Scholar]
- Schneider E., Müller H. W., Rittinghaus K., Thiele V., Schwuléra U., Dose K. Properties of the F0F1 ATPase complex from Rhodospirillum rubrum chromatophores, solubilized by Triton X-100. Eur J Biochem. 1979 Jul;97(2):511–517. doi: 10.1111/j.1432-1033.1979.tb13139.x. [DOI] [PubMed] [Google Scholar]
- Schäfer H. J., Scheurich P., Rathgeber G., Dose K. Fluorescent photoaffinity labeling of F1 ATPase from Micrococcus luteus with 8-azido-1,N6-etheno-adenosine 5'-triphosphate. Anal Biochem. 1980 May 1;104(1):106–111. doi: 10.1016/0003-2697(80)90282-1. [DOI] [PubMed] [Google Scholar]
- Sebald W., Friedl P., Schairer H. U., Hoppe J. Structure and genetics of the H+-conducting F0 portion of the ATP synthase. Ann N Y Acad Sci. 1982;402:28–44. doi: 10.1111/j.1749-6632.1982.tb25730.x. [DOI] [PubMed] [Google Scholar]
- Sebald W., Machleidt W., Wachter E. N,N'-dicyclohexylcarbodiimide binds specifically to a single glutamyl residue of the proteolipid subunit of the mitochondrial adenosinetriphosphatases from Neurospora crassa and Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Feb;77(2):785–789. doi: 10.1073/pnas.77.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda M., Kanazawa H., Tsuchiya T., Futai M. Conformational change of the alpha subunit of Escherichia coli F1 ATPase: ATP changes the trypsin sensitivity of the subunit. Arch Biochem Biophys. 1983 Feb 1;220(2):398–404. doi: 10.1016/0003-9861(83)90429-0. [DOI] [PubMed] [Google Scholar]
- Senior A. E. Divalent metals in beef heart mitochondrial adenosine triphosphatase. Demonstration of the metals in membrane-bound enzyme and studies of the interconversion of the "1-Mg" and "2-Mg" forms of the enzyme. J Biol Chem. 1981 May 25;256(10):4763–4767. [PubMed] [Google Scholar]
- Senior A. E., Downie J. A., Cox G. B., Gibson F., Langman L., Fayle D. R. The uncA gene codes for the alpha-subunit of the adenosine triphosphatase of Escherichia coli. Electrophoretic analysis of uncA mutant strains. Biochem J. 1979 Apr 15;180(1):103–109. doi: 10.1042/bj1800103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. E., Fayle D. R., Downie J. A., Gibson F., Cox G. B. Properties of membranes from mutant strains of Escherichia coli in which the beta-subunit of the adenosine triphosphatase is abnormal. Biochem J. 1979 Apr 15;180(1):111–118. doi: 10.1042/bj1800111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. E., Langman L., Cox G. B., Gibson F. Oxidative phosphorylation in Escherichia coli. Characterization of mutant strains in which F1-ATPase contains abnormal beta-subunits. Biochem J. 1983 Feb 15;210(2):395–403. doi: 10.1042/bj2100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior A. E., Richardson L. V., Baker K., Wise J. G. Tight divalent cation-binding sites of soluble adenosine triphosphatase (F1) from beef heart mitochondria and Escherichia coli. J Biol Chem. 1980 Aug 10;255(15):7211–7217. [PubMed] [Google Scholar]
- Senior A. E., Wise J. G. The proton-ATPase of bacteria and mitochondria. J Membr Biol. 1983;73(2):105–124. doi: 10.1007/BF01870434. [DOI] [PubMed] [Google Scholar]
- Serrano R., Kanner B. I., Racker E. Purification and properties of the proton-translocating adenosine triphosphatase complex of bovine heart mitochondria. J Biol Chem. 1976 Apr 25;251(8):2453–2461. [PubMed] [Google Scholar]
- Shavit N. Energy transduction in chloroplasts: structure and function of the ATPase complex. Annu Rev Biochem. 1980;49:111–138. doi: 10.1146/annurev.bi.49.070180.000551. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. B., Sternweis P. C. Subunit specific antisera to the Escherichia coli ATP synthase: effects on ATPase activity, energy transduction, and enzyme assembly. Arch Biochem Biophys. 1982 Aug;217(1):376–387. doi: 10.1016/0003-9861(82)90514-8. [DOI] [PubMed] [Google Scholar]
- Sone N., Yoshida M., Hirata H., Kagawa Y. Adenosine triphosphate synthesis by electrochemical proton gradient in vesicles reconstituted from purified adenosine triphosphatase and phospholipids of thermophilic bacterium. J Biol Chem. 1977 May 10;252(9):2956–2960. [PubMed] [Google Scholar]
- Sone N., Yoshida M., Hirata H., Kagawa Y. Resolution of the membrane moiety of the H+-ATPase complex into two kinds of subunits. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4219–4223. doi: 10.1073/pnas.75.9.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitsberg V., Haworth R. The crystallization of beef heart mitochondrial adenosine triphosphatase. Biochim Biophys Acta. 1977 May 27;492(1):237–240. doi: 10.1016/0005-2795(77)90231-8. [DOI] [PubMed] [Google Scholar]
- Sternweis P. C., Smith J. B. Characterization of the purified membrane attachment (beta) subunit of the proton translocating adenosine triphosphatase from Escherichia coli. Biochemistry. 1977 Sep 6;16(18):4020–4025. doi: 10.1021/bi00637a013. [DOI] [PubMed] [Google Scholar]
- Sternweis P. C. The epsilon subunit of Escherichia coli coupling factor 1 is required for its binding to the cytoplasmic membrane. J Biol Chem. 1978 May 10;253(9):3123–3128. [PubMed] [Google Scholar]
- Takeda K., Hirano M., Kanazawa H., Nukiwa N., Kagawa Y., Futai M. Hybrid ATPase's formed from subunits of coupling factor F1's of Escherichia coli and thermophilic bacterium PS3. J Biochem. 1982 Feb;91(2):695–701. doi: 10.1093/oxfordjournals.jbchem.a133742. [DOI] [PubMed] [Google Scholar]
- Tamura F., Kanzawa H., Tsuchiya T., Futai M. Structural gene coding for the dicyclohexylcarbodiimide-binding protein of the protein-translocating ATPase from Escherichia coli. Locus of the gene in the F1--F0 gene cluster. FEBS Lett. 1981 May 5;127(1):48–52. doi: 10.1016/0014-5793(81)80338-9. [DOI] [PubMed] [Google Scholar]
- Thayer W. S., Hinkle P. C. Kinetics of adenosine triphosphate synthesis in bovine heart submitochondrial particles. J Biol Chem. 1975 Jul 25;250(14):5336–5336. [PubMed] [Google Scholar]
- Thipayathasana P. Isolation and properties of Escherichia coli ATPase mutants with altered divalent metal specificity for ATP hydrolysis. Biochim Biophys Acta. 1975 Oct 10;408(1):47–57. doi: 10.1016/0005-2728(75)90157-7. [DOI] [PubMed] [Google Scholar]
- Ting L. P., Wang J. H. Functional groups at the catalytic site of BF1 adenosinetriphosphatase from Escherichia coli. Biochemistry. 1982 Jan 19;21(2):269–275. doi: 10.1021/bi00531a011. [DOI] [PubMed] [Google Scholar]
- Todd R. D., Douglas M. G. A model for the structure of the yeast mitochondrial adenosine triphosphatase complex. J Biol Chem. 1981 Jul 10;256(13):6984–6989. [PubMed] [Google Scholar]
- Todd R. D., Douglas M. G. Structure of the yeast mitochondrial adenosine triphosphatase. Results of trypsin degradation. J Biol Chem. 1981 Jul 10;256(13):6990–6994. [PubMed] [Google Scholar]
- Tzagoloff A., Macino G., Sebald W. Mitochondrial genes and translation products. Annu Rev Biochem. 1979;48:419–441. doi: 10.1146/annurev.bi.48.070179.002223. [DOI] [PubMed] [Google Scholar]
- Verheijen J. H., Postma P. W., van Dam K. Specific labelling of the (Ca2+ + Mg2+)-ATPase of Escherichia coli with 8-azido-ATP and 4-chloro-7-nitrobenzofurazan. Biochim Biophys Acta. 1978 May 10;502(2):345–353. doi: 10.1016/0005-2728(78)90054-3. [DOI] [PubMed] [Google Scholar]
- Viebrock A., Perz A., Sebald W. The imported preprotein of the proteolipid subunit of the mitochondrial ATP synthase from Neurospora crassa. Molecular cloning and sequencing of the mRNA. EMBO J. 1982;1(5):565–571. doi: 10.1002/j.1460-2075.1982.tb01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter E., Schmid R., Deckers G., Altendorf K. Amino acid replacement in dicyclohexylcarbodiimide-reactive proteins from mutant strains of Escherichia coli defective in the energy-transducing ATPase complex. FEBS Lett. 1980 May 5;113(2):265–270. doi: 10.1016/0014-5793(80)80606-5. [DOI] [PubMed] [Google Scholar]
- Wagenvoord R. J., Van der Kraan I., Kemp A. Specific photolabelling of beef-heart mitochondrial ATPase by 8-azido-ATP. Biochim Biophys Acta. 1977 Apr 11;460(1):17–24. doi: 10.1016/0005-2728(77)90147-5. [DOI] [PubMed] [Google Scholar]
- Wakabayashi T., Kubota M., Yoshida M., Kagawa Y. Structure of ATPase (coupling factor TF1) from a thermophilic bacterium. J Mol Biol. 1977 Dec 5;117(2):515–519. doi: 10.1016/0022-2836(77)90140-1. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Eberle A., Gay N. J., Runswick M. J., Saraste M. Conservation of structure in proton-translocating ATPases of Escherichia coli and mitochondria. Biochem Soc Trans. 1982 Aug;10(4):203–206. doi: 10.1042/bst0100203. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Runswick M. J., Saraste M. Subunit equivalence in Escherichia coli and bovine heart mitochondrial F1F0 ATPases. FEBS Lett. 1982 Sep 20;146(2):393–396. doi: 10.1016/0014-5793(82)80960-5. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. E. coli F1-ATPase interacts with a membrane protein component of a proton channel. Nature. 1982 Aug 26;298(5877):867–869. doi: 10.1038/298867a0. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb M. R., Grubmeyer C., Penefsky H. S., Trentham D. R. The stereochemical course of phosphoric residue transfer catalyzed by beef heart mitochondrial ATPase. J Biol Chem. 1980 Dec 25;255(24):11637–11639. [PubMed] [Google Scholar]
- Wise J. G., Latchney L. R., Senior A. E. The defective proton-ATPase of uncA mutants of Escherichia coli. Studies of nucleotide binding sites, bound aurovertin fluorescence, and labeling of essential residues of the purified F1-ATPase. J Biol Chem. 1981 Oct 25;256(20):10383–10389. [PubMed] [Google Scholar]
- Witt H. T. Energy conversion in the functional membrane of photosynthesis. Analysis by light pulse and electric pulse methods. The central role of the electric field. Biochim Biophys Acta. 1979 Mar 14;505(3-4):355–427. doi: 10.1016/0304-4173(79)90008-9. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Arfsten A. E., Nomura M. In vitro expression of Escherichia coli ribosomal protein genes: autogenous inhibition of translation. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1837–1841. doi: 10.1073/pnas.77.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Allison W. S., Esch F. S., Futai M. The specificity of carboxyl group modification during the inactivation of the Escherichia coli F1-ATPase with dicyclohexyl[14C]carbodiimide. J Biol Chem. 1982 Sep 10;257(17):10033–10037. [PubMed] [Google Scholar]
- Yoshida M., Okamoto H., Sone N., Hirata H., Kagawa Y. Reconstitution of thermostable ATPase capable of energy coupling from its purified subunits. Proc Natl Acad Sci U S A. 1977 Mar;74(3):936–940. doi: 10.1073/pnas.74.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Poser J. W., Allison W. S., Esch F. S. Identification of an essential glutamic acid residue in the beta subunit of the adenosine triphosphatase from the thermophilic bacterium PS3. J Biol Chem. 1981 Jan 10;256(1):148–153. [PubMed] [Google Scholar]
- Yoshida M., Sone N., Hirata H., Kagawa Y. Reconstitution of adenosine triphosphatase of thermophilic bacterium from purified individual subunits. J Biol Chem. 1977 May 25;252(10):3480–3485. [PubMed] [Google Scholar]
- Yoshida M., Sone N., Hirata H., Kagawa Y., Ui N. Subunit structure of adenosine triphosphatase. Comparison of the structure in thermophilic bacterium PS3 with those in mitochondria, chloroplasts, and Escherichia coli. J Biol Chem. 1979 Oct 10;254(19):9525–9533. [PubMed] [Google Scholar]
- Zurawski G., Bottomley W., Whitfeld P. R. Structures of the genes for the beta and epsilon subunits of spinach chloroplast ATPase indicate a dicistronic mRNA and an overlapping translation stop/start signal. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6260–6264. doi: 10.1073/pnas.79.20.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenburg K., Hansen F. G., Riise E., Bergmans H. E., Meijer M., Messer W. Origin of replication, oriC, of the Escherichia coli K12 chromosome: genetic mapping and minichromosome replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):121–128. doi: 10.1101/sqb.1979.043.01.018. [DOI] [PubMed] [Google Scholar]
- von Meyenburg K., Jørgensen B. B., Nielsen J., Hansen F. G., Michelsen O. The membrane bound ATP synthase of Escherichia coli: a review of structural and functional analyses of the atp operon. Tokai J Exp Clin Med. 1982;7 (Suppl):23–31. [PubMed] [Google Scholar]
- von Meyenburg K., Jørgensen B. B., Nielsen J., Hansen F. G. Promoters of the atp operon coding for the membrane-bound ATP synthase of Escherichia coli mapped by Tn10 insertion mutations. Mol Gen Genet. 1982;188(2):240–248. doi: 10.1007/BF00332682. [DOI] [PubMed] [Google Scholar]


