Abstract
Array-based comparative genomic hybridisation (aCGH) has diverse applications in cancer gene discovery and translational research. Currently, aCGH is performed primarily using high molecular weight DNA samples and its application to formalin-fixed and paraffin-embedded (FFPE) tissues remains to be established. To explore how aCGH can be reliably applied to archival FFPE tissues and whether it is possible to apply aCGH to small numbers of cells microdissected from FFPE tissue sections, we have systematically performed aCGH on 15 pairs of matched frozen and FFPE glioblastoma tissues using a well established in-house human 1Mb BAC/PAC genomic array. By spiking glioblastoma DNA with normal DNA, we demonstrated that at least 70% of tumour DNA was required for reliable aCGH analysis. Using aCGH data from frozen tissue as a reference, it was found that only FFPE glioblastoma tissues that supported PCR amplification of >300bp DNA fragment provided high quality, reproducible aCGH data. The presence of necrosis in a tissue specimen had an adverse effect on the quality of aCGH, while fixation in formalin for up to 96 hours of fresh tissue did not appear to affect the quality of the result. As little as 10-20ng DNA from frozen or FFPE tissues could be readily used for aCGH analysis following whole genome amplification. Furthermore, as few as 2000 microdissected cells from haematoxylin stained slides of archival FFPE tissues could be successfully used for aCGH investigations when whole genome amplification was used. By careful assessment of DNA integrity and review of histology, to exclude necrosis and select specimens with a high proportion of tumour cells, it is feasible to pre-select archival FFPE tissues adequate for aCGH analysis. With the help of microdissection and whole genome amplification, it is also possible to apply aCGH to histologically defined lesions, such as carcinoma in situ.
Keywords: array CGH, archival fixed tissue, microdissection, whole genome amplification, glioblastoma
Comparative genomic hybridisation (CGH) is a relatively new technology designed to allow high throughput screening of chromosomal gains and losses in diseases, particularly cancer. Traditionally, CGH was carried out by competitive in situ hybridisation of differentially labelled tumour and normal DNA to normal metaphase chromosome spreads. The advent of array-based CGH (aCGH), which uses genome-mapped and sequence-verified genomic clones arrayed on glass slides as the hybridisation target, has radically transformed this technique. aCGH not only confers high resolution but is also exceptionally versatile in its design and application. Essentially, the resolution of aCGH is determined by the size and/or gaps between genomic clones used for array construction. Genome-wide BAC arrays at megabase (1Mb) or submegabase (100kb) size are now available for screening of genomic gains/losses (1, 2). Genomic copy number changes can be further characterised by use of chromosome- specific or customised tiled arrays (3, 4), which can be tailor-made at any desired resolution. In addition, chromosome-specific tiled arrays can be used for characterisation of chromosomal translocation breakpoints when the derivative chromosome is isolated and used as the source of DNA (5). Clearly, aCGH has immense potential in disease gene discovery and translational research.
aCGH analyses were initially developed using high molecular weight DNA samples from fresh/frozen tissues or cells. The use of FFPE tissues as a source of DNA has many advantages. Such tissues are available as a huge resource, with detailed histological and phenotypic characterisation and valuable clinical-pathological follow-up data. It would be immensely beneficial if aCGH could be applied to such materials. So far, a few studies have used FFPE-derived DNA for aCGH (6-9), and it may be realistic to use small amounts of DNA extracted from whole tissue sections of frozen or FFPE specimens for aCGH following whole genome amplification (WGA) (10-12). Nonetheless, several critical issues remain to be investigated. Firstly, because DNA quality from FFPE tissues is variable, is it possible to reliably identify suitable archival FFPE tissues for aCGH? Secondly, can aCGH be applied to small numbers of cells microdissected from archival FFPE tissue samples? Thirdly, is it possible to distinguish true tumour-derived alterations from experimental artefacts? To address these issues, we have systematically performed aCGH on 15 pairs of matched frozen and FFPE glioblastoma tissues and developed a practical protocol which enables pre-selection of archival FFPE cases adequate for aCGH analysis and the successful application of the technique to DNA samples extracted from as few as 2,000 microdissected cells.
MATERIALS AND METHODS
Tumour tissues and fixation
The study was based on 15 pairs of matched fresh-frozen and FFPE glioblastoma tissues (11 xenografts archived for six years and 4 primary glioblastomas archived for fifteen years). All FFPE tissues were fixed overnight in 10% buffered formalin, routinely processed and paraffin-embedded. Use of these archival tissues for research was approved by the Ethics Committee of the Karolinska Hospital and Cambridge Local Research Ethics Committee.
In addition, to examine the effect of tissue fixation on aCGH, a glioblastoma xenograft tissue sample, previously archived at −80°C for six years, was divided into five equal pieces, fixed in 10% buffered formalin at room temperature for various times (12, 24, 48, 72 and 96 hrs), then routinely processed and paraffin-embedded.
Microdissection and DNA extraction
DNA was extracted from fresh-frozen tumour tissues and blood samples as previously described. FFPE tissue sections were de-waxed twice in xylene, washed in 100% ethanol, and digested overnight with 1mg/ml proteinase K (Qiagen, Valencia, CA) at 56°C in a 200μl reaction mixture containing 30mM Tris-Cl (pH 8.0), 10mM EDTA and 1% sodium dodecyl sulphate. The sample was then mixed with an equal volume of phenol-chloroform-isoamyl (25:24:1) (pH 7.9) (Ambion, TX, USA) and centrifuged through a phase-lock heavy gel (Eppendorf AG, Hamburg, Germany). DNA was ethanol precipitated overnight at −20°C and dissolved in 20μl of 10mM Tris/1mM EDTA.
Crude microdissection was performed on freshly prepared haematoxylin stained slides to avoid necrotic areas or to isolate various numbers of tumour cells. Microdissected cells were similarly digested with proteinase K in a 25μl reaction mixture as above and DNA was purified using a DNA micro-kit (Qiagen).
DNA concentration was determined using PicoGreen™ (Molecular Probes, Oregon, USA).
Assessment of DNA integrity
The integrity of DNA samples from FFPE tissue was assessed by PCR of variable sized DNA fragments (100bp, 200bp, 300bp, 400bp and 600bp) in separate reactions using 10ng of template DNA as described previously (14). PCR products were analysed by electrophoresis on 2% agarose gels.
Whole Genome Amplification (WGA)
Various amounts (10-20ng) of DNA from fresh-frozen or FFPE tissue was amplified using GenomePlex™ WGA kit (Rubicon Genomics, Ann Arbor, MI). Amplified genomic DNA was purified using DNA Clean and Concentrator (Zymo Research, CA, USA) and quantified using PicoGreen.
Array CGH
CGH arrays were constructed in-house based on protocols used by the Wellcome Trust Sanger Institute, Cambridge, UK (2) and comprised 3038 analysable BAC clones spaced at approximately 1Mb intervals across the whole genome. Construction and validation of the 1Mb genomic array has been previously described (2, 15). Briefly, clone DNA was extracted and amplified using three DOP primers which were subsequently mixed and amplified using a 5′-amine modified universal primer. Amine-linked PCR products were arrayed onto amine-binding slides (CodeLink, Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) in duplicate. Each array was composed of 24 blocks; Drosophila clones and clones from individual chromosomes were evenly distributed throughout all blocks.
Labelling of test (tumour) and reference (control) DNA, array hybridisation and washing were carried out essentially as previously described (3, 15). Briefly, 400ng of test (tumour) and reference DNA were labelled using a Bioprime Labelling Kit (Invitrogen, Carlsbad, CA) with a modified dNTP reaction mixture. Test DNA was hybridised with sex-mismatched reference DNA from samples of pooled blood from 20 normal males or 20 normal females. Labelled and purified test and reference DNA were pooled and co-precipitated with 45μg Cot-1 DNA (Roche Diagnostics, Mannheim, Germany) and 400μg herring sperm DNA (Sigma-Aldrich, St Louis, MO). The precipitated DNA was dissolved in hybridisation buffer and hybridised to an array that had been pre-hybridised with 80μg Cot-1 DNA and 400μg herring sperm DNA for 2 hours. Arrays were allowed to hybridise overnight at 37°C then washed as described (3).
aCGH data analysis
aCGH slides were scanned using an Axon 4100A scanner (Axon Instruments, Burlingame, CA). Scanned images were quantified using GenePix Pro 5.1 software (Axon Instruments) and primary data analysis and normalisation carried out using Microsoft Excel (3). An average between the duplicate spotted BACs was calculated and results were analysed using plots of log2-transformed normalised Cy5:Cy3 intensity ratios against clone position.
The mean and standard deviation (SD) used as the threshold value for identifying genomic gains and losses were determined from 4 normal male/female hybridisations. Mean ± 3SD (equivalent to log2 value ± 0.19) was used as the threshold for aCGH using DNA from frozen tissue. Systematic analysis of the mean ± various SD (3, 3.2, 3.4, 3.6, 3.8, 4) on 9 good quality aCGH FFPE glioblastoma samples (5 xenograft and 4 primary) data was used to determine the threshold value for aCGH with and without WGA. Altered clones were identified and percentages of concordance and non-concordance with those derived from the corresponding frozen tissue were calculated. The mean ± 3.2SD (log2 value ± 0.20) and mean ± 3.4SD (log2 value ± 0.22) was chosen as the cut-off for FFPE tissue and FFPE with WGA respectively. The three cut-off values chosen allowed confident detection of single copy gain or loss since the imbalance involving single copy change as demonstrated by chromosome X clones between normal male / female hybridisations typically showed log2 ratio changes in the range of 0.55-0.67. The suitability of these cut-off values was further verified by detection of single copy gains/losses even when the percentage of tumour cells was at 75% (detailed in Results).
Alterations that involve consecutive clones are likely to represent true genomic alterations which commonly affect a relatively large chromosomal region (16). However, it is more difficult to distinguish changes affecting single clones that may result from random or experimental variations, such as mismapped clones or cross hybridisation due to repetitive sequences. To test this, we compared the reproducibility of aCGH changes in four replicate aCGH hybridisations, using a DNA sample from frozen xenograft tissue.
Statistics
Comparison of the aCGH data of the same specimen among different experimental conditions (Pearson’s correlation test) and unsupervised hierarchical clustering was carried out using stats package in R version 2.1.1 (17) http://www.R-project.org.
RESULTS
Reproducibility of aCGH changes involving consecutive clones or single clones
To compare the reproducibility of aCGH changes involving consecutive or single clones, we performed 4 independent aCGH analyses using the same DNA sample from a frozen xenograft tissue. The concordant clones from all 4 independent experiments were used as reference to assess the reproducibility of each independent hybridisation. As expected, aCGH changes affecting two or more consecutive clones were highly reproducible (mean 88%), while those involving single clones were not (mean 16%) (Table 1). Similar results were seen when using DNA from frozen and FFPE tissues of the same cases (Supplemental table 1). This finding indicated that most of the changes affecting single clones were likely to be the result of experimental artefacts. For this reason, assessment of concordance between frozen and FFPE tissue under various experimental conditions focused on changes involving two or more consecutive clones.
Table 1.
aCGH using DNA from frozen tissue: reproducibility between consecutive and single clones affected
| Total no. clones affected (cut-off value mean +/− 3SD) |
Consecutive clones affected | Single clones affected | |||
|---|---|---|---|---|---|
| No. clones affected |
Percentage reproducibility |
No. clones affected |
Percentage reproducibility |
||
| Reference* | 292 | 283 | 9 | ||
| A951 Replicate 1 | 372 | 338 | 84% | 34 | 26% |
| A951 Replicate 2 | 476 | 359 | 79% | 117 | 8% |
| A951 Replicate 3 | 363 | 305 | 93% | 58 | 16% |
| A951- Replicate 4 | 353 | 291 | 97% | 62 | 15% |
| mean | 391 | 323 | 88% | 68 | 16% |
reference was calculated by extracting concordant consecutive and single clones in all four individual aCGH experiments, as these are less likely to be due to experimental artefacts.
aCGH: minimum proportion of tumour DNA required for reliable aCGH analysis
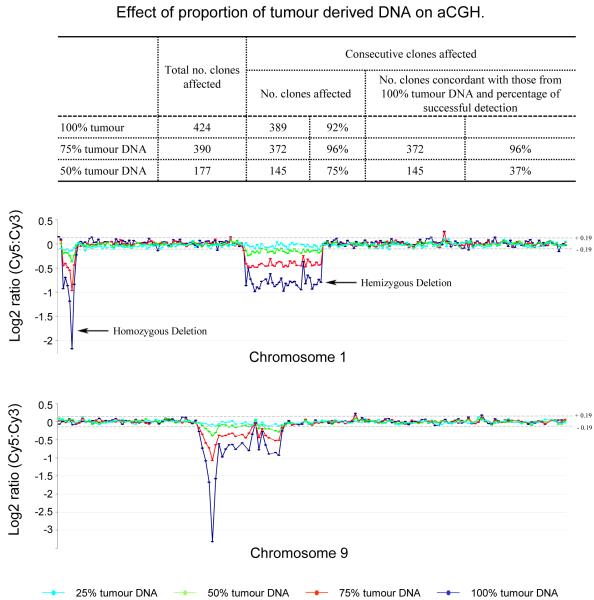
Tumour tissues invariably contain non-neoplastic cells. To examine the minimal proportion of tumour DNA required for reliable aCGH analysis, a DNA sample with known homozygous and hemizygous deletions from one frozen glioblastoma xenograft (X1) was mixed with various amounts of non-tumour DNA and then subjected to aCGH analyses. The ability of the aCGH technology to detect genomic gains/losses was shown to critically depend on the proportion of tumour DNA (Figure 1). When tumour DNA was ≤50%, both chromosomal gain and loss (including those affecting relatively large regions) became difficult to recognise (Figure 1) and their concordance with aCGH data from 100% tumour DNA was poor. Nonetheless, reliable CGH results could be obtained when the proportion of tumour DNA was ≥ 75%.
Figure 1.
Sensitivity of aCGH. High molecular DNA from a frozen glioblastoma xenograft was diluted with DNA from normal blood samples at various ratios and subjected to 1Mb aCGH analysis. The log2 ratio of tumour/reference fluorescence is shown for chromosomes 1 and 9. Clones are arranged in genomic order from pter to qter. The data indicates that at least 75% of tumour DNA is required for reliable aCGH analysis.
aCGH using DNA from FFPE tissues – effect of DNA integrity and necrosis
To examine to what extent archival FFPE tissues could be reliably used for aCGH analysis, DNA samples from whole tissue sections of archival FFPE glioblastoma xenografts were subjected to aCGH analysis and the resulting data compared to that from corresponding frozen tissues. The quality of aCGH data from FFPE tissues was then compared to the integrity of DNA samples.
Of the 11 FFPE glioblastoma xenografts with matched frozen tissues, sufficient DNA for aCGH analysis could be extracted from 9 cases. All 9 cases yielded a quality DNA sample (as assessed spectrophotometrically) with a similar smear pattern on 1% agarose gels, but gave rise to dramatically variable aCGH data quality when compared to that from the corresponding frozen tissue (Table 2, Figure 2). In 4 cases (X1, X3-5), a good correlation (r = 0.83-0.98) between data generated from FFPE and frozen tissues was obtained and 79-98% of CGH changes involving 2 or more consecutive clones identified from DNA samples of frozen tissues were seen when using DNA samples from FFPE tissue (Table3). It has to be stated that the true reproducibility of aCGH on FFPE tissue was most likely underestimated as 12% of aCGH changes involving 2 or more consecutive clones seen from frozen tissues were not reproducible (Table 1). In the remaining 5 cases, the quality of aCGH data from FFPE tissues was poor (Table 2).
Table 2.
aCGH using DNA from FFPE tissue: minimum requirement for DNA integrity, DNA quantity or number of cells
| Sample | Maximum DNA fragment (bp) amplified |
Quality of aCGH by visual inspection |
Pearson’s correlation to aCGH data from corresponding frozen tissue | ||
|---|---|---|---|---|---|
| 400ng DNA from FFPE tissue |
20ng DNA from FFPE tissue with WGA |
2000 cells from FFPE tissue with WGA |
|||
| Xenografts | |||||
| X1 | 400 | good | 0.98 | 0.96 | 0.91 |
| X2 | 400 | poor | 0.51* | 0.90 | 0.91 |
| X3 | 400 | good | 0.88 | 0.88 | 0.93 |
| X4 | 400 | good | 0.96 | 0.89 | |
| X5 | 300 | good | 0.83 | 0.82 | |
| X6 | 100 | poor | 0.72 | ||
| X7 | 100 | poor | 0.77 | ||
| X8 | 100 | poor | 0.50 | ||
| X9 | 100 | poor | 0.51 | ||
| X10 | 100 | Failed to label | |||
| X11 | 100 | Failed to label | |||
| Primary glioblastoma |
|||||
| P1 | 400 | good | 0.91 | ||
| P2 | 400 | good | 0.87 | 0.91 | |
| P3 | 300 | good | 0.86 | ||
| P4 | 300 | good | 0.91 | ||
poor correlation was due to the presence of >20% necrosis.
Figure 2.
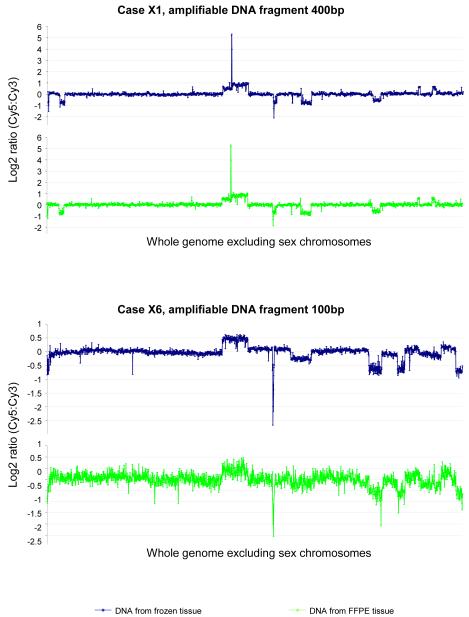
Effect of DNA integrity on aCGH analysis. DNA samples from matched frozen and FFPE glioblastoma xenograft are subjected to 1Mb aCGH analysis. The log2 ratio of tumour/reference fluorescence is shown for all BAC clones, which are arranged in genomic order from 1pter to 22qter. In case X1, the DNA sample from the FFPE glioblastoma xenograft is of good integrity (amplifiable for 400bp fragment) and shows a highly reproducible CGH profile as compared with the DNA from corresponding frozen tissue. Whereas in case X6, the DNA sample from the FFPE glioblastoma xenograft is of poor integrity (only amplifiable for 100bp fragment), displays a CGH profile with high background noise and fails to demonstrate the genomic aberrations seen from the DNA sample from the corresponding frozen tissue.
Table 3.
Correlation of aCGH between DNA samples from frozen tissues, FFPE tissue and small numbers of cells microdissected from FFPE tissue sections
| DNA from frozen tissue (cut-off value mean ± 3SD) |
400ng DNA from FFPE tissue (cut-off value mean ± 3.2SD) |
20ng DNA from FFPE tissue with WGA (cut-off value mean ± 3.4SD) |
2000 cells from FFPE tissue with WGA (cut-off value mean ± 3.4SD) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. clones affected |
No. consecutive clones affected |
Total no. clones affected |
Consecutive clones affected | Total no. clones affected |
Consecutive clones affected | Total no. clones affected |
Consecutive clones affected | |||||||
| No. clones affected |
No. clones concordant with those from the corresponding frozen tissue and percentage of successful detection |
No. clones affected |
No. clones concordant with those from the corresponding frozen tissue and percentage of successful detection |
No. clones affected |
No. clones concordant with those from the corresponding frozen tissue and percentage of successful detection |
|||||||||
| X1 | 480 | 408 85% | 458 | 405 88% | 399 | 98% | 486 | 396 81% | 386 | 95% | 743 | 446 60% | 334 | 82% |
| X2 | 398 | 376 94% | 441 | 394 89% | 368 | 98% | 588 | 439 75% | 370 | 98% | 519 | 398 77% | 354 | 94% |
| X3 | 947 | 765 81% | 1338 | 1117 83% | 637 | 83% | 1212 | 951 78% | 618 | 81% | 1215 | 939 77% | 642 | 84% |
| X4 | 761 | 664 87% | 1011 | 771 76% | 550 | 79% | 1014 | 742 73% | 503 | 76% | ||||
| X5 | 1578 | 1373 87% | 1702 | 1509 89% | 1155 | 85% | 1563 | 1276 82% | 1097 | 80% | ||||
| P1 | 779 | 608 78% | 1029 | 745 72% | 423 | 70% | ||||||||
| P2 | 410 | 401 98% | 805 | 580 72% | 358 | 90% | 661 | 447 68% | 369 | 92% | ||||
| P3 | 783 | 743 95% | 958 | 726 76% | 523 | 80% | ||||||||
| P4 | 1048 | 888 85% | 1504 | 1265 84% | 717 | 82% | ||||||||
| mean | 798 | 692 88% | 1027 | 835 81% | 570 | 85% | 973 | 761 78% | 595 | 86% | 785 | 558 70% | 425 | 88% |
The quality of aCGH from FFPE tissues corresponded well to the integrity of the DNA sample as shown by PCR of variable sized gene fragments. All 4 FFPE DNA samples that yielded representative aCGH data also supported amplification of a DNA fragment >300bp. Only 1 FFPE tissue specimens (X2) that produced poor quality data yielded a DNA sample that showed PCR amplification of a DNA fragment >300bp (Table 2).
To examine why specimen X2, with relatively good DNA integrity, failed aCGH analysis, we reviewed the histology. X2 contained a prominent necrotic area (~20%), while all other cases displayed minimal necrotic regions (<5%) or no necrosis. To test whether the presence of necrosis had an adverse effect on aCGH, we repeated aCGH using a DNA sample prepared from the microdissected “viable” tumour cells, which resulted in high quality aCGH results (Supplement figure 1), comparable to those from the corresponding frozen tissue (r = 0.94).
aCGH using DNA from FFPE tissues – effect of formalin fixation and storage time
To examine the effect of the length of formalin fixation on aCGH, tissue samples from one xenograft were fixed in 10% buffered formalin for various times, routinely processed and paraffin-embedded and subjected to aCGH. The aCGH data obtained was compared with that from the corresponding frozen tissue (Supplement table 2). The results showed that formalin fixation up to 96 hours did not have any major adverse effect on the quality of aCGH. DNA samples prepared from these FFPE specimens showed PCR amplification of a 400bp DNA fragment and a good correlation to aCGH results using DNA from the corresponding frozen tissue.
To further examine the effect of storage time on aCGH, we performed aCGH on 4 archival FFPE primary glioblastomas that had been stored for 15 years, and compared the data to the corresponding frozen tumours. A good correlation between FFPE and frozen tissues was seen in each case (Tables 2 and 3).
aCGH using minimum amount of DNA
To explore the potential applications of aCGH to small tissue biopsies and microscopic lesions, we investigated the minimum amount of DNA required for aCGH analysis. We first tested this by using DNA samples from the frozen glioblastoma xenografts. 10ng DNA from case X2 was amplified using GenomePlex™ WGA kit and subjected to aCGH. Correlation with data from non-amplified DNA of the same source was high (r = 0.99) and no chromosome or gene specific bias could be observed from the linear plots.
We next tested the minimum amount of DNA from FFPE tissue required for representative aCGH. This was carried out using 5 xenograft specimens that showed good quality aCGH data from non-amplified DNA samples. In each case, representative data was obtained from a 20ng DNA template, as judged by comparison with data from the corresponding frozen tissue (Tables 2 and 3, Figure 3).
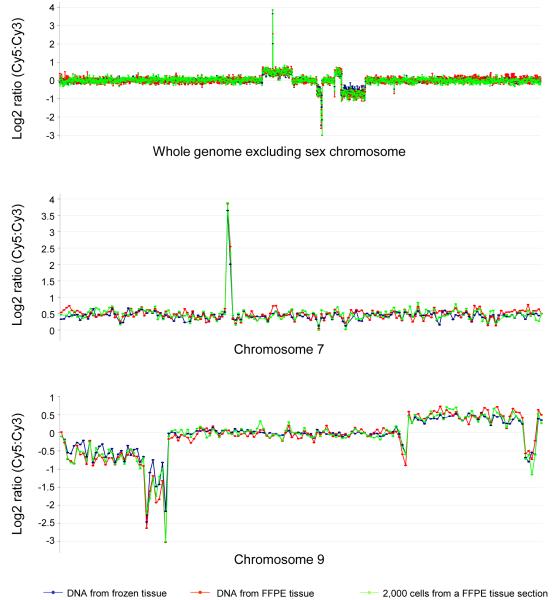
Figure 3.
aCGH using a minimum amount of DNA and ~2000 cells microdissected from a FFPE glioblastoma xenograft tissue slide. In case X2, 20ng DNA from the FFPE glioblastoma xenograft and the DNA sample prepared from 2000 cells microdissected from a haematoxylin stained FFPE xenograft tissue slide are amplified using GenomePlex™ WGA kit and subjected to aCGH analysis. The log2 ratio of tumour/reference fluorescence is shown for all BAC clones, which are arranged in genomic order from 1pter to 22qter and high-resolution correlation is also shown for chromosomes 7 and 9. The data shows a highly reproducible CGH profile from 20ng DNA from the FFPE xenograft tissue and the DNA sample from 2,000 cells microdissected from a FFPE xenograft tissue slide as compared with DNA sample from the corresponding frozen tumour tissue.
aCGH using minimum number of cells microdissected from FFPE tissue sections
To investigate whether aCGH could be applied to small numbers of cells microdissected from FFPE tissue sections, we microdissected and extracted DNA from various numbers (10,000, 5000, 2000 and 1000 cells) of cells from haematoxylin stained tissue sections, avoiding necrotic regions. Representative aCGH data was obtained from DNA samples extracted from 2000 cells or above, as judged by comparison with data from the corresponding frozen tissue (Tables 2 and 3, Figure 3).
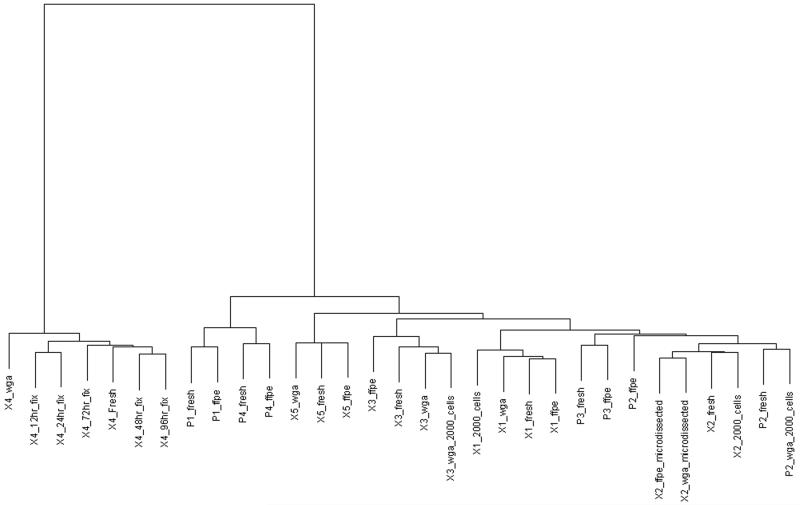
Clustering of aCGH data from frozen, FFPE tissue and microdissected cells
Unsupervised hierarchical clustering analysis showed that all data sets from the same specimen clustered together. This further demonstrated the similarity, thus reproducibility, of aCGH data between frozen, FFPE tissues and microdissected cells.
DISCUSSION
Archival FFPE tissues represent a rich resource of well-characterised pathological specimens for genetic study of human disease, particularly cancer. By systematic investigations of matched frozen and FFPE glioblastoma tissues, we developed a practical protocol to apply archival FFPE tissues to aCGH. As the quality of aCGH critically depends on the integrity of DNA samples, it is possible to predict whether an archival FFPE tissue specimen is suitable for aCGH analysis when extracted DNA supports PCR amplification of >300bp gene fragments. PCR-based DNA integrity assessment is superior to direct examination of the DNA sample on agarose gels, as it is more objective and uses only a minute amount of DNA. In our experience, direct analysis of DNA samples on agarose gels did not offer any value in the prediction of the suitability of a FFPE tissue sample for aCGH.
In general, aCGH alterations involving consecutive BAC clones are easily recognisable and most likely derive from tumour cells rather than experimental artefacts. However, interpretation of aCGH changes affecting single clones is difficult and these sporadic changes occur for DNA samples from both frozen and FFPE tissues, particularly the latter. In theory, alterations as a result of true genomic events should be reproducible and those highly associated with tumourigenesis and progression are likely recurrent. Conversely, changes due to experimental artefacts are largely random, although those resulting from mismapped clones or cross hybridisation of repetitive sequences may be reproducible. Such assumptions were well supported by our experimental data, demonstrating that aCGH changes involving two or more consecutive clones were highly reproducible, while those involving single sporadic clones were not. In view of this and the vast amount of data potentially derived from an aCGH study, it is pertinent to focus on alterations involving consecutive clones for any follow-up investigations. Nonetheless, genomic amplification/deletion occasionally involves a single gene locus: gene amplification can be readily identified by visual inspection of the aCGH profile, while deletion, if recurrent, should not be neglected.
One of the other major factors affecting the quality of aCGH was the presence of necrosis in tissue specimens. It is likely that the adverse effect of necrosis on aCGH is due to DNA degradation of necrotic cells. However, such DNA degradation contributed by necrosis could not be revealed by PCR-based assessment of DNA integrity, as it would be masked by the relatively intact DNA from “viable” cells. However, necrosis is easily identified by histological review and its adverse effect on aCGH can be eliminated by microdissection.
In line with loss of heterozygosity (LOH) analysis, sensitivity of aCGH in detection of DNA copy number changes heavily depends on the proportion of tumour cells in a tissue specimen (18). By spiking xenograft tumour DNA with normal DNA, we showed that at least 70% tumour cells are required for reliable aCGH analysis. Together with the adverse effect of necrosis, this emphasises the importance of routine histological review of tissue specimens selected for aCGH.
With adequate quality control, it was also possible to apply aCGH to small numbers of cells from FFPE tissue sections with a combination of microdissection and WGA. As shown in the present study, we were able to use as few as 2000 cells microdissected from haematoxylin stained tissue sections for aCGH and to obtain results comparable to those from frozen tissues. Therefore, it is possible to perform aCGH analysis on archival small tissue biopsies and potentially histologically defined microscopic lesions, such as carcinoma in situ. With the advantage of genomic arrays, particularly its versatile design, high throughput and high resolution, application of aCGH on archival FFPE tissues, an immense resource of well-documented pathological specimens, will no doubt play a significant role in disease gene discovery and translational research.
Supplementary Material
Figure 4.
Unsupervised hierarchical clustering. The dendogram demonstrates that aCGH data generated from the same tissue source is clustered together irrespective of whether DNA used is from frozen or FFPE tissue, or amplified from small quantities of DNA or cells. wga, whole genome amplification; 12hr-96hr_fix denotes hours of fixation in formalin; ffpe, formalin-fixed paraffin-embedded, X, glioblastoma xenograft; P, primary glioblastoma.
Acknowledgements
The Du Lab was supported by research grants from Leukaemia Research Fund, United Kingdom. The Collins Lab was supported by research grants from Cancer Research UK. We wish to thank Martin McCabe for providing data partially used to generate the reference data set.
The Du lab (M-Q Du, N. Johnson, R. Hamoudi) was supported by research grants from Leukaemia Research Fund, United Kingdom. The Collins Lab (V. Peter Collins, K. Ichimura, L. Liu, D. Pearson) was supported by research grants from Cancer Research UK.
Footnotes
Duality of interest: None declared.
Supplementary information is available at Laboratory Investigation's website.
References
- 1.Ishkanian AS, Malloff CA, Watson SK, deLeeuw RJ, Chi B, Coe BP, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nature Genetics. 2004;36(3):299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 2.Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Smith J, et al. DNA microarrays for comparative genomic hybridisation based on DOP-PCR amplification of BAC and PAC clones. Genes, Chromosomes and Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 3.Seng TJ, Ichimura K, Liu L, Tingby O, Pearson DM, Collins P. Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis. Genes, Chromosomes and Cancer. 2005;43:181–193. doi: 10.1002/gcc.20181. [DOI] [PubMed] [Google Scholar]
- 4.Ichimura K, Mungall AJ, Fiegler H, Pearson DM, Dunham I, Carter NP, et al. Small regions of overlapping deletions on 6q26 in human astrocytic tumours identified using chromosome 6 tile path array-CGH. Oncogene. 2006;25(8):1261–1271. doi: 10.1038/sj.onc.1209156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gribble SM, Fiegler H, Burford DC, Prigmore E, Yang F, Carr P, et al. Applications of combined DNA microarray and chromosome sorting technologies. Chromosome Research. 2004;12(1):35–43. doi: 10.1023/b:chro.0000009325.69828.83. [DOI] [PubMed] [Google Scholar]
- 6.Nessling M, Richter K, Schwaenen C, Roerig P, Wrobel G, Wessendorf S, et al. Candidate Genes in Breast Cancer Revealed by Microarray-Based Comparative Genomic Hybridization of Archived Tissue. Cancer Res. 2005;65(2):439–447. [PubMed] [Google Scholar]
- 7.Harvell JD, Kohler S, Zhu S, Hernandez-Boussard T, Pollack JR, van de Rijn M. High-resolution array-based comparative genomic hybridization for distinguishing paraffin-embedded Spitz nevi and melanomas. Diagnostic Molecular Pathology. 2004;13(1):22–25. doi: 10.1097/00019606-200403000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Ried T, Just K, Holtgreve-Grez H, du Manoir S, Speicher M, Schrock E, et al. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;55(22):5415–5423. [PubMed] [Google Scholar]
- 9.Loo LWM, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, et al. Array Comparative Genomic Hybridization Analysis of Genomic Alterations in Breast Cancer Subtypes. Cancer Res. 2004;64(23):8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 10.Bredel M, Bredel C, Juric D, Kim Y, Vogel H, Harsh GR, et al. Amplification of Whole Tumor Genomes and Gene-by-Gene Mapping of Genomic Aberrations from Limited Sources of Fresh-Frozen and Paraffin-Embedded DNA. J Mol Diagn. 2005;7(2):171–182. doi: 10.1016/S1525-1578(10)60543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Little SE, Vuononvirta R, Reis-Filho JS, Natrajan R, Iravani M, Fenwick K, et al. Array CGH using whole genome amplification of fresh-frozen and formalin-fixed, paraffin-embedded tumor DNA. Genomics. 2006;87(2):298–306. doi: 10.1016/j.ygeno.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Liu C, DeVries S, Waldman F, Cote RJ, Datar RH. LM-PCR permits highly representative whole genome amplification of DNA isolated from small number of cells and paraffin-embedded tumor tissue sections. Diagnostic Molecular Pathology. 2004;13(2):105–115. doi: 10.1097/00019606-200406000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Ichimura K, Schmidt EE, Goike HM, Collins VP. Human glioblastomas with no alterations of the CDKN2A (p16INK4A, MTS1) and CDK4 genes have frequent mutations of the retinoblastoma gene. Oncogene. 1996;13(5):1065–1072. [PubMed] [Google Scholar]
- 14.van Dongen JJ, Langerak AW, Bruggeman M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 15.McCabe MG, Ichimura K, Liu L, Plant K, Pearson DM, Collins VP. High resolution array-based comparative genomic hybridisation of medulloblastomas and supra-tentorial primitive neuroectodermal tumours. Journal of Neuropathology and Experimental Neurology. 2006 doi: 10.1097/00005072-200606000-00003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Houldsworth J, Olshen AB, Nabjangud G, Chaganti S, Venkatraman ES, et al. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood. 2005;107(6):2477–2485. doi: 10.1182/blood-2005-07-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Development Core Team . R: A Language and Environment for Statistical Computing. 2005. [Google Scholar]
- 18.Liu J, Zabarovska VI, Braga E, Alimov A, Klein G, Zabarovsky ER. Loss of heterozygosity in tumor cells requires re-evaluation: the data are biased by the size-dependent differential sensitivity of allele detection. FEBS Letters. 1999;462(1-2):121–128. doi: 10.1016/s0014-5793(99)01523-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.