Abstract
Phloem and xylem transport of amino acids involves two steps: export from one cell type to the apoplasm, and subsequent import into adjacent cells. High-affinity import is mediated by proton/amino acid cotransporters, while the mechanism of export remains unclear. Enhanced expression of the plant-specific type I membrane protein Glutamine Dumper1 (GDU1) has previously been shown to induce the secretion of glutamine from hydathodes and increased amino acid content in leaf apoplasm and xylem sap. In this work, tolerance to low concentrations of amino acids and transport analyses using radiolabeled amino acids demonstrate that net amino acid uptake is reduced in the glutamine-secreting GDU1 overexpressor gdu1-1D. The net uptake rate of phenylalanine decreased over time, and amino acid net efflux was increased in gdu1-1D compared with the wild type, indicating increased amino acid export from cells. Independence of the export from proton gradients and ATP suggests that overexpression of GDU1 affects a passive export system. Each of the seven Arabidopsis (Arabidopsis thaliana) GDU genes led to similar phenotypes, including increased efflux of a wide spectrum of amino acids. Differences in expression profiles and functional properties suggested that the GDU genes fulfill different roles in roots, vasculature, and reproductive organs. Taken together, the GDUs appear to stimulate amino acid export by activating nonselective amino acid facilitators.
Minerals and organic solutes cycle through the vascular conduits of higher plants (Marschner et al., 1996, 1997). Cycling requires multiple transport steps from apoplasm to cytosol (cellular import) and from cytosol to apoplasm (cellular export). Cellular import is typically mediated by proton cotransporters able to import solutes against a concentration gradient, such as for acquisition of amino acids from the rhizosphere (Hirner et al., 2006; Lee et al., 2007; Svennerstam et al., 2008), for phloem loading (Koch et al., 2003; Lalonde et al., 2003), or for import from the xylem or into developing embryos (Zhang et al., 2007). Export is required for xylem loading (Schobert and Komor, 1990; Gaymard et al., 1998; Takano et al., 2002), for efflux into the rhizosphere (Jaeger et al., 1999), and for the transfer of assimilates from leaf cells to the apoplasm before phloem loading (Lalonde et al., 2003) or from the seed coat into the apoplasm to supply developing embryos (Zhang et al., 2007).
Transport of metabolites across membranes is typically mediated by membrane proteins specific for a solute or a class of solutes. During the past two decades, numerous transporters of amino acids belonging to the amino acid transporter family 1 (ATF1) and amino acid-polyamine-organocation (APC) family have been isolated from plants. Several ATF1 members were shown to function as amino acid/proton cotransporters that are characterized by low amino acid selectivity (Rentsch et al., 2007). While the amino acid import process is well characterized at both the physiological level (Kinraide, 1981; Schobert and Komor, 1987) and molecular level (Li and Bush, 1990; Näsholm et al., 2009), the mechanism of amino acid export from plant cells is still elusive. The physiology of export has been addressed by a small number of studies that showed that export is independent of the proton-motive force or other source of energization (Jones and Darrah, 1994; De Jong et al., 1997). The identification of cellular exporters has been difficult, since yeast complementation assays that rely on auxotrophies may be unsuitable for identifying exporters, and export assays are more challenging in the context of screens (G. Pilot and W.B. Frommer, unpublished data). Recently, a putative amino acid exporter, bidirectional amino acid transporter 1 (BAT1), was isolated (Dundar and Bush, 2009). BAT1 shares weak sequence similarities with fungal γ-aminobutyrate transporters and with amino acid transporters of the APC family (Su et al., 2004). Unlike the APC proton-coupled amino acid importers, BAT1 mediates amino acid efflux (Glu and Lys) when expressed in yeast. Interestingly, yeast assays also showed that BAT1 mediates Ala and Arg uptake, suggesting that BAT1 may function as a facilitative uniporter (Dundar and Bush, 2009). Reverse transcription (RT)-PCR assays and microarray analyses suggested that BAT1 is expressed at high levels in all plant organs, especially the vasculature (Dundar, 2009).
Previous work had shown that the activation-tagged glutamine dumper1 mutant gdu1-1D secretes Gln from the hydathodes and is characterized by increased amino acid content in apoplasm and xylem sap (Pilot et al., 2004). Mutant gdu1-1D seedlings were tolerant to amino acids supplied at concentrations that are toxic to the wild type (Pratelli and Pilot, 2007a). The increased tolerance could result either from induction of a detoxification mechanism (Voll et al., 2004) or from reduced net amino acid uptake by roots (Lee et al., 2007). The features of the Gdu1 phenotype are consistent with an increased amino acid efflux.
The work presented here aimed at testing the hypothesis of GDU1 being an activator of amino acid efflux as well as evaluating the functional role of the GDU paralogs. Amino acid transport in gdu1-1D was studied by uptake and efflux analyses using radiolabeled compounds. The effects of the overexpression of the GDU paralogs on amino acid content and transport were examined. The results show that the seven GDU proteins are able to increase amino acid export by plant cells.
RESULTS
Increased Tolerance of gdu1-1D to Amino Acids
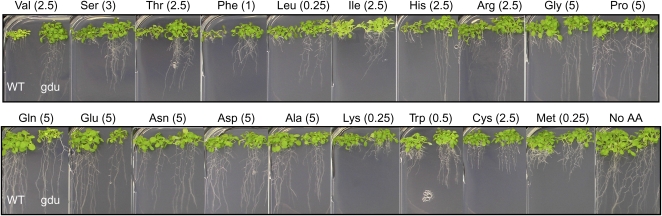
A sensitive, quantitative assay was developed to assess the tolerance of plants to toxic levels of amino acids. In contrast to previous systems that determined survival of seedlings at high external amino acid supply (10 mmol L−1; Pratelli and Pilot, 2007a), the new assay measures root growth at relatively low amino acid concentrations. To test the contribution of transport to amino acid tolerance in gdu1-1D, root growth was determined using the lowest amino acid concentration found to inhibit wild-type root growth (less than 5 mmol L−1). Length of wild-type roots was decreased by 80% to 95% on media containing Val, Ser, Thr, Phe, Leu, Ile, His, Arg, or Gly. By comparison, gdu1-1D roots showed only 0% to 40% root growth inhibition (Fig. 1, top). Tolerance of gdu1-1D to multiple amino acids synthesized from different metabolic pathways suggested a reduction of net amino acid uptake (i.e. amounts imported minus amounts exported) rather than elevated detoxification capacity.
Figure 1.
gdu1-1D root growth is tolerant to exogenously supplied toxic amino acids. Wild-type (WT) and gdu1-1D (gdu) plants were grown vertically for 10 d on solid medium containing the amino acid indicated above each image (the concentration in mmol L−1 is indicated in parentheses). Pro, Gln, Glu, Asn, Asp, and Ala had no effect on root growth; Lys, Trp, Cys, and Met inhibited equally the growth of wild-type and gdu1-1D roots; Val, Ser, Thr, Phe, Leu, Ile, His, Arg, and Gly inhibited strongly wild-type but not gdu1-1D root growth. [See online article for color version of this figure.]
Reduced Net Amino Acid Accumulation in gdu1-1D
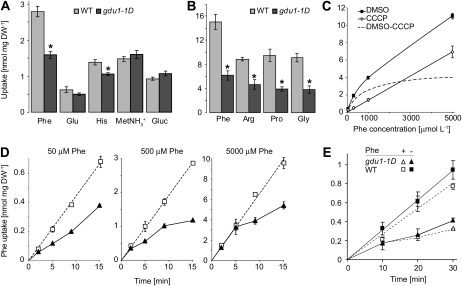
Uptake capacities of various radiolabeled compounds were compared in gdu1-1D and the wild type. Phe uptake was reduced by 55%, while Glc and methyl ammonium uptakes were unaffected or slightly increased. Uptake of other amino acids was also reduced in gdu1-1D: 50% to 60% for Arg, Pro, and Gly and 20% for His and Glu (Fig. 2, A and B). Reduced uptake of Phe, Arg, His, and Gly correlated with the tolerance of gdu1-1D to these toxic amino acids (Pro and Glu were not toxic to the wild type; Fig. 1).
Figure 2.
Analyses of the uptake of radiolabeled 14C-compounds by gdu1-1D and wild-type (WT) plants. A, Accumulation of Phe, Glu, His, methyl ammonium (MetNH3+), and Glc (Gluc) after 10 min (supplied at 1 mmol L−1) in whole plantlets. Wild type, light gray bars; gdu1-1D, dark gray bars. Means ± se of three biological replicates are shown. *, Significantly different from the wild type (t test, P < 0.02). DW, Dry weight. B, Accumulation of Phe, Arg, Pro, and Gly (supplied at 1 mmol L−1) after 1 h in plants. Means ± se of three biological replicates are shown. *, Significantly different from the wild type (t test, P < 0.01). C, Concentration dependence of Phe uptake into wild-type plantlets treated with dimethyl sulfoxide (DMSO; black squares) or CCCP (100 μm; white diamonds). CCCP inhibits the proton gradient-dependent high-affinity uptake system and reveals the activity of the low-affinity uptake system. The difference between the uptake of the dimethyl sulfoxide- and the CCCP-treated plants corresponds to activity of the high-affinity amino acid uptake system, indicated by the broken line. Plants were allowed to take up Phe for 15 min before counting the amount of absorbed radioactivity. Means ± se of three biological replicates are shown. D, Time-course kinetics of the uptake of Phe supplied at a concentration of 50, 500, or 5,000 μmol L−1 in wild-type (white squares) and gdu1-1D (black triangles) plants. Wild-type data points were fitted by the line. Means ± se of three biological replicates are shown. E, Effect of Phe pretreatment on time-course analysis of Phe uptake. Wild-type (squares) and gdu1-1D (triangles) plants were treated (dotted lines, white symbols) or not (solid lines, black symbols) with 1 mm Phe for 30 min prior to uptake analysis performed in the presence of 0.1 mm Phe. Means ± se of three biological replicates are shown.
Based on the large reduction of uptake and its effect in the tolerance assay, Phe was chosen for more detailed analyses. The uptake rate of radiolabeled Phe by wild-type plants followed a biphasic curve over the range 0.01 to 5 mmol L−1. Treatment with the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP) inhibited the high-affinity component (hyperbolic curve), revealing the activity of the low-affinity component (linear curve; Fig. 2C). Phe uptake was analyzed at three concentrations, each concentration revealing a different contribution of the high-affinity transport to the total uptake (80%, 60%, or 40%). After 2 min, Phe accumulated at the same level in gdu1-1D as in the wild type (Fig. 2D), suggesting that the initial import capacity of Phe is similar. Phe uptake in the wild type followed linear kinetics, corresponding to a constant uptake rate, as described for other amino acids (Schobert and Komor, 1987; Heremans et al., 1997; Hirner et al., 2006). On the contrary, the uptake rate in gdu1-1D decreased over time, leading to a lower Phe accumulation in gdu1-1D plants after 10 min (Fig. 2D, middle and right panels).
A decrease in uptake rates over time could result from (1) feedback inhibition of the uptake by accumulation of Phe or (2) increased amino acid export in gdu1-1D. When plants were pretreated with 1 mm Phe prior to uptake analyses, little difference in the time course of Phe accumulation for both the wild type and mutant was observed (Fig. 2E). This finding argues against a contribution of cytosolic Phe accumulation in inhibiting its own uptake. The unchanged initial Phe uptake, together with the unchanged uptake kinetics after Phe pretreatment, favors the second hypothesis (i.e. an increase in amino export as a result of GDU1 overexpression).
Increased Amino Acid Efflux from gdu1-1D
To further resolve Phe export capacity, seedlings were preloaded with [14C]Phe and the net efflux (i.e. export minus import) of radiotracer into the medium was measured. Thin-layer chromatography analysis of the medium proved that the radiolabeled compound released by the plants was Phe (Supplemental Fig. S1A). The amount of Phe released into the medium was higher in gdu1-1D than in wild-type plants (150 and 64 nmol mg−1 dry weight, respectively; Table I). In gdu1-1D, released Phe amount corresponded also to a higher fraction of the total radioactivity taken up, defined as the sum of Phe amounts left in the plant and released into the medium (approximately 60% in gdu1-1D versus approximately 15% in the wild type). Similar experiments showed that efflux of radiolabeled His and Glu was also increased in gdu1-1D, but the efflux of radiolabeled Glc was not (Supplemental Fig. S1B).
Table I.
Amounts of Phe taken up by the wild type, lht1-1, gdu1-1D, and the corresponding double mutant and subsequently released in the medium
After incubation with Phe supplied at the indicated concentration for 10 min, plantlets were quickly rinsed and placed in fresh medium for 1 h. Total Phe taken up was estimated by adding the amounts of radioactivity in the medium (efflux) and left in the plants after efflux analyses. Means ± se of three biological replicates are shown. Significant differences from the wild type are as follows (t test): ** P < 0.02, * P < 0.05.
| Plant | 1 mm |
10 μm |
||||
|---|---|---|---|---|---|---|
| Totala | Effluxa | Percentageb | Totala | Effluxa | Percentageb | |
| Wild type | 456 ± 45 | 64 ± 6 | 14 ± 2 | 20.8 ± 3.9 | 1.9 ± 0.3 | 9 ± 1 |
| gdu1-1D | 255 ± 15* | 150 ± 16* | 59 ± 3** | 3.2 ± 0.1* | 1.8 ± 0.1 | 55 ± 5** |
| lht1-1 | 354 ± 22 | 62 ± 5 | 18 ± 0 | 5.6 ± 0.6* | 0.6 ± 0.1* | 12 ± 1 |
| gdu1-1D/lht1-1 | 239 ± 28* | 135 ± 17** | 56 ± 1** | 2.7 ± 0.2* | 1.3 ± 0 | 50 ± 4** |
nmol mg−1 chlorophyll.
Percentage of Phe present in the medium relative to total Phe taken up.
It has been proposed that a fraction of the amino acids exported from roots is reimported by high-affinity transporters, controlling amino acid net efflux (Schobert and Komor, 1987; Jones and Darrah, 1993). The contribution of reimport activity to amino acid net efflux was assessed using the lysine histidine transporter1 (lht1-1) mutant, carrying a T-DNA insertion in the LHT1 gene, which encodes a dominant high-affinity amino acid importer (Hirner et al., 2006; Svennerstam et al., 2007). LHT1 contributed about 75% of Phe uptake when supplied with 10 μm Phe (Phe uptake was 5.6 versus 20.8 nmol mg−1 dry weight in lht1-1 and the wild type, respectively; Table I) but about 25% when supplied with 100 μm or 1 mm Phe (Table I; Supplemental Fig. S1C). The decreased import of lht1-1 was expected to lead to a decreased Phe reimport and hence an increase in Phe net efflux. Surprisingly, the fraction of labeled Phe exported after preloading with 10 μm or 1 mm Phe was comparable in lht1-1 and the wild type (approximately 15%; Table I), suggesting that Phe net efflux is not controlled by the high-affinity import system. The comparable Phe efflux from gdu1-1D and the gdu1-1D/lht1-1 double mutant further supported the hypothesis that Phe efflux is independent from LHT1-mediated import (Table I). The reductions in Phe uptake caused by the lht1-1 and gdu1-1D mutations were additive (Table I; Supplemental Fig. S1C), as expected if reduced import is cumulated with increased export.
To evaluate the selectivity of amino acid efflux, amino acid content in plants and conditioned medium (medium from plants grown for 2 d) was analyzed. Amino acid content was nearly identical or even decreased in gdu1-1D plants compared with wild-type plants (e.g. Pro, Tyr, Val, Met, Ile, Lys, and Leu; Table II). In contrast, gdu1-1D growth medium contained about four times as much amino acids as for the wild type (Table II). The content of most amino acids was increased in the medium, and Asn and Gln accounted for about 75% of the total increase, suggesting that the export mechanism stimulated by GDU1 overexpression is nonselective but shows a preference for Asn and Gln.
Table II.
Amino acid amounts in whole plantlets and released in the growth medium
Plants were grown for 6 d in modified MS medium containing no ammonium. Amino acids were quantitated by HPLC 3 d after transfer into fresh MS medium without ammonium. Means ± se of four biological replicates are shown. Significant differences from the wild type (t test) are as follows: * P < 0.05, ** P < 0.01.
| Amino Acid | Plant |
Medium |
||||||
|---|---|---|---|---|---|---|---|---|
| Wild Type |
gdu1-1D |
Wild Type |
gdu1-1D |
|||||
| Amounta | Percentage | Amounta | Percentage | Amounta | Percentage | Amounta | Percentage | |
| Asp | 1,699 ± 188 | 9.6 | 1,735 ± 272 | 10.8 | 41.9 ± 15.4 | 13.6 | 76.1 ± 15.8 | 4.1 |
| Glu | 5,024 ± 637 | 28.3 | 4,860 ± 811 | 30.1 | 39.4 ± 18.2 | 12.8 | 119.7 ± 28.2 | 6.5 |
| Asn | 776 ± 100 | 4.4 | 660 ± 109 | 4.1 | 15.6 ± 9.4 | 5.1 | 210.3 ± 36.8* | 11.4 |
| Ser | 2,030 ± 176 | 11.4 | 1,970 ± 273 | 12.2 | 13 ± 3.7 | 4.2 | 95.7 ± 17.4* | 5.2 |
| Gln | 2,174 ± 364 | 12.3 | 2,031 ± 295 | 12.6 | 72.3 ± 44.4 | 23.5 | 939.2 ± 114.8** | 50.7 |
| Gly | 1,720 ± 647 | 9.7 | 1,696 ± 481 | 10.5 | 17.9 ± 4.9 | 5.8 | 62.8 ± 10.6 | 3.4 |
| His | 91 ± 8 | 0.5 | 80 ± 14 | 0.5 | 5.1 ± 2.2 | 1.7 | 58.9 ± 11.2* | 3.2 |
| Thr | 149 ± 40 | 0.8 | 80 ± 27 | 0.5 | 41.6 ± 17.5 | 13.5 | 25.2 ± 9.3 | 1.4 |
| Ala | 2,738 ± 331 | 15.4 | 2,164 ± 399 | 13.4 | 47.1 ± 16.5 | 15.3 | 105.3 ± 27 | 5.7 |
| Pro | 407 ± 52 | 2.3 | 284 ± 73 | 1.8 | 4.8 ± 1.1 | 1.5 | 36.8 ± 7.2 | 2 |
| Tyr | 259 ± 56 | 1.5 | 185 ± 21 | 1.1 | 0.2 ± 0.1 | 0.1 | 4.2 ± 0.8* | 0.2 |
| Val | 265 ± 70 | 1.5 | 170 ± 30 | 1.1 | 6.2 ± 2.2 | 2 | 68.3 ± 9* | 3.7 |
| Met | 13 ± 2 | 0.1 | 7 ± 3 | 0 | 0 ± 0 | 0 | 3 ± 0.6* | 0.2 |
| Ile | 109 ± 42 | 0.6 | 45 ± 10 | 0.3 | 0.8 ± 0.6 | 0.3 | 16.9 ± 4.1 | 0.9 |
| Lys | 68 ± 25 | 0.4 | 37 ± 9 | 0.2 | 0.2 ± 0.1 | 0.1 | 5.6 ± 2.1 | 0.3 |
| Leu | 128 ± 51 | 0.7 | 62 ± 14 | 0.4 | 0.8 ± 0.3 | 0.3 | 12.8 ± 3 | 0.7 |
| Phe | 88 ± 20 | 0.5 | 58 ± 12 | 0.4 | 0.8 ± 0.4 | 0.3 | 10.2 ± 1.7* | 0.6 |
| Total | 17,738 ± 1,583 | 100 | 16,125 ± 2,266 | 100 | 307.6 ± 125 | 100 | 1,851.1 ± 278.9* | 100 |
nmol mg−1 chlorophyll.
Inhibitors as Tools for Dissecting GDU1-Mediated Efflux
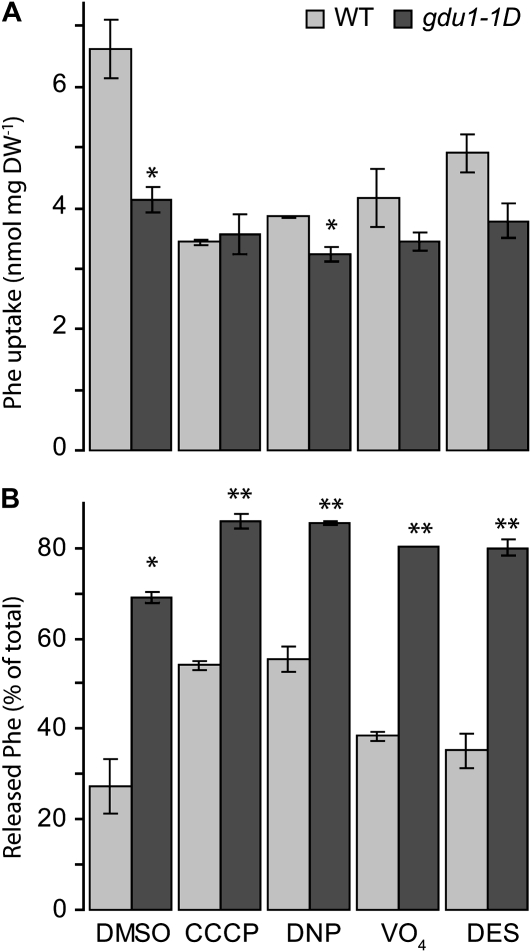
One may speculate that import and export may be differentially sensitive to inhibitors, specifically to protonophores. Net Phe efflux was thus measured in the presence of uncouplers (CCCP and 2,4-dinitrophenol [DNP]) or inhibitors of ATP hydrolysis (orthovanadate [VO4]) and proton pumps (diethylstilbestrol [DES]), shown to inhibit proton-coupled high-affinity amino acid import (Jones and Darrah, 1993). Uptake of 5 mm Phe in the presence of the inhibitors was decreased by up to 48% in the wild type (CCCP treatment) and 22% in gdu1-1D (DNP treatment; Fig. 3A). Independent of the conditions, Phe net efflux was always higher in gdu1-1D (Fig. 3B), suggesting that the GDU1-stimulated export mechanism is independent of ATP and proton gradient across the membrane, which is characteristic of a passive uniport system.
Figure 3.
Amino acid export sensitivity to inhibitors of the proton gradient or ATP hydrolysis. Wild-type (WT) and gdu1-1D plants were pretreated with dimethyl sulfoxide (DMSO), 100 μm CCCP, 100 μm DNP, 1 mm VO4, and 100 μm DES. The same amount of dimethyl sulfoxide was present in each sample. Uptake was performed for 10 min in the presence of 5 mm Phe; plants were allowed to release radioactivity for 10 min in the same medium without Phe. A, Total Phe taken up. DW, Dry weight. B, Percentage of Phe present in the medium after the efflux experiment, expressed as a percentage of the total Phe taken up. Means ± se of three biological replicates are shown. Significant differences from the wild type (t test) are as follows: * P < 0.05, ** P < 0.01.
Overexpression of GDU Paralogs
The Arabidopsis (Arabidopsis thaliana) genome encodes six GDU1-like proteins (hereafter called GDUs) that may share functions similar to that of GDU1. Five T-DNA insertions in the GDU genes caused more than 95% reduction in the content of the corresponding mRNA. Homozygous plants did not present any morphological abnormality when grown on soil, and no change in Phe uptake or efflux was observed (data not shown). Sequence conservation among the GDUs (Pratelli and Pilot, 2006, 2007b) and the absence of an obvious phenotype for the knockout lines may suggest that GDU proteins are functionally redundant.
To test whether all GDUs elicit amino acid efflux, each of the seven genes was expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35Sp), leading to a 100- to 50,000-fold increase in mRNA levels relative to their endogenous levels in the wild type. For each construct, six to eight transformant lines were selected that segregated 3:1 for the kanamycin resistance marker and whose kanamycin-resistant offspring were phenotypically identical. GDU mRNA accumulation, rosette diameter, and amino acid content were determined. Several lines containing the 35Sp-GDU2, 35Sp-GDU3, and 35Sp-GDU4 constructs also overaccumulated GDU1, GDU3, or GDU6 mRNA (circled symbols in Supplemental Figs. S2 and S3). No other GDU gene was overexpressed in the GDU1, GDU5, GDU6, and GDU7 overexpressors. The reason for the induction of other GDU genes is not clear at present, but it probably hints at complex regulations of the expression of the GDUs. Lines showing overaccumulation of the targeted GDU mRNA and not any other GDU were used for further characterization and were named GDU1-OE to GDU7-OE (Supplemental Table S1).
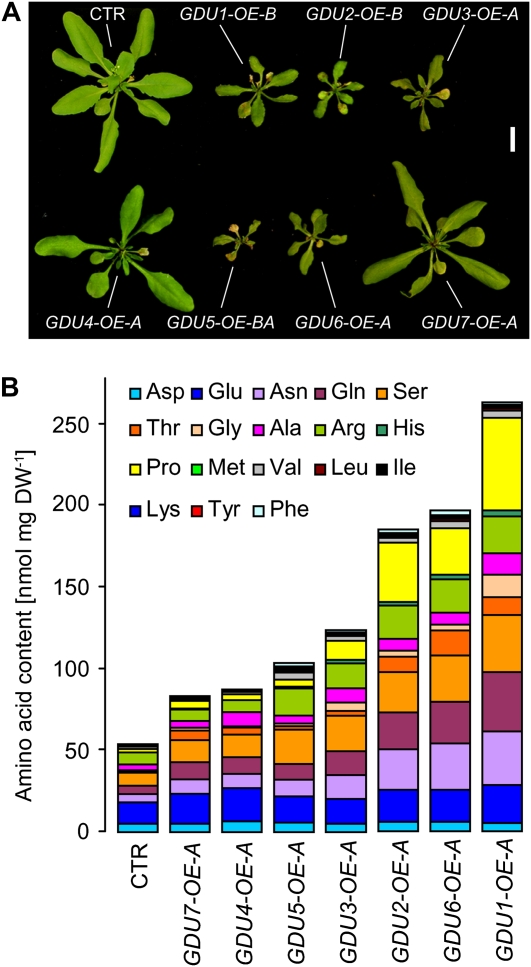
Except for GDU4 and GDU7, plant size decreased with mRNA accumulation of the GDU genes (Fig. 4A; Supplemental Fig. S2), and free amino acid content of all the GDU-OEs increased in correlation with the intensity of the overexpression (Supplemental Fig. S3), similar to what was observed with GDU1 overexpression in gdu1-1D (Pilot et al., 2004). HPLC analyses from two independent experiments showed that the content in nearly all free amino acids was increased in the GDU-OEs, except for Asp and Glu (Fig. 4B; Supplemental Table S2). The effects of the overexpression of the GDUs on plant size and amino acid content suggest that all the GDUs have similar functional properties, probably targeting a common mechanism for amino acid efflux.
Figure 4.
Size and amino acid profiling of plants overexpressing the GDU genes. A, Rosettes of representative plants from one of the two overexpressing lines used in this study (Supplemental Table S1). The amount of secreted crystals at the margin of the leaves was lower for GDU2-, GDU3-, GDU5-, and GDU6-OEs than for GDU1-OEs. Bar = 1 cm. CTR, Plants expressing GFP under the control of the 35S promoter. B, One GDU-OE line per gene (Supplemental Table S1) was selected on kanamycin-containing medium for 7 d, transferred to soil, and grown for 3 weeks more. Leaves from eight plants were pooled and freeze dried. The amino acids were extracted and quantitated by HPLC. DW, Dry weight. Increases in the content of Pro, Gln, Ser, and Asn accounted for 50% to 65% of the total augmentation in free amino acid content, while His and Thr contents were increased the most (5- to 14-fold and 2- to 19-fold, respectively).
GDU Expression Affects Amino Acid Export
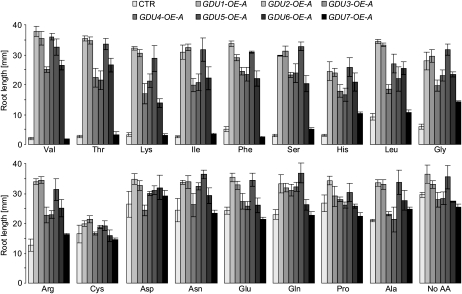
Amino acid tolerance of one GDU-OE line per gene was assessed using the root growth assay described previously. Root length was almost identical for the overexpressors and the wild type grown on Cys (Fig. 5), Met, and Trp (data not shown). Except for GDU7, the overexpression of the GDUs led to a tolerance to most of the toxic amino acids (Fig. 5, top), similar to what was found in the case of gdu1-1D.
Figure 5.
Tolerance of the seven GDU overexpressors toward exogenously supplied amino acids. The GDU-OEs were grown vertically for 10 d on solid MS medium supplemented with 5 mm Ala, Asn, Asp, Gln, Glu, Gly, or Pro; 3 mm Ser; 2.5 mm Arg, Cys, His, Ile, Thr, or Val; 1 mm Phe; or 0.25 mm Leu or Lys. Means ± se of the length of five to 10 roots are shown. CTR, Plants expressing GFP under the control of the 35S promoter.
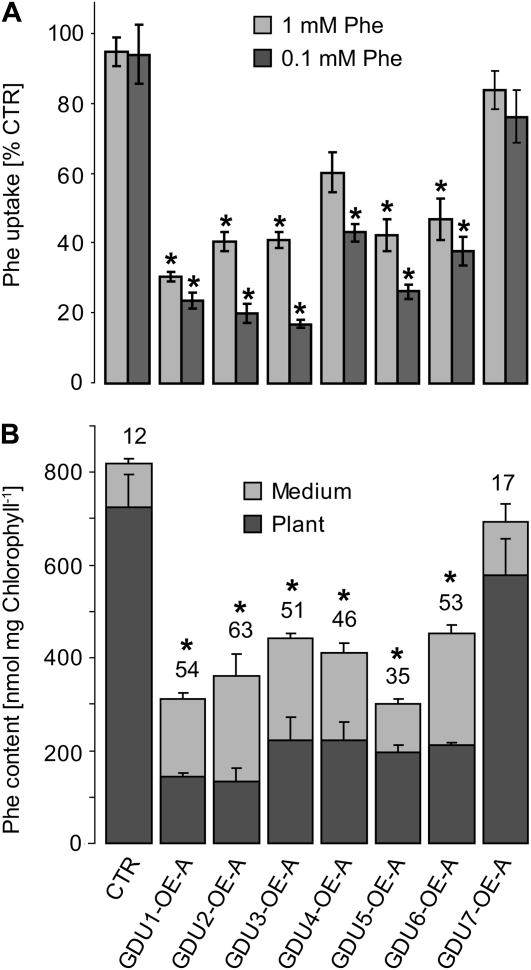
Phe net uptake was reduced for all GDU-OEs compared with the control (Fig. 6A). Phe accumulation kinetics in two GDU7-OE lines was nearly linear, with an uptake rate approximately 10% lower than the control. Radioactivity accumulation kinetics measured for two lines overexpressing the five other genes was characterized by a decrease in the uptake rate over time, as observed for gdu1-1D (Supplemental Fig. S4). Amino acid efflux analyses showed that a higher fraction of the incorporated radioactivity is exported by the GDU-OEs (approximately 35%–50%) compared with the control (approximately 12%), except for GDU7-OE (approximately 17%; Fig. 6B), indicating that Phe export is increased in almost all GDU-OEs.
Figure 6.
Analysis of the Phe uptake and efflux by the GDU overexpressors. A, Seven-day-old plantlets were assayed for uptake of 1 mm Phe (light gray bars) or 0.1 mm Phe (dark gray bars) for 1 h. Phe accumulation is expressed as a percentage of the control (CTR) uptake (15.1 ± 1.1 and 2.9 ± 0.5 nmol mg−1 dry weight for 1 and 0.1 mm Phe, respectively). Error bars represent the se of three biological replicates. *, Significantly different from the control (t test, P < 0.05). B, Analysis of Phe efflux for the GDU-overexpressing lines, performed as described in Table I. Amounts of Phe remaining in plants or released into the medium were calculated from the respective amounts of radioactivity. The number above each bar represents the percentage of incorporated radioactivity that was released into the medium. Error bars represent the se of three biological replicates. *, Percentage significantly different from the wild type (t test, P < 0.05). Uptake and efflux analyses were performed for two overexpressor lines per GDU gene with similar results, only one of which is presented for clarity and simplicity. CTR, Plants expressing GFP under the control of the 35S promoter.
The culture medium in which the GDU-OEs had been grown for 2 d contained two to 60 times more amino acids compared with control plants grown in the same conditions. Gln and Asn accounted for approximately 50% to 70% of the increase (Supplemental Table S3). The amino acid profiles were similar for all GDU-OEs and different from the control, especially concerning the amino acids Asp, Glu, Asn, and Gln (Supplemental Fig. S5). Amino acid contents of the plants employed for this in vitro assay were slightly increased (by approximately 30%–50%) for GDU3-, GDU4-, GDU5-, GDU6-, and GDU7-OEs but not for GDU1-OE and GDU2-OE (Supplemental Table S4), while all GDU-OEs exhibited elevated amino acid contents when grown on soil (Fig. 4B). The increased release of amino acids into the growth medium suggested that similar nonselective amino acid export systems are stimulated by overexpression of any of the seven GDUs.
The GDU Genes Display Specific Expression Patterns That Overlap Partially in the Vascular Tissues
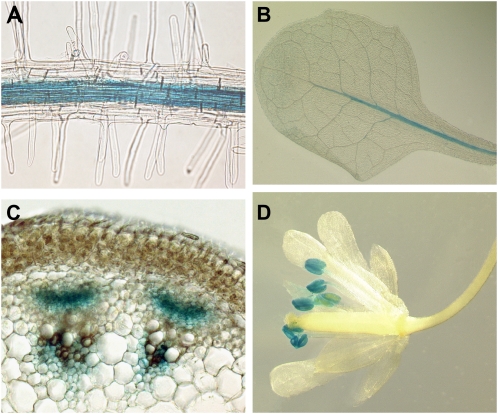
Different roles of the GDUs in the plant could be revealed by specific expression patterns for each of the genes. Analysis of the expression of the GDU genes showed that the mRNA accumulation levels varied greatly, from 12 ng of genomic DNA equivalent (see “Materials and Methods”) for GDU6 to 1.9 μg of genomic DNA equivalent for GDU3, in agreement with an analysis of Arabidopsis gene expression using tiling arrays (Table III; Laubinger et al., 2008). Both RT-PCR and tiling array showed that all GDUs were mainly expressed in roots and stems (Table III). To analyze the cell specificity of the expression, stable transformants expressing GDU promoter-GUS fusions were generated. Promoter activity of all GDUs was detected in the vascular tissues of the roots (Fig. 7A), and, except for GDU6, in vascular tissues of leaves and stems (Fig. 7, B and C). While the expression patterns overlapped in the vasculature, each GDU gene showed expression specificity for other cell types (Supplemental Table S5; Supplemental Text S1; Supplemental Fig. S6). For instance, GDU5 promoter drove GUS activity in guard cells, GDU3 promoter was active in anthers (Fig. 7D), and GDU4 and GDU7 promoters were the only ones to be active in the minor veins of the leaves, suggesting that the differences in localization of the GDU proteins could account for specific roles in different tissues.
Table III.
Absolute mRNA levels of the GDU genes in various plant organs
GDU mRNA abundance was determined by quantitative RT-PCR. Tiling array data are from Laubinger et al. (2008). Unless otherwise mentioned, organs were harvested from 6-week-old plants.
| Organ | GDU1 | GDU2 | GDU3 | GDU4 | GDU5 | GDU6 | GDU7 |
|---|---|---|---|---|---|---|---|
| Roots | 246a | 33 | 1,239 | 123 | 81 | 12 | 180 |
| 2-week-old leaves | 15 | 35 | 404 | 395 | 64 | 1 | 22 |
| 4-week-old leaves | 4 | 3 | 288 | 285 | 33 | 0.5 | 9 |
| Not senescing leaves | 14 | 5 | 513 | 174 | 49 | 0 | 5 |
| Senescing leaves | 9 | 4 | 1,783 | 126 | 72 | 0 | 1 |
| Cauline leaves | 6 | 2 | 126 | 27 | 13 | 0.2 | 2 |
| 4-week-old rosette core | 76 | 49 | 1,893 | 944 | 84 | 4 | 50 |
| Stems | 86 | 13 | 1,642 | 700 | 96 | 0.1 | 30 |
| Flowers | 10 | 18 | 562 | 94 | 4 | 0.4 | 18 |
| Young siliques | 9 | 2 | 120 | 66 | 30 | 0.1 | 64 |
| Old siliques | 1 | 0 | 7 | 6 | 3 | 0 | 0.4 |
| RT-PCR maximum | 14b | 2 | 100 | 39 | 5 | 1 | 10 |
| RT-PCR average | 6b | 1 | 100 | 25 | 6 | 0.2 | 5 |
| Tiling array maximum | 20b | 10 | 100 | 50 | 28 | 1 | 10 |
| Tiling array average | 14b | 10 | 100 | 45 | 26 | 1 | 9 |
ng genomic DNA equivalent 250 ng−1 total RNA extracted (see “Materials and Methods”).
mRNA content expressed as a percentage of GDU3 transcript levels.
Figure 7.
Localization of the activity of the GDU promoters in Arabidopsis organs. GUS activity was revealed by histochemical staining of plants expressing GUS under the control of the GDU promoters. A, Root stele, GDU1 promoter. Similar staining was obtained for all other GDUs. B, Leaves, GDU5 promoter. Similar staining was obtained for GDU1, GDU2, and GDU3. C, Stem cross section, GDU3 promoter. Similar staining was obtained for GDU1 and GDU4. D, Flowers, GDU3 promoter.
DISCUSSION
Overexpression of the GDU Genes Stimulates Amino Acid Efflux
Plant cell growth is sensitive to high levels of exogenously supplied amino acids (Heremans and Jacobs, 1994; Bonner et al., 1996; Voll et al., 2004) due to feedback inhibition of amino acid biosynthesis pathways (Less and Galili, 2008). Amino acid-resistant mutants have been found to express feedback-insensitive enzymes (Mourad and King, 1995; Li and Last, 1996; Heremans and Jacobs, 1997), show reduced amino acid uptake activity (Bright et al., 1983; Heremans et al., 1997; Lee et al., 2007), or display a deregulation of amino acid metabolism like the pig1-1 mutant (Voll et al., 2004). By analogy to pig1-1, tolerance of gdu1-1D to exogenously supplied amino acids was proposed to result from a perturbation in amino acid metabolism (Pratelli and Pilot, 2007a). The elevated amino acid content of gdu1-1D apoplasm (root xylem sap and leaf apoplasm wash fluid) also suggested an increased net efflux of amino acids from the cells (Pilot et al., 2004).
Phe, Arg, Pro, Gly, Glu, and His net uptake was lower in gdu1-1D compared with the wild type, while time-course analyses indicated that the high-affinity uptake capacity of Phe was identical in gdu1-1D and the wild type (Fig. 2D). Direct determination of amino acid efflux and analysis of amino acid content in the growth medium showed that amino acid export is enhanced in gdu1-1D. The observed decrease in Phe uptake rate shown in Figure 2D can then be explained as follows. At the beginning of the uptake experiment, Phe is more concentrated in the medium than in the cytosol (1 mm Phe in the medium and less than 0.5 mm Phe in the cytosol; Farré et al., 2001). Proton-coupled uptake systems concentrate Phe in the cytosol. When cytosolic concentration of Phe exceeds the external concentration, Phe diffuses out of the cell along the concentration gradient via the passive export system enhanced in gdu1-1D. This futile cycle of successive import and export would result in a decreased net uptake rate. Lower amino acid uptake has been shown to lead to amino acid tolerance (Bright et al., 1983; Heremans et al., 1997; Lee et al., 2007). A decreased amino acid uptake (observed for Phe, Arg, Gly, Pro, Glu, and His) consequent to an increased amino acid export would result in the amino acid tolerance described in Figure 1. Enhanced amino acid export would also explain the increased amino acid content in the xylem and apoplasm and the secretion of Gln at the hydathodes of gdu1-1D.
The overexpression of the GDU genes using the CaMV 35S promoter led to plants that were phenotypically similar to the activation-tagged mutant gdu1-1D. The GDU-OEs displayed increased free leaf amino acid content when grown on soil (Fig. 4B), enhanced amino acid tolerance, decreased Phe uptake rate over time, and increased amino acid export into the medium. All the GDU-OEs exported a similar set of amino acids, different from the wild type (Supplemental Fig. S5). All GDUs have similar functional properties and are able to increase amino acid export upon overexpression. The weaker phenotype of the GDU7-OEs (Figs. 4 and 6) is possibly related to the sequence divergence of GDU7 from the other genes (Pratelli and Pilot, 2007b).
Mechanisms of Amino Acid Efflux in Plants
Tolerance of gdu1-1D to 11 out of 14 toxic amino acids (Fig. 1; Pratelli and Pilot, 2007a), reduction of Pro and Glu uptake, and increased release into the medium of most amino acids (Table II) suggested that at least 16 amino acids are substrates of the export mechanism enhanced by the GDU1 overexpression. The transport appeared not specific to any amino acid but seemed to be more efficient with Gln and Asn. The observed similarity of the amino acid composition of apoplasm and cytosol further indicates that the amino acid export system is poorly selective (Lohaus et al., 1995; Pilot et al., 2004; Hirner et al., 2006).
The GDU1-stimulated amino acid export mechanism was not dependent on the proton gradient or ATP hydrolysis (Fig. 3). Consequently, vesicular trafficking and ATP-binding cassette transporters, requiring ATP for energization, and putative amino acid/proton antiporters are not candidates for the export mechanism. Another possibility would be a facilitator, which allows passive transport of amino acids along the concentration gradient. The increased activity of a facilitator in gdu1-1D is expected to lead to increased Phe uptake when external Phe concentration is higher than in the cytosol (e.g. 5 mmol L−1; Fig. 3A, CCCP), which was not observed. The GDU1-stimulated amino acid export then appears to be mediated by a system able to transport amino acids only toward the outside of the cell. Such a system and the corresponding export mechanism still remain to be identified in plants.
Amino acid exporters from other organisms have been found to be (1) highly selective, like the vesicular γ-aminobutyrate and Glu exporters of neurons (McIntire et al., 1997; Takamori et al., 2000); (2) selective for a class of amino acids, such as the amino acid exporters from microbes (Eggeling and Sahm, 2003); or (3) poorly selective, like an amino acid exporter cloned from yeast (Velasco et al., 2004). The cloning and the characterization of the transporter BAT1 has recently shed light on plant amino acid export systems (Dundar and Bush, 2009). Amino acid tolerance and uptake experiments have suggested that BAT1 can both import and export amino acids when expressed in yeast and might be poorly selective for amino acids. BAT1 is expressed at high levels throughout the plants, especially in vascular tissues (Dundar, 2009), compatible with an important role in amino acid export from cells.
Because of its expression and functional properties, BAT1 would be a candidate for the export system stimulated by GDU1 overexpression. By analogy to the effects of subunits of animal transporters, GDU1 would modulate the trafficking and/or the functional properties of BAT1. Mammalian heteromeric amino acid transporters require the activity of a single-transmembrane domain protein for correct targeting to the plasma membrane (Palacin and Kanai, 2004; Verrey et al., 2004). Transporter functional properties have been also shown to be modified by regulatory subunits. For instance, association of β-subunits to potassium channels can modify the opening properties of the complex (McCormack et al., 1999). In this context, GDU1 could be an accessory subunit of BAT1, controlling its cellular trafficking and/or functional properties: upon GDU1 overexpression, BAT1 could accumulate, or be more active, at the plasma membrane and function as an exporting-only amino acid transporter. This hypothesis could be tested by analyzing the phenotype of a gdu1-1D/bat1 double mutant or the trafficking of BAT1 protein in a gdu1-1D background.
Differences in the Roles of the GDU Genes
The absence of an apparent phenotype of the GDU knockout mutants, the overlapping expression patterns, and the overall similar phenotypes of the GDU overexpressors suggest functional redundancy among the GDU proteins. GDUs are expressed in regions where amino acid transport is known to occur. Phloem has been shown to unload in seeds at the chalaza/funiculus region (Stadler et al., 2005), where GDU1 and GDU2 are expressed (Supplemental Fig. S6). Amino acid efflux has been detected from roots (Schobert and Komor, 1987; Jones and Darrah, 1993; Phillips et al., 2004), and it has been suggested that efflux takes place mainly at the root tip, where GDU2 and GDU3 are expressed. Amino acids constitutively exported from roots are recovered by active transporters (Schobert and Komor, 1987; Phillips et al., 2004). A similar efflux/import cycle is also suspected to occur along the root stele, where high-affinity amino acid importers (Okumoto et al., 2002, 2004) and the GDU proteins are expressed. The GDU proteins could also be involved in apoplasmic transport of amino acids from one cell to another, necessary for xylem loading and phloem reloading (Schobert and Komor, 1990; Lalonde et al., 2003).
The phenotypes of the GDU-OEs are not identical in every respect. Several differences were noted for the amount of secretion crystals on the leaves (Fig. 4A) and the magnitude of the size reduction and amino acid content upon overaccumulation of the GDU mRNAs (Supplemental Figs. S2 and S3). In addition to the specificities of expression patterns (Supplemental Fig. S6; Supplemental Table S5), the phenotypical differences suggest that the GDUs play different roles in the plant and are endowed with similar but not identical functional properties.
CONCLUSION
Uptake of radiolabeled amino acids by gdu1-1D was reduced and gdu1-1D plants excreted more amino acids in the growth medium than the wild type, indicating an increased amino acid export. The overexpression of the GDU1-like proteins from Arabidopsis led to amino acid tolerance and increased amino acid export, suggesting that the GDU proteins have a similar function. Differential expression of the seven GDU genes suggests specific roles in amino acid export in different cell types. The data presented here provide direct evidence that the overexpression of the GDU genes specifically stimulates amino acid export and that they potentially act as regulators of amino acid exporters. It is expected that the study of the GDUs and interacting proteins will shed light on the poorly understood export mechanisms in plants.
MATERIALS AND METHODS
Plant Growth and Transformation
Arabidopsis (Arabidopsis thaliana ecotype Columbia-7) plants were grown in soil (Floragard type B without clay from Floradur) in the greenhouse or in a growth room (16 h of light, 120 μmol m−2 s−1, at 23°C) and were watered from below. Fertilizer was applied once at the time of bolting.
Constructs were introduced into plants using Agrobacterium tumefaciens GV3101 (pMP90) and the floral dip method (Clough and Bent, 1998). Transgenic plant selection and segregation analyses for kanamycin resistance were performed in vitro on Murashige and Skoog (MS) medium (Phytotechnology Laboratories) containing 50 μg mL−1 kanamycin, 1% (w/v) Suc, and 0.7% (w/v) agar (16 h of light, 120 μmol m−2 s−1, at 22°C).
For amino acid tolerance experiments, surface-sterilized seeds were sown using 0.1% (w/v) agar on half-strength MS medium containing 0.5% (w/v) Suc, 0.5 g L−1 MES, pH 5.7, 0.8% (w/v) agar, and various concentrations of amino acids. Plants were grown vertically for 10 d in a growth cabinet (16 h of light, 80 μmol m−2 s−1, at 22°C). Plates were scanned and root lengths were calculated using ImageJ (http://rsbweb.nih.gov/ij/).
Expression Analysis
Total RNA extraction and northern blotting were performed as described previously (Pratelli and Pilot, 2006). Overaccumulated mRNAs were quantitated by reference to a dot blot. For this purpose, amounts from 2 to 500 amol of PCR fragments corresponding to the full coding sequence of the GDU genes were dotted on a nylon membrane using 0.4 n NaOH. These membranes were hybridized at the same time and in the same hybridization bottle as the membranes used for the RNA gel blots. Signals were quantitated with the Typhoon 9400 scanner and ImageQuant software (GE Healthcare). Pixel intensities on RNA gel blots were converted in amol DNA using the standard curve obtained with the dot blots. Probes corresponded to the coding sequence of the GDU genes.
For real-time RT-PCR, RNA (10 μg) was treated with DNAse I (Invitrogen) for 30 min at room temperature and precipitated in the presence of ethanol. After solubilization in 10 μL of water, 2.5 μg of RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen) in a 10-μL reaction. Real-time PCR was performed on 5 μL of a 50-fold dilution of the RT product using the Lightcycler 480 SYBR Green I Master Mix and detected with a Roche 480 Lightcycler (Roche Applied Science). Threshold cycle values were determined by the Roche Lightcycler 480 SW 1.5 software. Amplification efficiencies were determined using amounts from 2.5 pg to 2.5 ng of genomic DNA as template and used for absolute quantitation of mRNA levels. Absolute quantities of mRNA are given in genomic DNA equivalents, corresponding to the amount of genomic DNA that would be needed as template to get the same threshold cycle as the RT template. Sequences of the oligonucleotides used as primers for quantitative PCR are available upon request.
Constructs
pPTkan3 and pUTkan binary vectors are derivatives of pJHA212K (Yoo et al., 2005). The promoterless pUTkan contains, in this order, a multiple cloning site, the Escherichia coli β-glucuronidase coding sequence, and the Rubisco terminator from pea (Pisum sativum). pPTkan3 contains the CaMV 35S promoter, a multiple cloning site, and the Rubisco terminator from pea. Promoters of the GDU genes (between 1,300 and 3,050 bp) were amplified by PCR (Pfx accuprime; Invitrogen) from genomic DNA and cloned into pUTkan using restriction enzymes. DNA fragments starting at the start codon and ending close to the poly(A) tail of the GDU mRNAs were amplified by PCR as above and cloned into pPTkan3. All amplified fragments were sequenced after cloning to ensure the absence of any error. Cloning strategies, primer sequences, and resulting plasmids are available upon request.
Localization of Gene Expression
Histochemical staining of GUS activity was performed on in vitro- and soil-grown plants as described (Lagarde et al., 1996). Histochemical staining reactions were performed at 37°C for times ranging from 2 to 24 h, depending on the organ and the studied gene. After staining and clearing in ethanol, stems were embedded in 4% (w/v) agarose and cut into thin sections by hand. For each gene, the localization of the staining was studied on about 20 independent lines. Histochemical staining of plant organs was more precisely investigated in parallel on six lines representative of the most frequently observed staining pattern and showing a similar expression level.
Amino Acid Quantitation
Tissues were frozen in liquid nitrogen, freeze dried, and ground with a 5-mm steel ball in a Tissue-lyzer (Qiagen). Amino acids were extracted from the dry powder by 500 μL of 80% (v/v) ethanol at 80°C for 30 min. The pellets obtained by centrifugation at 16,000g were extracted a second time in the same way. The supernatants were pooled and dried under vacuum. Chlorophyll content was estimated by spectrophotometry of a 5-fold dilution of the extracts in 80% ethanol according to Lichtenthaler (1987).
Total amino acid content was determined by reaction with ninhydrin. Dried samples were solubilized in 250 μL of 20% ethanol. After centrifugation, 30 μL of the supernatant was mixed with 200 μL of ninhydrin reagent (3.34 m propionic acid, 2.1 n NaOH, 50% [v/v] 2-ethoxy-ethanol, and 2% [w/v] ninhydrin) and 100 μL of 0.1% (w/v) ascorbic acid. The mixture was heated for 10 min at 95°C and cooled for 5 min on ice, and the optical density at 570 nm was determined after addition of 500 μL of 60% (v/v) ethanol. Reactions with increasing amounts of Gly were used to establish a standard curve.
For determination of amino acid content in plants, extracts were prepared as above. For determination of amino acids present in the medium, growth solutions were filtered (0.2-μm pore size) and dried under vacuum. Amino acids were then derivatized with fluorophore 6-aminoquinolyl-N-hydroxysuccimidyl carbamate (AccQ Taq; Waters), and the amino acid derivatives were separated at a flow rate of 1 mL min−1 at 37°C on a Dionex Summit HPLC system essentially as described (van Wandelen and Cohen, 1997) using the eluents A (140 mm sodium acetate, pH 6, and 7 mm triethanolamine), B (acetonitrile), and C (water) and detected by fluorescence (excitation at 300 nm, detection at 400 nm).
Amino Acid Transport Analysis in Plantlets
Arabidopsis plantlets were grown for 7 d (16 of h light, 80–120 μmol m−2 s−1, at 21°C) on solid (0.7% agar) half-strength MS medium containing 1% (w/v) Suc, pH 5.7. Plates contained 50 μg mL−1 kanamycin for the selection of plants harboring the 35Sp-GDU constructs or the 35Sp-GFP construct, used as a control. About six plants were immersed into 3 mL of half-strength MS medium containing 1% (w/v) Suc, pH 5.7, and grown on a 12-well plate under gentle shaking (40 rpm) for 4 to 5 d with the same light regime. One hour before transport analyses, plants were transferred to a 24-well plate, into 1 mL of the same medium, and shaken for 1 h at room temperature under incandescent light for acclimatization to the uptake conditions. The solution was replaced with fresh medium containing between 3.7 and 7.4 kBq U-14C-radiolabeled substrate (GE Healthcare) and unlabeled substrate supplied at the concentrations indicated in the figures. Plantlets were washed three times with 5 mL of 0.2 mm CaSO4. For efflux analyses, plants were allowed to take up the radiolabeled substrate for 10 min, then rinsed three times with 0.2 mm CaSO4 and transferred to 1 mL of half-strength MS medium without Suc, pH 5.7. Radioactivity released in the medium was quantitated by scintillation counting of the solution, after addition of 5 mL of scintillation cocktail (Ultima Gold XR; Perkin-Elmer). Plants were then dried for 3 h at 70°C, weighed, and digested overnight in 1 mL of 7% NaClO. Radioactivity in the samples was measured by scintillation counting several hours after the addition of 5 mL of scintillation cocktail. For treatments with proton gradient and ATP hydrolysis inhibitors, preincubation (40 min), uptake, and efflux were performed in the presence of 100 μm CCCP, DNP, or DES (stock solutions at 40 mmol L−1 in dimethyl sulfoxide) or 1 mm VO4 (stock solution at 100 mmol L−1 in water).
For thin-layer chromatography analyses of amino acid extracts, the efflux medium (half-strength MS medium, pH 5.7, without any Suc) was dried under vacuum. Metabolites were solubilized in 30 μL of 70% ethanol, and 15 μL was loaded on thin-layer chromatography silica plates (GF; Analtech), 1 μL at a time. The plates were developed for 2 h at room temperature with a 1-butanol:acetic acid:water (3:1:1) mix. After drying under air flow, the plates were placed for 7 d against a storage phosphor screen. The screen was then scanned by a Typhoon 9400 scanner.
The locus numbers of the GDU genes are as follows: GDU1, At4g31730; GDU2, At4g25760; GDU3, At5g57685; GDU4, At2g24762; GDU5, At5g24920; GDU6, At3g30725; GDU7, At5g38770.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of Phe accumulation and export in wild-type, gdu1-1D, and lht1-1 plants.
Supplemental Figure S2. Correlation between the mRNA levels and the size of plants overexpressing the GDU genes.
Supplemental Figure S3. Correlation between the mRNA levels and the free amino acid content of plants overexpressing the GDU genes.
Supplemental Figure S4. Time-course analysis of Phe accumulation in wild-type and GDU-overexpressing plants.
Supplemental Figure S5. Amino acid content of plants grown in liquid culture.
Supplemental Figure S6. Localization of the activity of the GDU promoters in the organs of Arabidopsis.
Supplemental Table S1. Summary of the GDU-overexpressing lines from Supplemental Figures S3 and S4 chosen for further analyses.
Supplemental Table S2. Free amino acid accumulation in leaves of greenhouse-grown plants overexpressing GDU1, GDU2, GDU3, GDU4, GDU5, and GDU6.
Supplemental Table S3. Amino acid content of medium from GDU-OEs and the control, grown in liquid medium.
Supplemental Table S4. Amino acid profiles of plants from GDU-OEs and the control, grown in liquid medium.
Supplemental Table S5. Summary of the localization of the promoter activity of the GDU genes.
Supplemental Text S1. Analysis of the expression pattern of the GDU genes.
Supplementary Material
Acknowledgments
We thank Prof. Uwe Sonnewald (Friedrich-Alexander University, Erlangen-Nuremberg, Germany) for providing HPLC facilities for amino acid analysis.
This work was supported by the National Science Foundation (“Arabidopsis 2010” grant no. 0618402 to W.B.F.) and the Deutsche Forschungsgemeinschaft (grant nos. PI 607/2–1 to G.P. and VO 985/1–1 to L.M.V.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Guillaume Pilot (gpilot@vt.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bonner CA, Williams DS, Aldrich HC, Jensen RA (1996) Antagonism by L-glutamine of toxicity and growth inhibition caused by other amino acids in suspension cultures of Nicotiana sylvestris. Plant Sci 113 43–58 [Google Scholar]
- Bright SWJ, Kueh JSH, Rognes SE (1983) Lysine transport in two barley mutants with altered uptake for basic amino acids in the root. Plant Physiol 72 821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- De Jong A, Koerselman-Kooij JW, Schuurmans J, Borstlap AC (1997) The mechanism of amino acid efflux from seed coats of developing pea seeds as revealed by uptake experiments. Plant Physiol 114 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundar E (2009) Multiple GUS expression patterns of a single Arabidopsis gene. Ann Appl Biol 154 33–41 [Google Scholar]
- Dundar E, Bush DR (2009) BAT1, a bidirectional amino acid transporter in Arabidopsis. Planta 229 1047–1056 [DOI] [PubMed] [Google Scholar]
- Eggeling L, Sahm H (2003) New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch Microbiol 180 155–160 [DOI] [PubMed] [Google Scholar]
- Farré EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L (2001) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiol 127 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferrière N, Thibaud JB, Sentenac H (1998) Identification and disruption of a plant Shaker-like outward channel involved in K+ release into the xylem sap. Cell 94 647–655 [DOI] [PubMed] [Google Scholar]
- Heremans B, Borstlap AC, Jacobs M (1997) The rtl1 and raec1 mutants of Arabidopsis thaliana lack the activity of a basic-amino-acid transporter. Planta 201 219–226 [DOI] [PubMed] [Google Scholar]
- Heremans B, Jacobs M (1994) Selection of Arabidopsis thaliana (L.) Heynh. mutants resistant to aspartate-derived amino acids and analogues. Plant Sci 101 151–162 [Google Scholar]
- Heremans B, Jacobs M (1997) A mutant of Arabidopsis thaliana (L.) Heynh. with modified control of aspartate kinase by threonine. Biochem Genet 35 139–153 [DOI] [PubMed] [Google Scholar]
- Hirner A, Ladwig F, Stransky H, Okumoto S, Keinath M, Harms A, Frommer WB, Koch W (2006) Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18 1931–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger CH III, Lindow SE, Miller W, Clark E, Firestone MK (1999) Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol 65 2685–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Darrah PR (1993) Influx and efflux of amino acids from Zea mays L. roots and their implications for N nutrition and the rhizosphere. Plant Soil 155/156 87–90 [Google Scholar]
- Jones DL, Darrah PR (1994) Amino-acid influx at the soil-root interface of Zea mays L. and its implications in the rhizosphere. Plant Soil 163 1–12 [Google Scholar]
- Kinraide TB (1981) Interamino acid inhibition of transport in higher plants. Plant Physiol 68 1327–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W, Kwart M, Laubner M, Heineke D, Stransky H, Frommer WB, Tegeder M (2003) Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J 33 211–220 [DOI] [PubMed] [Google Scholar]
- Lagarde D, Basset M, Lepetit M, Conejero G, Gaymard F, Astruc S, Grignon C (1996) Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J 9 195–203 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Tegeder M, Throne-Holst M, Frommer WB, Patrick JW (2003) Phloem loading and unloading of sugars and amino acids. Plant Cell Environ 26 37–56 [Google Scholar]
- Laubinger S, Zeller G, Henz SR, Sachsenberg T, Widmer CK, Naouar N, Vuylsteke M, Scholkopf B, Ratsch G, Weigel D (2008) At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol 9 R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Foster J, Chen J, Voll LM, Weber AP, Tegeder M (2007) AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J 50 305–319 [DOI] [PubMed] [Google Scholar]
- Less H, Galili G (2008) Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol 147 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Last RL (1996) The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble tryptophan. Plant Physiol 110 51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bush DR (1990) D-pH dependent amino acid transport into plasma membrane vesicles isolated from sugar beet leaves. Plant Physiol 94 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148 350–382 [Google Scholar]
- Lohaus G, Winter H, Riens B, Heldt HW (1995) Further studies of the phloem loading process in leaves of barley and spinach: the comparison of metabolite concentrations in the apoplastic compartment with those in the cytosolic compartment and in the sieve tubes. Bot Acta 108 270–275 [Google Scholar]
- Marschner H, Kirkby EA, Cakmak I (1996) Effect of the nutritional status on shoot-root partitioning of photoassimilates and cycling of mineral nutrients. J Exp Bot 47 1255–1263 [DOI] [PubMed] [Google Scholar]
- Marschner H, Kirkby EA, Engels C (1997) Importance of cycling and recycling of mineral nutrients within plants for growth and development. Bot Acta 4 265–274 [Google Scholar]
- McCormack T, McCormack K, Nadal MS, Vieira E, Ozaita A, Rudy B (1999) The effects of Shaker beta-subunits on the human lymphocyte K+ channel Kv1.3. J Biol Chem 274 20123–20126 [DOI] [PubMed] [Google Scholar]
- McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM (1997) Identification and characterization of the vesicular GABA transporter. Nature 389 870–876 [DOI] [PubMed] [Google Scholar]
- Mourad G, King J (1995) l-O-Methylthreonine-resistant mutant of Arabidopsis defective in isoleucine feedback regulation. Plant Physiol 107 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näsholm T, Kielland K, Ganeteg U (2009) Uptake of organic nitrogen by plants. New Phytol 182 31–48 [DOI] [PubMed] [Google Scholar]
- Okumoto S, Koch W, Tegeder M, Fischer WN, Biehl A, Leister D, Stierhof YD, Frommer WB (2004) Root phloem-specific expression of the plasma membrane amino acid proton co-transporter AAP3. J Exp Bot 55 2155–2168 [DOI] [PubMed] [Google Scholar]
- Okumoto S, Schmidt R, Tegeder M, Fischer WN, Rentsch D, Frommer WB, Koch W (2002) High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J Biol Chem 277 45338–45346 [DOI] [PubMed] [Google Scholar]
- Palacin M, Kanai Y (2004) The ancillary proteins of HATs: SLC3 family of amino acid transporters. Pflugers Arch 447 490–494 [DOI] [PubMed] [Google Scholar]
- Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR (2004) Microbial products trigger amino acid exudation from plant roots. Plant Physiol 136 2887–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VP, Frommer WB (2004) Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Pilot G (2006) The plant-specific VIMAG domain of Glutamine Dumper1 is necessary for the function of the protein in Arabidopsis. FEBS Lett 580 6961–6966 [DOI] [PubMed] [Google Scholar]
- Pratelli R, Pilot G (2007. a) Altered amino acid metabolism in glutamine dumper1 plants. Plant Signal Behav 2 171–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Pilot G (2007. b) Corrigendum to “The plant-specific VIMAG domain of Glutamine Dumper1 is necessary for the function of the protein in Arabidopsis,” FEBS Lett 580 (2006) 6961–6966. FEBS Lett 581 1248–1249 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M (2007) Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 581 2281–2289 [DOI] [PubMed] [Google Scholar]
- Schobert C, Komor E (1987) Amino acid uptake by Ricinus communis roots: characterization and physiological significance. Plant Cell Environ 10 493–500 [Google Scholar]
- Schobert C, Komor E (1990) Transfer of amino acids and nitrate from the roots into the xylem of Ricinus communis seedlings. Planta 181 85–90 [DOI] [PubMed] [Google Scholar]
- Stadler R, Lauterbach C, Sauer N (2005) Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol 139 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Frommer WB, Ludewig U (2004) Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol 136 3104–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennerstam H, Ganeteg U, Bellini C, Näsholm T (2007) Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiol 143 1853–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennerstam H, Ganeteg U, Näsholm T (2008) Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease 5. New Phytol 180 620–630 [DOI] [PubMed] [Google Scholar]
- Takamori S, Rhee JS, Rosenmund C, Jahn R (2000) Identification of a vesicular glutamate transporter that defines a glutamatergic phenotype in neurons. Nature 407 189–194 [DOI] [PubMed] [Google Scholar]
- Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, Hayashi H, Yoneyama T, Fujiwara T (2002) Arabidopsis boron transporter for xylem loading. Nature 420 337–340 [DOI] [PubMed] [Google Scholar]
- van Wandelen C, Cohen SA (1997) Using quaternary high-performance liquid chromatography eluent systems for separating 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate derivatized amino acid mixtures. J Chromatogr A 763 11–22 [Google Scholar]
- Velasco I, Tenreiro S, Calderon IL, André B (2004) Saccharomyces cerevisiae Aqr1 is an internal-membrane transporter involved in excretion of amino acids. Eukaryot Cell 3 1492–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrey F, Closs EI, Wagner CA, Palacin M, Endou H, Kanai Y (2004) CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch 447 532–542 [DOI] [PubMed] [Google Scholar]
- Voll LM, Allaire EE, Fiene G, Weber AP (2004) The Arabidopsis phenylalanine insensitive growth mutant exhibits a deregulated amino acid metabolism. Plant Physiol 136 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SY, Bomblies K, Yoo SK, Yang JW, Choi MS, Lee JS, Weigel D, Ahn JH (2005) The 35S promoter used in a selectable marker gene of a plant transformation vector affects the expression of the transgene. Planta 221 523–530 [DOI] [PubMed] [Google Scholar]
- Zhang WH, Zhou Y, Dibley KE, Tyerman SD, Furbank RT, Patrick JW (2007) Nutrient loading of developing seeds. Funct Plant Biol 34 314–331 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.