Abstract
Multimeric protein complexes are required during development to regulate transcription and orchestrate cellular proliferation and differentiation. The Arabidopsis (Arabidopsis thaliana) SEUSS (SEU) gene encodes a transcriptional adaptor that shares sequence similarity with metazoan Lim domain-binding transcriptional adaptors. In Arabidopsis, SEU forms a physical complex with the LEUNIG transcriptional coregulator. This complex regulates a number of diverse developmental events, including proper specification of floral organ identity and number and the development of female reproductive tissues derived from the carpel margin meristem. In addition to SEU, there are three Arabidopsis SEUSS-LIKE (SLK) genes that encode putative transcriptional adaptors. To determine the functions of the SLK genes and to investigate the degree of functional redundancy between SEU and SLK genes, we characterized available slk mutant lines in Arabidopsis. Here, we show that mutations in any single SLK gene failed to condition an obvious morphological abnormality. However, by generating higher order mutant plants, we uncovered a degree of redundancy between the SLK genes and between SLK genes and SEU. We report a novel role for SEU and the SLK genes during embryonic development and show that the concomitant loss of both SEU and SLK2 activities conditions severe embryonic and seedling defects characterized by a loss of the shoot apical meristem. Furthermore, we demonstrate that SLK gene function is required for proper development of vital female reproductive tissues derived from the carpel margin. We propose a model that posits that SEU and SLK genes support organ development from meristematic regions through two different pathways: one that facilitates auxin response and thus organ initiation and a second that sustains meristematic potential through the maintenance of SHOOTMERISTEM-LESS and PHABULOSA expression.
The control of transcriptional programs during development requires the action of multimeric protein complexes that interact with DNA regulatory regions and alter transcriptional efficiency. These complexes contain sequence-specific DNA-binding proteins that directly bind to the DNA as well as coregulatory proteins (coregulators or adaptors) that bridge interactions between the sequence-specific DNA-binding proteins and the general transcriptional machinery.
Lim domain-binding (Ldb) proteins are metazoan transcriptional adaptors required for a diversity of developmentally important events, including specification of chick neuronal identity and function of the amphibian Spemann organizer during dorsal/ventral polarity specification (Agulnick et al., 1996; Thaler et al., 2002; Matthews and Visvader, 2003). Ldb proteins have no known DNA-binding or enzymatic activities; rather, they function as adaptor proteins that provide protein interaction surfaces required for the assembly of multimeric regulatory complexes. Metazoan Ldb proteins contain a LIM-interaction domain (LID) required for interactions with LIM domain-containing proteins as well as a dimerization domain (DD) that supports the formation of higher order protein complexes (Fig. 1A; Agulnick et al., 1996; Jurata et al., 1996; Jurata and Gill, 1997). Ldb proteins also contain an Ldb1/Chip conserved domain (LCCD) that is contiguous with the dimerization domain. The LCCD was originally identified in the Drosophila Chip protein, a member of the Ldb adaptor family (van Meyel et al., 2003). The LCCD mediates the physical interaction of the Chip protein with another Drosophila transcriptional coregulator, Ssdp (for single-stranded DNA-binding protein).
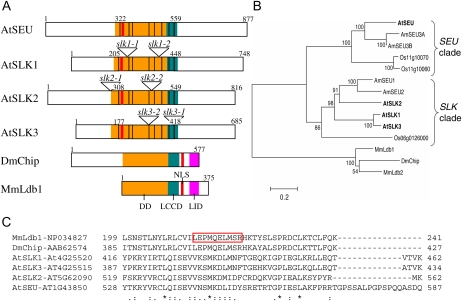
Figure 1.
Structural and evolutionary relationships of SEU, SLK, and Ldb proteins. A, Conserved protein domains in AtSEU, AtSLK, and select metazoan Ldb proteins. The Arabidopsis proteins share sequence similarity to the DD of Ldb1 (orange) and the LCCD of Chip (teal). The LID domain in metazoan Ldb proteins (purple) does not appear to be conserved in the AtSEU and AtSLK proteins. Numbers indicate amino acid positions, and black dividing lines indicate the locations of exon/exon boundaries that are conserved in the Arabidopsis proteins. Mutant allele insertion sites are indicated for SLK genes. B, The evolutionary history of full-length SLK proteins inferred using the neighbor-joining method (Saitou and Nei, 1987). The optimal tree (shown) suggests that the AtSLK proteins fall into a clade that is distinct from that containing AtSEU. The percentages of bootstrap support are shown next to the branches (Felsenstein, 1985). Evolutionary distances are in units of number of amino acid substitutions per site. C, ClustalW2 analysis of the LCCD region of metazoan Ldb and Arabidopsis SLK proteins. LCCD spans amino acids 201 to 249 in MmLdb1 and 387 to 435 in DmChip; the red boxed area indicates 10 amino acids deleted from Ldb1 that specifically disrupt activity of LCCD (van Meyel et al., 2003). At, Arabidopsis thaliana; Am, Antirrhinum majus; Dm, Drosophila melanogaster; Mm, Mus musculus; NLS, nuclear localization signal; Os, Oryza sativa. Protein identifiers not listed above are as follows: MmLdb2, NP034828; Os06g0126000, NP_001056655; Os11g10060, ABA91996; Os11g10070, ABA91997; AmSEU1, AJ620907; AMSEU2, AJ620908; AmSEU3A, AJ620909; AmSEU3B, AJ620910.
The Arabidopsis (Arabidopsis thaliana) SEUSS (SEU) gene (AT1G43850) encodes a transcriptional adaptor that shares sequence similarity with the metazoan Ldb proteins throughout the DD and LCCD (Franks et al., 2002; Fig. 1A). However, SEU does not contain a recognizable LID, and there is no evidence of physical interactions between SEU and plant LIM domain-containing proteins. In Arabidopsis, the SEU LCCD likely supports a physical interaction between SEU and two paralogous transcriptional coregulators: LEUNIG (LUG) and LEUNIG_HOMOLOGUE (LUH; Sridhar et al., 2004, 2006; Sitaraman et al., 2008). The Arabidopsis LUG and LUH proteins share a conserved domain with the metazoan Ssdp proteins, and this domain is required for physical interactions between LUG and SEU in Arabidopsis and Ssdp and Ldb1 in metazoans (Conner and Liu, 2000; van Meyel et al., 2003; Sridhar et al., 2004). Thus, the LCCD portion of the SEU protein likely mediates a set of protein-protein interactions that are functionally conserved across the plant and animal kingdoms (van Meyel et al., 2003).
The Arabidopsis SEU gene encodes a nucleus-localized protein that is expressed widely throughout many developmental stages and tissues (Franks et al., 2002; Azhakanandam et al., 2008). Functional analyses indicate that SEU plays multiple roles during Arabidopsis development. The most well characterized of these is a role for SEU in the repression of AGAMOUS (AG) during floral organ identity specification (Franks et al., 2002). Within the developing flower, the SEU/LUG protein complex physically interacts with pairs of MADS domain DNA-binding proteins (i.e. SEPALATA3, APETALA1, SHORT VEGETATIVE PHASE1, and AGAMOUS-LIKE24) that bind to AG regulatory sequences (Gregis et al., 2006; Sridhar et al., 2006). LUG then recruits HDA19, a class 1 histone deacetylase, as well as components of the Mediator complex to bring about repression of AG transcription in the perianth (nonreproductive) floral organs (Gonzalez et al., 2007).
SEU also functions during the development of the carpel margin meristem (CMM; Azhakanandam et al., 2008). The CMM (also termed the medial ridge of the gynoecium) is a vital meristematic structure that is located on the margins of the fused Arabidopsis carpels and gives rise to several critical female reproductive structures including the ovules, the septum, and the transmitting tract (Bowman et al., 1999). SEU functions in a partially redundant manner with a number of other transcriptional regulators, including AINTEGUMENTA (ANT), FILAMENTOUS FLOWER/YABBY1 (FIL/YAB1), and LUG, during the development of the CMM (Liu et al., 2000; Nole-Wilson and Krizek, 2006; Azhakanandam et al., 2008). The analysis of lug ant and seu ant double mutant gynoecia indicates that the disruption of CMM development in these genotypes is not caused by ectopic AG expression (Liu et al., 2000; Azhakanandam et al., 2008). Rather, these experiments support a model in which SEU, LUG, and ANT maintain the expression of the adaxial fate determinant PHABULOSA (PHB) and thus reinforce polarity specification within the gynoecium. Thus, the SEU and LUG coregulators participate in a diversity of transcriptional events during varied developmental processes in a manner analogous to that of the Ldb and SSDP metazoan proteins (van Meyel et al., 2003).
SEU is also required for phenotypic and transcriptional responses to the auxin class of plant hormones (Pfluger and Zambryski, 2004). The seu mutant seedlings display a reduced sensitivity to applied auxin as well as a variety of auxin-resistant growth phenotypes. Furthermore, the expression of the auxin-sensitive DR5:GUS reporter (Ulmasov et al., 1997) was reduced in seu mutant roots relative to wild-type roots (Pfluger and Zambryski, 2004). SEU was shown to physically interact with ETTIN/AUXIN RESPONSE FACTOR3 (ETT/ARF3) in a yeast two-hybrid assay (Pfluger and Zambryski, 2004). ETT/ARF3 is a member of a family of transcription factors that bind to auxin response elements located in the regulatory regions of auxin-responsive genes (Sessions et al., 1997). Thus, the physical interaction between SEU and ETT suggests a potential direct role for SEU during auxin response.
In addition to SEU, there are three Arabidopsis SEUSS-LIKE (SLK) genes that encode putative transcriptional adaptors containing a Ldb-type DD followed by an LCCD: SLK1 (AT4G25520), SLK2 (AT5G62090), and SLK3 (AT4G25515; Franks et al., 2002; Fig. 1A). The sequence similarity between SEU and the SLK genes suggests that they may share functional redundancy. Putative Antirrhinum orthologs of SEU and the SLK genes physically interact with STYLOSA, the Antirrhinum LUG ortholog; however, mutations in the Antirrhinum SLK genes and SEU have not been isolated (Navarro et al., 2004).
To determine the functions of the SLK genes and investigate the degree of functional redundancy between SEU and SLK genes, we characterized available slk mutant lines in Arabidopsis. Here, we show that mutations in any single SLK gene failed to condition an obvious morphological abnormality. However, by generating higher order mutant plants, we uncovered a degree of redundancy between the SLK genes and between SLK genes and SEU. Notably, the concomitant loss of both SEU and SLK2 activities results in severe embryonic and seedling defects that are characterized by a loss of all structures derived from the shoot apical meristem (SAM). These observations suggest a previously unrecognized role for SEU and the SLK genes during embryonic development. We also demonstrate that SLK genes function in the development of the CMM in a manner that is similar to that of SEU. We propose a model that suggests that SEU and SLK genes support organ development from meristematic regions through two different pathways: one that facilitates auxin response and thus organ initiation and a second that sustains meristematic potential through the maintenance of SHOOTMERISTEM-LESS (STM) and PHB expression.
RESULTS
Structural and Phylogenetic Analyses of SEU, SLK, and Ldb Proteins
The three Arabidopsis SLK genes and SEU all encode putative transcriptional adaptors that share sequence similarity to the Ldb family of transcriptional regulators within the DD and LCCD (Franks et al., 2002; Fig. 1A). Outside of the DD and LCCD, the three SLK genes share significant sequence similarity with each other, but their sequences diverge from SEU. Thus, the three SLK genes are more similar to each other than they are to SEU. This became more apparent when we employed a neighbor-joining method to generate a phylogeny for this group of transcriptional regulators including members from other monocot and dicot species. This analysis supports the separation of the SLK genes into a phylogenetic clade (SLK clade) that is distinct from that of SEU (SEU clade; Fig. 1B). A protein sequence similarity/identity matrix also supports the differentiation of the SEU and SLK clades (Table I). The percentage identity between AtSEU and the putative rice (Oryza sativa) SEU ortholog Os11g10060 is higher than it is between AtSEU and the Arabidopsis SLK genes (Table I). The apparent sequence divergence in the N- and C-terminal portions suggests that these protein regions may support functional differences between the SEU and SLK clade members. The SLK1 and SLK3 genes appear to have arisen from a tandem duplication event on chromosome 4 and encode highly similar proteins (greater than 80% similarity). For this article, we use the term “SLK genes” to describe genes in the SLK clade (i.e. SLK1, SLK2, and SLK3 in Arabidopsis) and the term “SEUSS-related genes” to describe the larger gene family that includes members of both clades. Interestingly, the rice and snapdragon (Antirrhinum majus) genomes encode at least two paralogs in the SEU clade, while AtSEU is the only paralog found in the SEU clade in Arabidopsis. We carried out ClustalW2 analysis to more closely examine the sequence similarity within the LCCD of SEU, the SLK proteins, and the metazoan Ldb proteins (Fig. 1C). Our analysis indicates that the LCCD sequence is conserved in SEU as well as in the three SLK proteins and suggests that the SLK proteins may physically interact with LUG as well as with the LUH protein, another Arabidopsis coregulator that is structurally related to LUG (Sitaraman et al., 2008). Recently, Stahle et al. (2009) reported that SEU and the SLK proteins can interact with LUG and LUH in a yeast two-hybrid assay.
Table I.
Amino acid similarity and identity (% similarity/% identity) across three conserved portions of selected SEU-related proteins
| Portion | SEU | SLK1 | SLK2 | SLK3 | Os11g10060 | Os06g0126000 |
|---|---|---|---|---|---|---|
| N-terminal portiona | ||||||
| SEU | XXX | 39/27 | 45/25 | 35/25 | 58/41 | 36/24 |
| SLK1 | 39/27 | XXX | 48/37 | 83/79 | 34/26 | 58/34 |
| SLK2 | 45/25 | 48/37 | XXX | 44/33 | 41/26 | 47/34 |
| SLK3 | 35/25 | 83/79 | 44/33 | XXX | 30/24 | 55/33 |
| Os11g10060 | 58/41 | 34/26 | 41/26 | 30/24 | XXX | 36/21 |
| Os06g0126000 | 36/24 | 58/34 | 47/34 | 55/33 | 36/21 | XXX |
| Dimerization and LCCD portionb | ||||||
| SEU | XXX | 76/55 | 76/58 | 76/54 | 89/82 | 72/49 |
| SLK1 | 76/55 | XXX | 86/72 | 99/97 | 73/55 | 75/54 |
| SLK2 | 76/58 | 86/72 | XXX | 85/71 | 74/57 | 76/53 |
| SLK3 | 76/54 | 99/97 | 85/71 | XXX | 74/55 | 76/54 |
| Os11g10060 | 89/82 | 73/55 | 74/57 | 74/55 | XXX | 71/49 |
| Os06g0126000 | 72/49 | 75/54 | 76/53 | 76/54 | 71/49 | XXX |
| C-terminal portionc | ||||||
| SEU | XXX | 40/23 | 39/23 | 38/24 | 52/32 | 41/17 |
| SLK1 | 40/23 | XXX | 48/32 | 80/75 | 36/21 | 42/26 |
| SLK2 | 39/23 | 48/32 | XXX | 50/33 | 35/20 | 40/25 |
| SLK3 | 38/24 | 80/75 | 50/33 | XXX | 35/21 | 42/27 |
| Os11g10060 | 52/32 | 36/21 | 35/20 | 35/21 | XXX | 37/18 |
| Os06g0126000 | 41/17 | 42/26 | 40/25 | 42/27 | 37/18 | XXX |
Amino acids 1 to 322 in SEU.
Amino acids 323 to 570 in SEU.
Amino acids 571 to 877 in SEU.
SLK1 Shares Redundant Functions with SEU during Flower and Gynoecium Development
To investigate the functions of the SLK genes in Arabidopsis, we characterized three independent mutant alleles of SLK1 (slk1-1, slk1-2, and slk1-3) and two mutant alleles of SLK2 (slk2-1 and slk2-2) and SLK3 (slk3-1 and slk3-2; see “Materials and Methods”; Fig. 1; Supplemental Table S1; Alonso et al., 2003). Our analysis indicated that homozygous loss-of-function mutations in these alleles do not condition any obvious morphological abnormalities (Fig. 2; data not shown). The slk1-1 allele is a strong loss-of-function allele that expresses SLK1 mRNA at 30% of wild-type levels and is predicted to encode a truncated protein product containing 242 of 748 amino acids that would lack most of the DD and LCCD (Fig. 1; Supplemental Fig. S2). The slk1-2 allele did not display reduced levels of mRNA but is predicted to truncate the protein at amino acid 387. Our quantitative real-time PCR (qRT-PCR) analysis indicated that the slk2-1 allele was a near null mutant, expressing only 0.4% of the wild-type level of SLK2 mRNA. The slk2-2, slk3-1, and slk3-2 alleles are not RNA null alleles but are predicted to be hypomorphic alleles with insertions after amino acid 455 of SLK2 and in intron 8 and intron 4 of SLK3, respectively. Except where otherwise noted, we used slk1-1 and slk2-1 alleles for phenotypic analysis. Our preliminary data suggest that the slk3-1 mutant line contains a reciprocal chromosomal translocation that can be found in a subset of the T-DNA mutant lines (Curtis et al., 2009), complicating the construction of higher order mutants. We report data for only the slk3-1 and slk3-2 homozygous single mutants in this paper.
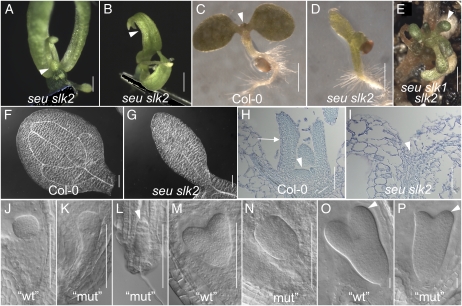
Figure 2.
SLK1 shares redundant function with SEU during gynoecium and ovule development. A to F, Scanning electron micrograph images of flowers of the indicated genotypes. Bars = 1 mm. Some organs from the front of the flower have been removed to image internal whorls. D to F show three seu slk1 double mutant flowers that display enhanced splitting of the gynoecial apices and narrow or filamentous petals (arrowheads). Arrows indicate the basal extent of the carpel valves that is shifted toward the apex of the gynoecium. G, Whole plant phenotypes of slk1, seu, and seu slk1. Bar = 10 cm. H to M, Cleared ovules of the indicated genotypes. The extent of outer integument development is indicated with arrowheads. The majority (67%) of seu mutant ovules (H; Table III) display a wild-type extent of outer integument development (arrowhead). A minority (33%) of seu mutant ovules display a reduction in outer integument development (I; Table III). J shows enhanced ovule defects in the seu slk1 double mutant. In K, the slk1 slk2 double mutant appears morphologically wild type. In L and M, the disruption of ovule development is enhanced in seu/+ slk1 slk2 gynoecia (M) relative to seu/+ slk1 gynoecia (L). [See online article for color version of this figure.]
To investigate functional redundancy between SEU and members of the SLK gene family, we created a collection of higher order mutants. Like the slk1, slk2, and slk3 single mutant plants, the slk1 slk2 double mutants also did not display an observable morphological phenotype (Fig. 2, B and K). Interestingly, the seu slk1 and seu slk2 double mutants did display phenotypic enhancements (relative to the seu single mutant phenotype) in several aspects of plant development.
The seu slk1 plants are shorter in stature than the seu and slk1 single mutants and are sterile (Fig. 2G). The seu slk1 double mutants also displayed enhanced disruptions of floral development. This was characterized by a reduction in organ numbers in whorls 2, 3, and 4 and weak homeotic organ identity transformations in 2% of whorl 1 organs (Table II). The majority (74%) of whorl 2 organs were filaments or filamentous petals (Fig. 2, D and F, arrowheads). Disruption of gynoecial morphology in the seu slk1 double mutant was somewhat variable but was consistently more severe than in the seu and slk1 single mutants, with a greater degree of splitting at the gynoecial apex. In 18% of the seu slk1 flowers, the gynoecium was made up of a single carpel that was fused along its margins into a tube (Fig. 2E). Additionally, we observed a short-valve phenotype in 41% (n = 128) of the carpels in which the basal boundary of the valve was shifted toward the gynoecial apex (Fig. 2, E and F, arrows). This short-valve phenotype is less penetrant in the first 10 flowers to arise from the inflorescence meristem and was observed in 6% of the carpels from these early-arising flowers. The short-valve phenotype and the loss of floral organs is reminiscent of similar phenotypes correlated with auxin homeostasis defects in the ett, short valve1, pinoid, pin-formed1, and monopteros (mp) mutants (Okada et al., 1991; Bennett et al., 1995; Sessions and Zambryski, 1995; Przemeck et al., 1996; Nishimura et al., 2005). These phenotypic enhancements were observed in both the seu-3 slk1-1 and seu-3 slk1-2 genotypes and similar but weaker enhancements were observed in the seu-3 slk1-3 genotype, suggesting that slk1-3 is an intermediate strength loss-of-function allele (data not shown).
Table II.
Floral organ counts for seu and slk mutants
| Genotype | Whorl 1 | Whorl 2 | Whorl 3 | Whorl 4 |
|---|---|---|---|---|
| Col-0 (n = 33) | 4.0 (±0.0) | 4.0 (±0.0) | 5.9 (±0.24) | 2.0 (±0.0) |
| slk1-1 (n = 33) | 4.0 (±0.0) | 4.0 (±0.0) | 5.9 (±0.38) | 2.0 (±0.0) |
| seu-3 (n = 30) | 4.0 (±0.0) | 4.0 (±0.0) | 6.0 (±0.18) | 2.0 (±0.0) |
| seu-3 slk1-1 (n = 49) | 4.0 (±0.43) | 2.2 (±1.1) | 4.2 (±1.2) | 1.4 (±0.54) |
SLK1 and SLK2 Function in Ovule Outer Integument Development
To determine if SLK1 functions during ovule development, we examined ovule development in the seu, slk1, and seu slk1 genotypes. The slk1 mutant plants display morphologically normal ovules and female gametophytes (Fig. 2K; data not shown). The seu single mutant displays a partially penetrant disruption of ovule development in which the outer integument fails to properly surround the nucellus and inner integument (Franks et al., 2002; Fig. 2, H and I). As a semiquantitative measure of outer integument growth, we determined the number of ovules that displayed wild-type outer integument development (i.e. greater than 90% coverage) as well as the number that displayed an intermediate disruption (between 90% and 50% coverage) or a severe disruption (less than 50% coverage) of outer integument development (Table III). In wild-type (ecotype Columbia [Col-0]) siliques, 100% of the ovules displayed wild-type outer integument development. In the seu mutant plants, 67% displayed wild-type outer integument development, while in the seu slk1 double mutant, only 8% displayed wild-type development. Thus, the disruption of outer integument development was enhanced in the seu slk1 double mutant relative to either single mutant (Fig. 2J).
Table III.
Outer integument and female gametophyte defects in seu and slk mutant ovules
| Genotype | Percentage with Disrupted Female Gametophyte Development | Outer Integument Near Wild Type > 90% | Outer Integument Intermediate Disruption > 50% | Outer Integument Severe Disruption < 50% | n |
|---|---|---|---|---|---|
| Col-0 | 0 | 100 | 0 | 0 | n = 76 |
| seu-3/+ | 0 | 100 | 0 | 0 | n = 97 |
| seu-3 | 18 | 67 | 32 | 1 | n = 87 |
| slk1-1 | 0 | 100 | 0 | 0 | n = 125 |
| slk1-2 | 2 | 100 | 0 | 0 | n = 43 |
| slk2-1 | 0 | 100 | 0 | 0 | n = 94 |
| slk3-1 | 2 | 97 | 3 | 0 | n = 111 |
| slk1-1 slk2-1 | 0 | 100 | 0 | 0 | n = 136 |
| seu-3 slk1-1 | 94 | 8 | 60 | 32 | n = 50 |
| seu-3 slk1-2 | 44 | 9 | 86 | 5 | n = 270 |
| seu-3 slk-1–1/+ | 37 | 16 | 75 | 9 | n = 105 |
| seu-3 slk1-1/+ slk2-1/+ | 77 | 1 | 53 | 46 | n = 104 |
| seu-3/+ slk1-1 | 0 | 90 | 10 | 0 | n = 59 |
| seu-3/+ slk1-1 slk2-1 | 81 | 17 | 81 | 2 | n = 268 |
| seu-3/+ slk2-1 | 2 | 98 | 2 | 0 | n = 44 |
| lug-1 | 7 | 91 | 9 | 0 | n = 395 |
| slk1-1 lug-1 | 74 | 91 | 9 | 0 | n = 350 |
| lug-1 slk2-1 | 73 | 24 | 69 | 6 | n = 62 |
| slk1-1 lug-1 slk2-1 | 93 | 5 | 49 | 45 | n = 226 |
Female gametophyte development in the slk1 mutants did not deviate from that of the wild type (Table III). However, the seu mutants displayed a partially penetrant (18%) disruption of female gametophyte development. The female gametophytes in these ovules displayed a range of developmental abnormalities including missing, arrested, and morphologically abnormal female gametophytes. The seu slk1 double mutant plants displayed an enhanced frequency of female gametophyte disruption (greater than 90%) relative to the seu single (Fig. 2J; Table III). As segregation ratios of progeny were not distorted from the predicted Mendelian values, the loss of the female gametophyte is likely due to an indirect effect of the maternal (sporophytic) tissues and not a haploinsufficiency during female gametophyte development (data not shown). To further investigate this, we examined ovules from seu-3/+ and slk1-1 seu-3/+ and found that the morphology of the female gametophyte was indistinguishable from that of the wild type (Table III). Examination of later developmental stages revealed normal endosperm and embryonic development. These results indicate that the seu and seu slk1 chromosomes can be passed efficiently through the female gametophyte.
We also examined the role of SLK2 during ovule development. Ovule development in the slk2 single mutant and the slk1 slk2 double mutant appeared morphologically normal (Fig. 2K; Table III). We could not examine seu slk2 double mutant ovules because this genotype fails to make gynoecia (see below). However, seu-3/+ slk1 slk2 plants displayed an enhanced disruption of the female gametophyte development and outer integument growth relative to the seu-3/+ slk1 mutants (Table III; Fig. 2, L and M). Similarly, seu-3 slk1/+ slk2/+ plants displayed enhanced ovule development disruptions relative to seu-3 slk1/+ (Table III). These results indicate that SLK2 functions during outer integument development, at least when SEU and SLK1 activities are compromised.
SLK2 Shares Redundant Functions with SEU in the Early Embryo
In order to determine if SLK2 shares redundant function with SEU during other stages of Arabidopsis development, we attempted to generate seu slk2 double mutant plants. We examined the progeny of a self-cross from the parental genotype of seu/+ slk2 and seu/+ slk2/+ by planting seeds directly to soil. When using the strong slk2-1 allele (a near RNA null), we were unable to recover seu slk2-1 double mutant plants even after several hundred seeds were planted on soil. When the intermediate strength slk2-2 allele was examined, we did recover seu slk2-2 double mutant plants from slk2-2/+ seu-3/+ parents, but these represented just 2% (three of 149) of the progeny, less than the 6.25% expected. These plants were very late flowering, they were very reduced in stature, and they exhibited severe floral phenotypes (Fig. 3; data not shown). The floral phenotypes were characterized by a reduction in floral organ number and a striking loss of the fourth whorl gynoecium. In these flowers, the fourth whorl was replaced by a small mound of morphologically indistinct tissue (Fig. 3A, arrowhead). In early-arising flowers from the seu slk2-2 double mutants, the whorl 1 organs were narrow sepals (Fig. 3A). However, in late-arising flowers, the whorl 1 organs were carpelloid but failed to develop ectopic ovules (Fig. 3B, arrowhead). The seu slk2-2 carpelloid whorl 1 organs and the overall floral phenotype are morphologically very similar to the previously described seu lug double mutant flowers (Franks et al., 2002).
Figure 3.
Floral, seedling, and embryonic phenotypes of seu slk2 double mutants. A, Early-arising flower from seu slk2 “escaper” plant. The arrowhead indicates a reduced gynoecial mound in whorl 4. B, Late-arising flower from seu slk2 escaper plant. The arrowhead indicates carpelloid whorl 1 sepals. C to H, Seedlings at 5 d post germination. C, Wild-type (Col-0) seedling. The arrowhead indicates leaves initiating from the SAM. D, The seu slk2 double mutant displays narrow cotyledons and a lack of true leaf development. E, A seu slk1 slk2 triple mutant seedling that has escaped embryonic lethality displays bulbous and very reduced rosette leaves (arrowhead). F, Cleared wild-type (Col-0) cotyledon shows vascular loops. G, The seu slk2 double mutant displays a very narrow cotyledon with a single unbranched central vascular element. H, Longitudinal section of a Col-0 seedling. SAM (arrowhead) and rosette leaf (arrow) are indicated. I, SAM and rosette leaves are not detected in the seu slk2 seedling. The arrowhead indicates the expected location of the SAM if wild type. J to P, Embryos segregating from a slk2/slk2 seu/+ parental self-cross. Embryos were classified as morphologically wild type (wt) or mutant (mut). See text for details. J to L, Globular-stage sibling embryos displayed wild-type (J), weakly disrupted (K), or severely disrupted (L) morphologies. The arrowhead in L indicates a globular domain with reduced cell number. M and N, Heart-stage embryos. While cotyledon primordia are apparent in morphologically wild-type embryos (M), morphologically mutant sibling embryos (N) lack obvious cotyledon primordia. O and P, Reduced cotyledon development (arrowhead) is apparent at the torpedo stage in those embryos displaying mutant morphologies (compare P with O). Bars = 100 μm in A, E, F to I, and J to N, 200 μm in B, 1 mm in C and D, and 10 μm in O and P. [See online article for color version of this figure.]
The reduced segregation of the seu slk2 double mutants suggested that seedling or embryonic lethality might be conditioned in the seu slk2 double mutant. To examine seedling phenotypes, we planted seeds from the seu/+ slk2-1 and seu/+ slk2-2 parents onto Murashige and Skoog (MS) agar plates. In both of these populations, we observed a subset of seedlings that displayed a mutant phenotype. This phenotype was characterized by the development of narrow and small cotyledons (Fig. 3D). We genotyped a subset of morphologically abnormal seedlings and determined that all were seu slk2 double mutants (11 of 11) using appropriate PCR-based markers (Supplemental Table S2). The extent of vascularization in the seu slk2 cotyledons was dramatically reduced most often to a single centrally located vascular element (Fig. 3G). In sectioned material, the cotyledon mesophyll cells were larger in the seu slk2 double mutant relative to the wild type, indicating that the reduction of cotyledon size was likely due to a reduction in cell number and not to a reduced cell size (Supplemental Fig. S4). In the majority of the seu slk2 cotyledons examined, the morphology of the mesophyll cells and the vascular elements indicated that patterning along the adaxial/abaxial axis was relatively unaltered. However, in about 30% of the cotyledons examined, intermediate or severe disruptions of adaxial/abaxial patterning were observed. In extreme cases, the cotyledons were radialized, lacked a morphologically distinct adaxial palisade layer, and displayed abnormal arrangements of vascular bundles (Supplemental Fig. S4). All seu slk2 seedlings we examined lacked true leaves and the SAM. We examined chloral hydrate-cleared seedlings and toluidine blue-stained seedling sections and could not detect any morphological manifestation of the SAM or true leaves in the seu slk2 double mutant seedlings (Fig. 3I). The seu slk2 mutants displayed slightly shorter roots at 5 d post germination but appeared normal on a gross morphological level (data not shown). The seu slk2 double mutant seedling phenotype is expected in 25% of the progeny; it was observed in 24% of the progeny from a self-cross of seu/+ slk2-2 (n = 457) and 19% of the seu/+ slk2-1 self-cross progeny (n = 216).
To look for defects during embryonic development, we examined siliques (seed pods) from seu/+ slk2-2 parents. We observed deviations from the wild-type patterns of embryonic development in a subset of the embryos. The earliest defect that we identified was observed in three of 28 (11%) of the globular-stage embryos. We expect that 25% of the embryos in these siliques will be homozygous slk2 seu double mutants, suggesting that this early phenotype may not be fully penetrant. This globular-stage phenotype varied in its severity from mild to severe. Mild disruptions were characterized by fewer cells in the globular domain when compared with the wild type and less clearly organized tiers of cells (Fig. 3K). Severe disruptions displayed very few cells in the globular domain (eight cells in Fig. 3L, arrowhead) and some irregularity in the development of the suspensor. We also observed mutant phenotypes in 14 of 59 (24%) heart-stage embryos (Fig. 3N) and in torpedo-stage embryos (Fig. 3P) that were characterized by much smaller cotyledon primordia. While this paper was under review, Stahle et al. (2009) independently reported similar phenotypes in seu slk2 double mutant seedlings and embryos.
SLK1 Function in Embryo and Seedling Development Is Revealed When SLK2 and SEU Activity Is Compromised
We also examined seedling phenotypes in progeny from a seu-3/+ slk1 slk2-1/+ self-cross. From 473 seeds plated to MS plates, we recovered 66 seedlings (14%) that displayed narrow cotyledons and a reduction of the SAM. These seedlings were phenotypically similar to the seu slk2 double mutant seedlings, but the majority of these had a less severe SAM phenotype. PCR analysis indicated that these seedlings were divided into two genotypes: seu slk1 slk2-1/+ and seu slk1 slk2 triple mutant seedlings. The seu slk1 slk2 triple mutant seedlings did survive for some time but remained very small. They produced few very small bulbous rosette leaves and appeared to be mostly lacking a clearly organized SAM (Fig. 3E). They accounted for less than 2% of the progeny (expected, 6.25%); thus, it is likely that the seu slk1 slk2 triple mutant also conditions an embryonic lethality in the majority of instances and that these seedlings represent escapers that occasionally successfully complete embryogenesis and appear as severely disrupted seedlings. The seu slk1 slk2-1/+ seedlings were found at the approximately expected frequency of 12%, and these plants grew weakly and slowly, were late flowering, and displayed flowers that were similar to those of the seu slk2 escaper plants (data not shown). These results support a role for SLK1 during embryo or seedling development that is uncovered as SEU and SLK2 activity is reduced.
SLK1 and SLK2 Are Required with ANT and SEU for Ovule Initiation from the CMM
We sought to determine if SLK genes functioned during CMM development. As a measure of CMM function, we determined the number of ovule primordia that initiated in gynoecia of different genotypes. The slk1 and slk2 single mutants did not show a reduction in the number of ovules initiated from the CMM. The slk1 slk2 double mutants showed a slight but statistically significant reduction in the number of ovules (89% of the wild type). As mutations in SEU enhance the loss of ovule primordia in ant mutants (Azhakanandam et al., 2008), we sought to determine if the slk mutants also enhanced the loss of ovules in the ant mutant background. The ant-1 single mutant initiates only 64% of the wild-type number of ovule primordia (Table IV). In the slk1 ant and ant slk2 double mutants, the numbers of initiated ovules fell further, to 41% and 47%, respectively. Finally, the slk1 ant slk2 triple mutants generated only 19% of the wild-type ovule number. We then generated seu/+ slk1 ant slk2 mutant plants and observed a complete absence of ovule primordia in these gynoecia (Table IV; Fig. 4K).
Table IV.
Ovule primordia counts from mature gynoecia
| Organ | Col-0 | slk1 slk2 | ant | slk1 ant | ant slk2 | slk1 ant slk2 | seu/+ slk1 ant slk2 |
|---|---|---|---|---|---|---|---|
| Ovule primordia | 47 ± 3.7 (n = 24) | 42 ± 3.2 (n = 24) | 30 ± 1.6 (n = 25) | 20 ± 4.1 (n = 25) | 22 ± 5.3 (n = 24) | 9 ± 6.0 (n = 24) | 0 ± 0.0 (n = 16) |
| Ovules (% of wild type) | 100 | 89 | 64 | 43 | 47 | 19 | 0 |
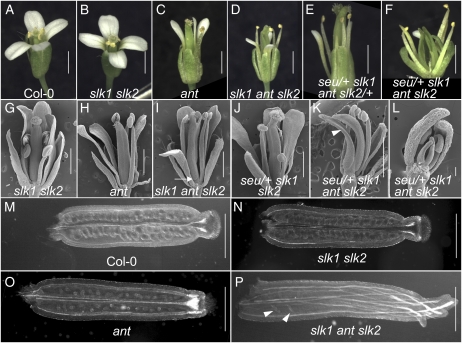
Figure 4.
SLK1 and SLK2 are required with ANT and SEU for ovule initiation from the CMM. A, Col-0 flower. B, Morphology of a slk1 slk2 mutant flower is near that of Col-0. C, ant mutant. D, Gynoecial disruptions are enhanced in slk1 ant slk2 triple mutant relative to the ant and slk1 slk2 mutants. E and F, More severe reductions of CMM-derived tissues are observed in seu/+ slk1 ant slk2 (F) relative to the seu/+ slk1 ant slk2/+ (E) genotype. G to L, Scanning electron micrographs of flowers from the indicated genotypes. In I, the arrow indicates the basal valve boundary. In K, the seu/+ slk1 ant slk2 genotype conditions complete loss of ovule primordia and severe reduction of CMM-derived tissues (arrowhead). L shows a stage 10 flower. Disruption of the CMM is already evident at this stage. M to P, Optical cross sections from cleared gynoecia of the indicated genotypes. Arrowheads in P indicate the few ovules that have initiated. Bars = 1 mm in A to K, 100 μm in L, and 0.5 mm in M to P. [See online article for color version of this figure.]
Decreasing the activity of SEU and the SLK genes in the ant mutant background also had additional effects on the overall development of the gynoecium and other tissues derived from the CMM. The slk1 slk2 (Fig. 4, B and G) and ant (Fig. 4, C and H) genotypes displayed nearly wild-type external gynoecial morphology. The ant single mutants displayed a slight reduction in stigmatic tissue and occasionally very slight splitting of the gynoecial apex (Fig. 4, C and H; Elliott et al., 1996; Klucher et al., 1996). The slk1 ant slk2 genotype conditioned an increased carpel splitting and a nearly complete loss of stigmatic tissue (Fig. 4, D and I). The basal valve boundary was often shifted apically on one or both valves of the gynoecial tube (Fig. 4I, arrow). Occasionally, trichome-like cells were observed at the apex of the slk1 ant slk2 mutant gynoecia (data not shown). These arose from the style-like cells that were present at the tips of the individual carpels. The seu/+ slk1 ant slk2 genotype was severely inhibited in its ability to form CMM tissues (Fig. 4, K and L). In addition to the complete loss of ovule primordia (Table IV), the gynoecia were dramatically split and stigmatic, stylar, and septal tissues were almost completely absent (Fig. 4K). In comparison with the seu/+ slk1 ant slk2 gynoecia, seu/+ slk1 slk2 gynoecia (Fig. 4J) exhibited a less severe disruption of overall gynoecial morphology that was characterized by a fused gynoecial tube topped by an extended stylar region and wild-type amounts of stigmatic tissue. These data together suggest that slk1 and slk2 play a partially redundant role with seu and ant during CMM and gynoecial development.
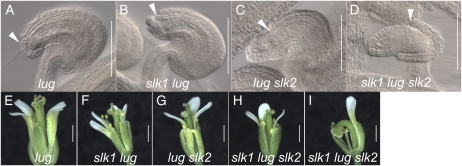
Mutations in slk1 and slk2 Enhance lug Ovule and Floral Defects
Mutations in SEU dramatically enhance the lug mutant phenotype and result in enhanced AG derepression and associated homeotic organ transformations, enhanced floral organ loss, and severe reductions of the gynoecial mound (Franks et al., 2002). In order to determine if mutations in the SLK genes enhanced the lug phenotype, we created a series of higher order mutant combinations employing the lug-1 allele (Liu and Meyerowitz, 1995) that was first introgressed into the Col-0 background. The lug-1 (Col-0) mutant flowers exhibit splitting of the gynoecium at the apex and slightly narrower floral organs relative to the wild type (Fig. 5E). However, homeotic transformations are only rarely observed in the Col-0 background. The loss of slk1 and slk2 in the lug-1 (Col-0) mutant background conditioned relatively subtle enhancements relative to the lug-1 (Col-0) single mutant. The most obvious of these was noted in ovules (Fig. 5; Table III). As slk1 and slk2 activity are reduced in the lug-1 (Col-0) mutant background, the extent and penetrance of the ovule outer integument disruption is increased (Fig. 5, A–D). The slk1 lug, lug slk2, and slk1 lug slk2 mutants also displayed slightly narrower sepals, reduced pollen development, and a smaller overall floral size than the lug single mutant (Fig. 5, E–I). The lug slk2 and slk1 lug slk2 mutants increased the percentage of gynoecia composed by a single carpel (Fig. 5I). However, none of the slk1 lug slk2 higher mutant combinations conditioned severe homeotic transformations within the flower, nor did they condition severe gynoecial disruptions. This was somewhat surprising, given the severe reduction of gynoecium extent and the enhanced carpelloidy previously reported in the seu lug double mutant (Franks et al., 2002).
Figure 5.
Mutations in SLK1 and SLK2 enhance the lug ovule and floral phenotypes. A to D, Cleared ovules from gynoecia of the indicated genotypes. Arrowheads indicate the extent of outer integument development. Bars = 100 μm. E to I, Floral phenotypes of the indicated genotypes. H and I show the two-carpel and one-carpel phenotypes observed in the slk1 lug slk2 triple mutant flowers, respectively. The one-carpel gynoecia are often curved and bent over. Bars = 1 mm. [See online article for color version of this figure.]
SLK2 Is Expressed in the CMM and Young Floral Organ Primordia
Evaluation of data available with the Genevestigator software tool suggests that the expression of the SLK genes is widespread throughout developmental stages and tissues (Zimmermann et al., 2004; Grennan, 2006). These data support a general difference in the level of expression of the SEU and SLK mRNAs, with the relative levels of expression being SEU > SLK2 > SLK1 (Supplemental Fig. S1). Our qRT-PCR data from inflorescence tissue supported the expression of these three genes as well as SLK3 in RNA prepared from inflorescence tissues (Supplemental Fig. S2).
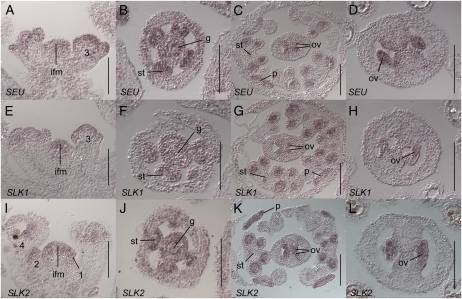
To determine the tissue-specific expression patterns of SEU, SLK1, and SLK2, we employed in situ hybridization using specific antisense probes (see “Materials and Methods”). We carefully examined expression patterns in the inflorescence meristem through stage 12 flowers and found that the patterns of expression of SEU, SLK1, and SLK2 were indistinguishable. However, the level of expression from the SEU probe was consistently higher than that of the SLK2 probe, which was generally higher than SLK1. The detailed description below describes the expression patterns of all three genes examined. We detected strong signal in the inflorescence meristem and throughout young floral meristems at stages 1 through early 3 (Fig. 6, A, E, and I). Stages are according to Smyth et al. (1990). During late stage 3 and stage 4, signal is more strongly detected in the adaxial and marginal portions of the sepal primordia (data not shown). Expression is detected throughout the stamen anlagen/primordia at stage 4 and then becomes restricted to the developing tepetum and microspore mother cells (locules) during later stages (Fig. 6, C, G, and K). Expression within the petals is seen throughout the primordia as they arise (data not shown) and remains in the expanding blade through at least stage 10 (Fig. 6, C, G, and K). Expression is detected throughout the gynoecium at stages 6 and 7 (Fig. 6, B, F, and J). By stage 8, gynoecial expression is most highly detected within the CMM (medial ridge) and in the developing ovule anlagen and primordia (Fig. 6, C, G, and K). Expression is detected throughout the ovules until at least floral stage 11, corresponding to ovule stage 3-I according to Schneitz et al. (1995; Fig. 6, D, H, and L; data not shown). No signal was detected with a control sense strand probe, and signal with the antisense probes was greatly reduced in each respective mutant tissue (Supplemental Fig. S3), further confirming the specificity of the hybridization conditions and probes.
Figure 6.
SEU, SLK1, and SLK2 are expressed in young flower meristems, ovules, and the CMM. In situ hybridization on wild-type (Col-0) tissue with a SEU (A–D), SLK1 (E–H), or SLK2 (I–L) antisense probe (A–E and G–H). A, E, and I, Inflorescence longitudinal sections. Hybridization signal is detected in the inflorescence meristem (ifm) and throughout stage 1, 2, and early 3 floral primordia. Numbers indicate floral stages (Smyth et al., 1990). B, F, and J, Stage 6 floral cross section. Expression is detected throughout gynoecial mound (g) and stamen (st) primordia. C, G, and K, Stage 9 or early 10 floral cross sections. Expression is detected in ovules (ov) and petals (p). Within the stamens, expression is detected most strongly in the locules, pollen mother cells, and the tapetum. D, H, and L, Gynoecial cross sections show expression throughout the ovules at floral stages 10 or 11. Bars = 100 μm.
SEU and SLK1 Are Required for Optimal Auxin Signaling during Root and Gynoecial Development
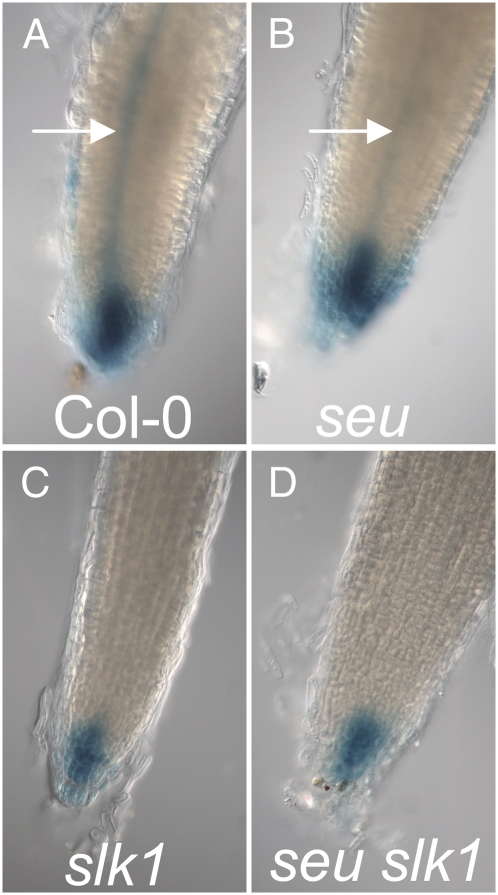
Work from Pfluger and Zambryski (2004) indicated that SEU is required for proper response to the plant phytohormone auxin. To determine if SLK1 is required for the ability to respond to auxin, we examined expression from the auxin-responsive DR5:GUS reporter (Ulmasov et al., 1997) in Col-0, slk1, seu, and seu slk1 seedling roots 7 d after germination. In Col-0 seedlings, GUS activity was observed strongly in the root tip and at intermediate levels in the stele (root vascular tissue; Fig. 7A). In the slk1 single mutant roots, the activity of DR5:GUS reporter appeared reduced relative to the wild type (Fig. 7C). This is similar to the reduced expression of DR5:GUS in the seu single mutant (Pfluger and Zambryski, 2004; Fig. 7B). The DR5:GUS reporter activity was also reduced in the seu slk1 double mutant relative to the wild type.
Figure 7.
The seu, slk1, and seu slk1 mutants display reduced DR5-GUS activity. DR5:GUS reporter activity is shown in primary root apices of the indicated genotypes. Arrows in A and B indicate stele (root vasculature). [See online article for color version of this figure.]
To determine if altered auxin signaling might contribute to the floral and gynoecial defects observed in the seu slk1 double mutants, we examined the expression of genes with known auxin synthesis (YUCCA4 [YUC4]) and auxin response (ETT and MP) functions as well as two genes known to be induced by auxin (INDOLE ACETIC ACID1 [IAA1] and IAA17; Abel et al., 1994; Sessions et al., 1997; Hardtke and Berleth, 1998; Zhao et al., 2001). Although not directly tied to auxin regulation, we also examined levels of PHB and REVOLUTA (REV) mRNA, as reductions in PHB and REV expression have been reported in the seu ant double mutant carpels (Azhakanandam et al., 2008). We examined the expression of these genes in two different RNA samples: inflorescence apices (containing the inflorescence meristem and floral stages 1–6) and staged dissected gynoecia from floral stages 8 to 10 (Tables V and VI). We were particularly interested to identify genes in which the expression in the seu slk1 double mutant was statistically different from each of the single mutants as well as the wild type. These genes might underlie the synergistic enhancement of phenotypes seen in the seu slk1 double mutant. In the inflorescence apices samples, IAA1, IAA17, MP, PHB, and REV fell into this statistically significant category (Table V). The levels of IAA1, IAA17, MP, PHB, and REV expression in the seu slk1 double mutant are reduced to 20%, 36%, 26%, 24%, and 43% of the wild-type level, respectively. The level of YUC4 expression was not statistically different between these genotypes. The expression of ETT was significantly reduced in the seu slk1 double mutant inflorescence apices relative to Col, but it was not statistically different from expression in the seu single mutant. In the dissected gynoecia (stages 8–10), we also detected a statistically significant reduction in expression of MP and PHB in the seu slk1 mutants relative to the wild type and both single mutants. Expression levels were reduced to 35% and 21% of wild-type levels, respectively (Table VI).
Table V.
qRT-PCR quantification in inflorescence meristem through stage 6 flowers
Late-arising flowers are flowers 20 to 35 from the apical meristem. ± values are se.
| Gene | Col-0 | slk1 | seu | seu slk1 |
|---|---|---|---|---|
| IAA1 | 0.10 ± 0.01 | 0.07 ± 0.01 | 0.05 ± 0.01 | 0.02 ± 0.002ab |
| IAA17 | 1.7 ± 0.0 | 1.1 ± 0.0 | 1.0 ± 0.0 | 0.62 ± 0.0ab |
| YUC4 | 0.09 ± 0.01 | 0.24 ± 0.04 | 0.09 ± 0.02 | 0.07 ± 0.006 |
| MP | 13.5 ± 1.7 | 6.4 ± 0.46 | 6.7 ± 0.50 | 3.5 ± 0.31ab |
| ETT | 2.5 ± 0.48 | 2.1 ± 0.30 | 1.6 ± 0.03 | 0.98 ± 0.19a |
| PHB | 0.96 ± 0.09 | 0.59 ± 0.05 | 0.50 ± 0.05 | 0.23 ± 0.02ab |
| REV | 2.8 ± 0.17 | 3.4 ± 0.12 | 1.9 ± 0.03 | 1.2 ± 0.08ab |
Statistically different from Col-0 at P < 0.05.
Statistically different from all other genotypes at P < 0.05.
Table VI.
qRT-PCR quantification in stage 8 to 10 carpels
Late-arising flowers are flowers 20 to 35 from the apical meristem. ± values are se.
| Gene | Col-0 | slk1 | seu | seu slk1 |
|---|---|---|---|---|
| IAA1 | 0.06 ± 0.006 | 0.03 ± 0.001 | 0.02 ± 0.002 | 0.02 ± 0.0007a |
| IAA17 | 1.0 ± 0.11 | 0.55 ± 0.05 | 0.66 ± 0.04 | 0.56 ± 0.03a |
| YUC4 | 0.14 ± 0.015 | 0.25 ± 0.03 | 0.09 ± 0.005 | 0.07 ± 0.007a |
| MP | 16.1 ± 1.0 | 11.8 ± 0.75 | 8.7 ± 0.81 | 5.6 ± 0.39ab |
| ETT | 2.85 ± 0.77 | 1.35 ± 0.11 | 0.92 ± 0.05 | 1.12 ± 0.04a |
| PHB | 1.17 ± 0.18 | 0.76 ± 0.04 | 0.76 ± 0.11 | 0.25 ± 0.01ab |
| REV | 4.5 ± 0.68 | 3.2 ± 0.23 | 2.6 ± 0.25 | 1.6 ± 0.20a |
Statistically different from Col-0 at P < 0.05.
Statistically different from all other genotypes at P < 0.05.
We also used qRT-PCR to characterize gene expression in 6-d-old seu slk2 double mutant seedlings. These experiments indicated that the levels of STM and PHB were reduced in the seu slk2 mutant seedlings to 20% and 30% of wild-type levels, respectively (Table VII). The levels of expression in the seu slk2 double mutants were statistically lower than in the wild type and all of the single mutants.
Table VII.
qRT-PCR quantification in 6-d-old seedlings
± values are se.
| Seedling | Col-0 | slk2-1 | slk2-2 | seu | seu slk2-1 | seu slk2-2 | Statistical Significance Yes/No (P) |
|---|---|---|---|---|---|---|---|
| STM | 2.2 × 10−4 ± 2.6 × 10−5 | 2.0 × 10−4 ± 1.8 × 10−5 | 2.1 × 10−4 ± 8.5 × 10−6 | 3.2 × 10−4 ± 4.2 × 10−5 | 5.6 × 10−5 ± 6.3 × 10−6 | 4.4 × 10−5 ± 6.1 × 10−6 | Yes (0.0001) |
| PHB | 0.33 ± 0.01 | 0.23 ± 0.005 | 0.31 ± 0.03 | 0.18 ± 0.01 | 0.11 ± 0.008 | 0.09 ± 0.005 | Yes (0.004) |
DISCUSSION
The SEU-related genes encode putative transcriptional adaptor proteins with sequence similarity to metazoan Ldb-type transcriptional adaptors. These transcriptional adaptors in both animals and plants function in many gene regulation events and are required for a diversity of developmental processes. In Arabidopsis, SEU functions with LUG in the repression of AG in perianth floral organs, with ETT for correct auxin response during floral organ patterning and with ANT, LUG, and FIL for the proper initiation of ovule primordia from the marginal domain of the carpel. Here, we demonstrate that SLK1 and SLK2 share a high degree of functional similarity to SEU during this diverse set of developmental events. However, our results also indicate that a degree of functional differentiation exists between the different members of the SEU-related gene family. We show that loss of SLK gene function leads to altered auxin responses, altered development of the CMM, but surprisingly little effect on floral organ identity specification. Additionally, we report a novel role for SEU and SLK2 during embryonic SAM development.
Role of SLK Genes in Auxin Signaling
The seu single mutants exhibit reduced expression of the DR5:GUS auxin-responsive reporter (Pfluger and Zambryski, 2004). Additionally, SEU physically interacts with ETT/ARF3 in a yeast two-hybrid assay and thus may directly participate in the regulation of auxin-responsive genes (Pfluger and Zambryski, 2004). Here, we show that slk1 and seu slk1 mutants also display reduced levels of expression from the DR5:GUS reporter in the root. Furthermore, the seu slk1 and seu slk2 flowers display a number of phenotypes that have been reported in mutant plants for auxin synthesis, transport, or signaling components (Okada et al., 1991; Bennett et al., 1995; Sessions and Zambryski, 1995; Przemeck et al., 1996; Friml et al., 2004; Nishimura et al., 2005; Cheng et al., 2006; Stepanova et al., 2008; Tao et al., 2008). These include shifts in the basal boundary of the gynoecial valve in seu slk1 plants and reductions in floral organ number in both seu slk1 and seu slk2 flowers. These results suggest that some of the seu slk1 and seu slk2 defects may be conditioned by altered auxin response or homeostasis. In further support of this hypothesis, we identified two genes previously shown to be induced by auxin (IAA1 and IAA17) and two genes with known roles in auxin signaling (MP and ETT) whose expression was reduced in seu slk1 inflorescences. A reduced ability to respond to auxin maxima in the seu slk1 and seu slk2 double mutants might in part underlie the loss of floral organs, as these organ initiation events are known to be marked by auxin maxima and require functioning auxin pathways (Benkova et al., 2003). In the case of ETT, the SEU-related genes may regulate ETT activity at two levels: at the level of mRNA abundance (as supported by qRT-PCR data) and through direct protein-protein interactions, as have been shown for SEU and ETT (Pfluger and Zambryski, 2004).
SEU and SLK Genes May Support CMM and SAM Development by Similar Mechanisms
Our analysis of SLK1 and SLK2 suggests that these two genes function during the development of the marginal domain of the carpel and the initiation of ovule primordia from the CMM. Our in situ hybridization data indicate that SEU, SLK1, and SLK2 are all expressed in the developing CMM and ovule primordia throughout their early development and thus likely function in a cell autonomous manner in these tissues. The seu/+ slk1 ant slk2 mutant conditions a dramatic loss of ovule primordia that is similar to that previously reported for seu ant double mutants. Thus, it appears that within the CMM, SLK1 and SLK2 also support ovule initiation in a manner similar to that of SEU. However, the fact that single mutations in seu, slk1, and slk2 all independently enhance the phenotypes of ant mutants suggests that either the SEU-related genes are not completely substitutable during CMM development or that the initiation of ovules in the ant mutant responds to activities of the SEU-related genes in a graded fashion.
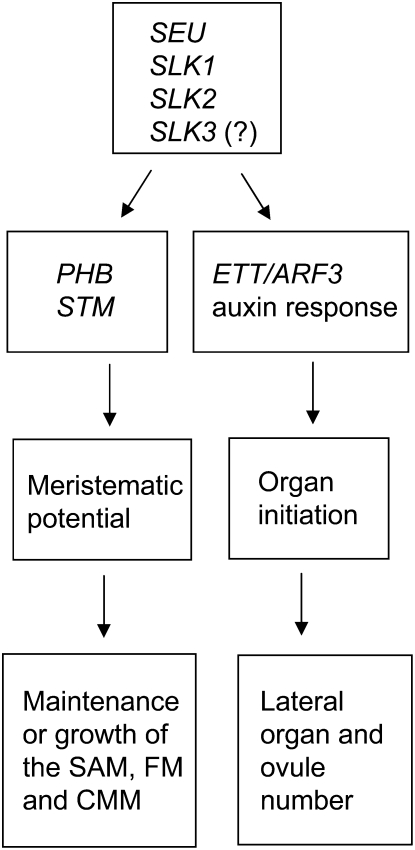
Within the seu ant CMM, the loss of ovule primordia has been correlated with a loss of PHB expression within the developing core of the gynoecium (Azhakanandam et al., 2008). Here, we report a reduction of PHB mRNA levels in seu slk2 mutant seedlings. We speculate that the CMM and SAM defects are mechanistically related to this reduction in PHB expression. Additionally, we report a reduction of STM levels in the seu slk2 seedlings. STM is known to play a key role in the maintenance of meristematic properties in both the SAM and the CMM (Long et al., 1996; Scofield et al., 2007). By supporting the expression of PHB and STM, SEU and SLK genes may function to maintain meristematic potential in both the CMM and the SAM. As PHB and STM expression is not known to be directly responsive to auxin and meristematic regions are typically maintained by lower auxin-cytokinin ratios (Shani et al., 2006), we suggest that the maintenance of meristematic potential that is supported by SEU, SLK1, and SLK2 is not due to their role in auxin response. Rather, we propose a model in which SEU and SLK genes support meristematic properties by potentiating the expression of PHB and STM either directly or indirectly in a manner independent from the above-mentioned role of SEU and SLK in the auxin response. We suggest that the dramatic loss of ovule primordia that is conditioned in the seu ant (Azhakanandam et al., 2008) and seu/+ slk1 ant slk2 mutants (Fig. 4; Table IV) is the result of a reduction in gynoecial meristematic potential coincident with a reduced auxin response in ovule anlagen during their earliest stages of initiation (Fig. 8). This results in a complete loss of ovule primordia as well as a reduction in other tissues derived from the CMM/medial ridge in these genotypes.
Figure 8.
Model of functional roles of SEU and SLK genes in the SAM and CMM. In this model, we propose that the SEU, SLK1, SLK2, and possibly SLK3 genes support the development of organs from the SAM, the floral meristem (FM), and the CMM through two gene regulatory events (right and left sides). SEU and the SLK genes support auxin responses that are required for organ initiation events in the CMM, floral meristem, and embryonic SAM. Additionally, SEU and SLK genes support the maintenance or growth of these meristematic regions by enabling the expression of PHB and STM.
Functional Redundancy and Diversification within the SEU-Related Gene Family
The slk1 slk2 double mutant and the seu single mutant both progress through embryogenesis without noticeable defects. However, the concomitant loss of both SLK2 and SEU conditions a severe embryonic defect in SAM and cotyledon development. Thus, during embryonic development, SEU and SLK2 appear to be functionally interchangeable and provide an important redundant function supporting embryonic SAM development. Our data indicate that SLK1 and SLK3 cannot provide a sufficient level of this activity to support embryonic SAM development, at least when expressed from their endogenous promoters in the seu slk2 double mutant background. Further characterization of expression patterns as well as ectopic expression or domain-swap constructs may help to elucidate the molecular mechanisms that underlie the functional diversity within the SEU-related genes. Interestingly, the lug luh double mutant genotype also conditions an embryonic lethality (Sitaraman et al., 2008). However, the degree of similarity between the molecular causes of the lug luh and seu slk2 embryonic defects remains to be determined.
Mechanism of Functional Redundancy within the SEU-Related Gene Family
In the developmental events that we have analyzed, members of the SEU-related gene family often share some degree of functional redundancy. One mechanistic explanation for this redundancy would be an overlap in the physical partners of the SEU-related proteins. SEU has been shown to work in complexes with several MADS domain proteins and with ETT, LUG, and LUH. It will be interesting to determine if the SLK proteins can also interact with these proteins. All three SLK genes appear to encode an LCCD, suggesting that they form physical complexes with either LUG or LUH proteins. Also supporting this proposition, SLK homologs in Antirrhinum can physically interact with the LUG Antirrhinum ortholog STYLOSA (Navarro et al., 2004), and recently, LUG and LUH have been reported to interact with Arabidopsis SLK proteins in a yeast two-hybrid assay (Stahle et al., 2009). An alternative, but not mutually exclusive, possibility is that SEU and SLK proteins form heterodimeric complexes. The conserved DD in the metazoan Ldb proteins mediates dimerization events that are required for the formation of higher order protein complexes (Agulnick et al., 1996; Jurata et al., 1996; Jurata and Gill, 1997; Morcillo et al., 1997). This domain may function similarly in the SEU and SLK proteins in Arabidopsis and mediate either homodimeric or heterodimeric interactions. Heterodimeric complexes between SEU and SLK proteins would provide a mechanism through which the SLK proteins could modulate the activity of transcriptional complexes that contain SEU. Furthermore, as the protein sequences of the SEU protein and the SLK proteins diverge outside of the central DD and LCCD, heterodimeric SEU/SLK complexes would likely contribute a more diverse protein interaction surface than homodimeric forms, allowing the SEU-related gene family to participate in a wider diversity of gene regulation events.
Role of SLK Genes in the Repression of AG
It is notable that slk1 and slk2 mutants only mildly enhance the floral phenotypes of the lug single mutant. We did not observe enhanced homeotic organ identity transformations, as has been reported for the seu lug and luh/+ lug mutants (Franks et al., 2002; Sitaraman et al., 2008). Additionally, the seu slk1, slk1 slk2, and seu slk2 double mutants rarely displayed carpelloidy that would indicate ectopic AG expression within the floral meristem. Only in late-arising seu slk2 and rarely in seu slk1 flowers were carpelloid whorl 1 organs observed. These results might be explained by SLK3 activity still present in these plants. Alternatively, these results may suggest that SLK1 and SLK2 play a lesser role in the repression of AG than does SEU. This may reflect a differential ability of SEU and SLK proteins to physically interact with LUG or other regulators in the flower.
MATERIALS AND METHODS
Mutant Alleles
The Arabidopsis (Arabidopsis thaliana) alleles of SLK genes used in this study are reported in Supplemental Table S1. We amplified and sequenced the DNA junction fragment using the appropriate border primers (Supplemental Table S2) to confirm each insertion site. Each mutant line was backcrossed to the parent ecotype three times before generating homozygous stocks for phenotypic and genetic analysis. The seu-3 and ant-1 alleles have been reported previously (Klucher et al., 1996; Pfluger and Zambryski, 2004). The lug-1 allele was originally isolated in the Landsberg erecta ecotype (Liu and Meyerowitz, 1995) and has subsequently been backcrossed into Col-0 for four generations before use in this study. Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Phylogenic Analysis
ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) was used to align amino acid sequences of the full-length proteins. The optimal tree with the sum of branch length = 4.91935227 was inferred using the neighbor-joining method (Saitou and Nei, 1987). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in units of the number of amino acid substitutions per site. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (pairwise deletion option). There were a total of 1,232 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007). Protein sequence identity and similarity matrix was calculated with the Matrix Global Alignment tool (Campanella et al., 2003).
In Situ Hybridization and Microscopy
In situ hybridizations were carried out as reported previously (Franks et al., 2002) with the following modifications: acetic anhydride and RNase treatment steps were omitted. A detailed protocol is available at http://www4.ncsu.edu/∼rgfranks/research/protocols.html. The SEU, SLK1, and SLK2 antisense probes were generated from pCRII_SEU_HFFL and Arabidopsis Biological Resource Center clones G66746 and G10219, respectively. Probes were made corresponding to the C-terminal domains and did not overlap highly conserved DD/LCCD regions. Scanning electron and light microscopy were performed as described previously (Azhakanandam et al., 2008).
qRT-PCR Analysis and GUS Analysis
qRT-PCR analysis was as described previously (Azhakanandam et al., 2008). Results shown in Tables V, VI, and VII are mean expression of the indicated gene normalized relative to ADENOSINE PHOSPHORIBOSYL TRANSFERASE (AT1G27450). Results are averages and se of the mean from three technical replicates of three biological replicates. Statistical analysis was done in JMP 7.0 (SAS Institute). Statistical differences of means were evaluated by pairwise t tests in all combinations. For GUS analysis, seedlings were grown on half-strength MS + 10% Suc under 16-h/8-h light/dark conditions and assayed 7 d after germination. For photography, seedlings were stained for either 2 or 4 h at room temperature and then cleared in 70% ethanol.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genevestigator expression data for SEU and SLK genes.
Supplemental Figure S2. qRT-PCR expression data for SLK1, SLK2, and SLK3 in slk1 and slk2 mutant inflorescences.
Supplemental Figure S3. Specificity of the SEU, SLK1, and SLK2 antisense in situ probes.
Supplemental Figure S4. Morphological alterations of cotyledon mesophyll cells and vascular elements in the seu slk2 double mutant.
Supplemental Table S1. Alleles of SLK transcriptional coregulators in Arabidopsis.
Supplemental Table S2. Oligonucleotides used in this study.
Supplemental Table S3. qRT-PCR quantification in inflorescence meristem through stage 6 flowers (from early-arising flowers).
Supplemental Table S4. qRT-PCR quantification in stage 8 to 10 carpels (from early-arising flowers).
Supplementary Material
Acknowledgments
We thank L. Mathies, A. Stepanova, and the anonymous reviewers for comments on the manuscript; the Arabidopsis Biological Resource Center for the distribution of mutant seed stocks; and the North Carolina State University Center for Electron Microscopy and Cellular and Molecular Imaging.
This work was supported by the National Science Foundation (grant nos. IOS 0416759 and IOS 0821896 to R.G.F.) and the U.S. Department of Agriculture Agricultural Research Service (grant no. NC06759).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert G. Franks (rgfranks@ncsu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abel S, Oeller PW, Theologis A (1994) Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA 91 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H (1996) Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384 270–272 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Azhakanandam S, Nole-Wilson S, Bao F, Franks RG (2008) SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol 146 1165–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8 505–520 [Google Scholar]
- Bowman JL, Baum SF, Eshed Y, Putterill J, Alvarez J (1999) Molecular genetics of gynoecium development in Arabidopsis. Curr Top Dev Biol 45 155–205 [DOI] [PubMed] [Google Scholar]
- Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner J, Liu Z (2000) LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci USA 97 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MJ, Belcram K, Bollmann SR, Tominey CM, Hoffman PD, Mercier R, Hays JB (2009) Reciprocal chromosome translocation associated with TDNA-insertion mutation in Arabidopsis: genetic and cytological analyses of consequences for gametophyte development and for construction of doubly mutant lines. Planta 229 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–791 [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z (2002) SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129 253–263 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865 [DOI] [PubMed] [Google Scholar]
- Gonzalez D, Bowen AJ, Carroll TS, Conlan RS (2007) The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol 27 5306–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM (2006) AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grennan AK (2006) Genevestigator: facilitating Web-based gene-expression analysis. Plant Physiol 141 1164–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Gill GN (1997) Functional analysis of the nuclear LIM domain interactor NLI. Mol Cell Biol 17 5688–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Kenny DA, Gill GN (1996) Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci USA 93 11693–11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Franks RG, Klink VP (2000) Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM (1995) LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121 975–991 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69 [DOI] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE (2003) LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep 4 1132–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D (1997) Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev 11 2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C, Efremova N, Golz JF, Rubiera R, Kuckenberg M, Castillo R, Tietz O, Saedler H, Schwarz-Sommer Z (2004) Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131 3649–3659 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA (2006) AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol 141 977–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J, Zambryski P (2004) The role of SEUSS in auxin response and floral organ patterning. Development 131 4697–4707 [DOI] [PubMed] [Google Scholar]
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T (1996) Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200 229–237 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–425 [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hulskamp M, Pruitt RE (1995) Wild-type ovule development in Arabidopsis thaliana: a light-microscope study of cleared whole-mount tissue. Plant J 7 731–749 [Google Scholar]
- Scofield S, Dewitte W, Murray JA (2007) The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J 50 767–781 [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC (1997) ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124 4481–4491 [DOI] [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC (1995) Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121 1519–1532 [DOI] [PubMed] [Google Scholar]
- Shani E, Yanai O, Ori N (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9 484–489 [DOI] [PubMed] [Google Scholar]
- Sitaraman J, Bui M, Liu Z (2008) LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol 147 672–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA 101 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133 3159–3166 [DOI] [PubMed] [Google Scholar]
- Stahle MI, Kuehlich J, Staron L, von Arnim AG, Golz JF (2009) YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21 3105–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL (2002) LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 110 237–249 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meyel DJ, Thomas JB, Agulnick AD (2003) Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development 130 1915–1925 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Evolving Genes and Proteins. Academic Press, New York
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.