The aerial tissues and organs of the plant are initiated by populations of undifferentiated, rapidly dividing cells called shoot apical meristems (SAMs). Within one species, there are different types of these meristems depending on developmental stages and environmental conditions: (1) vegetative meristems that give rise to the nonreproductive organs such as leaves or lateral branches, (2) inflorescence meristems producing flowers, and finally (3) the floral meristems producing the perianth and reproductive organs.
Although these meristems produce different organs and are characterized by specific gene activities, they have a number of common structural characteristics (e.g. Lyndon, 1998; Traas and Doonan, 2001). One of these features is the presence of one or several distinct cell layers that form the so-called tunica and cover the internal tissues, collectively called corpus. Superimposed on these layers, zones can be distinguished characterized by subtle differences in cytological properties and that have particular functions. At the meristem summit the central zone (CZ) contains the true stem cells that ensure meristem maintenance. Their relatively slow proliferation rates distinguish them from the cells at the peripheral zone where the new lateral organs are initiated.
In view of their importance in plant development, shoot meristems have received considerable attention and an impressive amount of information has accumulated regarding the molecular mechanisms that govern their function. Somewhat paradoxically, this wealth of knowledge has not proportionally increased our understanding.
There are several reasons for this. First, it has become more and more difficult to propose hypotheses that integrate and are compatible with all available data on meristem function. Second, we seem to have reached the limits of the current reductionist approaches aimed at analyzing the individual molecular and cellular components of the shoot meristem. This problem is not specific for meristem research but concerns all fields of research on plant and animal development. This is due to the fact that it is not sufficient to gather information at a single level of observation without considering the system also at a more global, integrated level. In this general context a novel approach, called systems biology was introduced.
SYSTEMS BIOLOGY: FROM SYST-OMICS TO COMPLEX SYSTEMS
Some Definitions
Initially, systems biology seemed to be nothing else than a variant of existing approaches, extending high-throughput genomics methods to a range of omics approaches, like the localosome (systematic localization of proteins in situ), the phenome (massive phenotyping of whole plants), etc. Gradually, however, systems biology has become associated with the concept of complex systems. This concept, which emerged from physics and chemistry, does not necessarily refer to living organisms only. Indeed, complex systems can be found in many fields including such remote examples as economical systems, traffic jams, or moving sand dunes. Formally, a complex system can be defined as a set of entities that interact according to simple local rules. These interactions lead to the emergence of new collective properties at a higher level of organization, through a process of self organization. Importantly, these so-called emergent properties are nonlinear, i.e. they cannot be deduced from simply adding up local behavior.
Multicellular organisms are typical examples of complex systems. They are composed of thousands of different molecules with particular properties, which interact to form cells with novel emergent characteristics. In turn, the cells, complex systems on their own, interact to generate tissues, organs, and finally the individual organism. The organisms themselves can then form higher level complex systems in the form of societies, ecosystems, etc.
An important property of complex systems is the presence of multiple feedbacks between the different levels of organization. For example, cells can generate gradients of signaling molecules spanning whole tissues, which can then feed back locally on the behavior of the same individual cells.
If we accept that living organisms are complex systems, it becomes clear why it is necessary but not sufficient to analyze the molecular mechanisms that are at the basis of their function: We need also to understand how the individual parts interact at different scales. To address this problem, approaches from a range of disciplines are required, from molecular genetics and biochemistry to physics. In particular, in view of the complexity of this task, approaches involving mathematical and computational modeling have become more and more important.
The Shoot Meristem as a Complex System
The SAM perfectly illustrates the problems discussed in the previous paragraphs. As mentioned, we have gained significant insight in the nature of the genetic networks and cellular processes during the last decades. Extensive genetic screens have helped to identify key elements in the regulatory networks that coordinate meristem function. In parallel, high-throughput approaches have helped to determine the expression patterns of thousands of genes in the different parts of the SAM, whereas cell biology has characterized basic cellular processes such as cell division and cell expansion. Nevertheless, major questions have remained unsolved. It has been difficult, for instance, to define the precise role of the regulatory networks in controlling morphogenesis and shape. We might understand the basics of cell division and growth, but it has been difficult to grasp how hundreds of cells can create such a highly dynamic structure as the meristem, able to maintain itself for years or even centuries. In the following sections, we will discuss three examples illustrating how systems biology has started to change our view on meristem function.
ANALYZING ORGAN INITIATION AT THE MERISTEM: TOWARD A SYSTEMS BIOLOGY APPROACH
A Major Challenge: Analyzing the Genetic Regulatory Network Controlling Meristem Function
The analysis of the genetic regulatory network controlling shoot meristems started decades ago with a set of extensive genetic screens, which have identified a series of key regulators (for review, see Traas and Doonan, 2001; Aida and Tasaka, 2006; Sablowski, 2007; Rast and Simon, 2008). Since the main aim of this article is rather to illustrate an approach than to provide a detailed review of meristem function, we will discuss here only the main lines. Since all angiosperm meristems seem to function in a similar manner, we will mainly limit ourselves to Arabidopsis (Arabidopsis thaliana), which is currently the best characterized system.
All meristematic cells share a number of common molecular characteristics, defined by homeodomain proteins of the KNOX family, in particular SHOOTMERISTEMLESS. In combination with several members of the so-called CUP SHAPED COTYLEDON family of transcription factors the KNOX genes determine what has been called the meristematic state of the cells. Within this population of meristematic cells, at the meristem summit (or CZ), shoot meristem maintenance involves the transcription factor WUSCHEL (WUS) that interacts with the CLAVATA (CLV) receptor kinase pathway to regulate meristem size (Laux et al., 1996). When a new organ is initiated at the peripheral zone of the meristem the KNOX genes are switched off. This implicates the transcription factor ASYMMETRIC LEAVES1, repressing meristem identity factors in the young primordia (Byrne et al., 2000). The identity of the lateral organs produced (leaves, lateral vegetative meristems, or flowers) depends on the activity of the transcription factor LEAFY, which, by interacting with other transcriptional regulators involved in meristem function such as WUS or AGAMOUS, plays a major role in plant and flower architecture (Lenhard et al., 2001; Lohmann et al., 2001).
Our knowledge on the successive outgrowth of the lateral organs is more limited. We do not understand in detail how cell division and cell expansion in the young growing organ are coordinated by the genetic regulators. The transcription factor AINTEGUMENTA, for instance, strongly expressed in the rapidly outgrowing organ, seems to act at least partially via CYCLIN D, a gene involved in cell proliferation (Mizukami and Fischer, 2000). Other examples include JAGGED, ARGOS, or BIGPETALp, which control petal growth by influencing cell proliferation and/or cell expansion (Dinneny et al., 2004; Hu et al., 2006; Szécsi et al., 2006).
The genes discussed above represent only a tip of the iceberg and currently the available data on the molecular basis of meristem function are scattered over hundreds of articles. So far relatively little attempts have been made to integrate this information, but it is obvious that databases alone will not suffice and that theoretical approaches including modeling will be indispensable. This problem does not only concern plant research, and it would be far beyond the scope of this article to review the entire literature concerning regulatory networks in biology. We will, therefore, only give a brief outline of the methods currently used in meristem research and provide a number of relevant examples (for further reading, see de Jong, 2002; Albert, 2007; Long et al., 2008; and refs. therein).
Intuitively it seems desirable to develop models where quantified gene activities or protein concentrations can be expressed as continuous functions, using sets of connected differential equations. Ideally, these should also be able to take into account variability and noise, frequently observed in biological systems. Unfortunately, such models also have the highest requirement for quantitative data, which are often lacking. In addition, they cannot handle large networks.
As an alternative, more qualitative, discrete models can be extremely useful. As a typical example we briefly mention here the so-called Boolean approaches. In their simplest form such models represent network interactions as either positive or negative, network components either switching on or off their targets. Although at first sight they might seem to be very rough representations of reality, there are several theoretical and practical reasons to favor such models. For example, they often represent a relatively intuitive and simple manner to describe the available data in a formal manner. In addition, they greatly facilitate the identification of potential contradictions or missing information and can help to construct the most parsimonious network consistent with all available information. Finally, these approaches are much better adapted for networks with multiple components. This does not preclude Boolean network models from posing a number of problems. For instance, when a particular gene receives both negative and positive inputs, how does one decide which of the two will dominate? To address this problem these models can be extended, and usually some level of quantified information (e.g. indicating that one input is stronger than the other one) is integrated. Since the strength of a particular interaction is often unknown (and the interactions often indirect) this has to be estimated, frequently from indirect evidence such as a double-mutant phenotype analysis. Once the network model has been constructed, it can be used to determine how coherent and complete the available information is. An important aspect of the model analysis involves computer simulations where specific gene combinations in the Boolean network are initially switched on or off. From this starting point, the network will reach ultimately a stable state. These stable states should logically correspond to combinations of gene expression patterns also found in real life in specific cell types. If so, we can at least conclude that the information expressed in the model is coherent, but in principle Boolean approaches can also be used to make predictions.
So far relatively little studies have addressed the modeling of regulatory networks involved in meristem function. Boolean-type models have been used to analyze flower development (Espinosa-Soto et al., 2004; Alvarez-Buylla et al., 2007). This Boolean network faithfully reproduced the gene activity states associated with the floral organs. Interestingly, minor alterations of the interaction rules did not significantly alter the outcome of the simulations, predicting that the regulatory network is very robust (Espinosa-Soto et al., 2004).
The limitations in reconstructing a coherent model of the regulatory network involved in SAM function do not only lie in the lack of quantitative data. In addition, our knowledge on the composition of the network is incomplete (Fig. 1). This is partially because the genetic screens do not give access to all genes involved in meristem function. A typical limitation of genetic dissection is redundancy, which makes it difficult to reveal gene function without the often time-consuming production of multiple mutants. To circumvent this problem at least partially, transcriptomic or proteomic approaches can provide useful information. With this purpose in mind, Yadav et al. (2009) performed a transcriptomic analysis of the meristem. Starting from an elegant method originally developed by Birnbaum et al. (2003) they isolated protoplasts from meristematic tissues expressing GFP markers in specific domains. The GFP-expressing protoplasts were subsequently harvested using a cell sorter and RNA was isolated from these cells. Using this approach, they were even able to identify rare transcripts expressed in specific subsets of cells. This method not only confirmed earlier results, but also led to the identification of novel gene expression patterns. Since transcriptomics give access to a wide range of gene activities, it seems logical to use these to further complete the network models. However, it is not that straightforward to infer network properties from this information. Often transcriptomic data lack sufficient spatial and temporal information, whereas low abundant transcripts can be missed. Finally, transcriptomics gives information on coexpression, not necessarily on functional relationships. The latter can often only be inferred when many independent experiments (e.g. different conditions or genetic backgrounds) are analyzed.
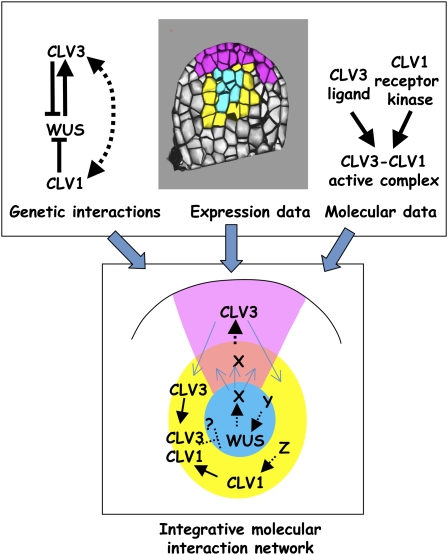
Figure 1.
Approach used for the reconstruction of an integrative molecular interaction network. We take as an example, the well-known WUS-CLV regulatory loop. Genetic interactions show that WUS is necessary for CLV3 activation and that it is itself repressed by CLV3 and CLV1, two genes acting in the same genetic pathway (for details see text). Expression data show that each of these three genes has a distinct and specific expression pattern within the meristem. From a molecular point of view, CLV3 encodes a ligand that is secreted and physically binds to the CLV1 receptor kinase. The protein complex formed by this interaction is necessary to restrict the expression of WUS in the organizing center. The structure of the molecular regulatory network is given by the molecular interactions between the entities of this network. For each element differentially localized among regions, the network must reproduce the active or inactive state of the corresponding element and the incident interactions must therefore be able to account for these different states. The network can then be used to run simulations and see which stable states can be obtained. In parallel, expression data for the molecular elements of the network can be compiled and superposed on a virtual meristem to identify zones of cells that coexpress the same set of elements. In principle, the stable states of the network should correspond to the coexpression zones observed. Lastly, genetic interaction data, which give information on the dynamic of the network but not on the molecular trajectories, should be explained by the molecular network. Due to the lack of available molecular interaction data, the problem is in fact to build the network. In the case of WUS-CLV, we not only lack molecular interactions (for example, is the repression of WUS by CLV3-CLV1 direct?) but we also cannot explain WUS and CLV1 expression pattern since their activators are unidentified yet. In addition, it is likely that there exists an intermediate messenger for the activation of CLV3 by WUS since their expression pattern does not overlap. We can therefore introduce hypothetical molecular interactions (dotted arrows) and see if it is consistent with both expression and genetic data. These hypothetical interactions can then be tested experimentally and used either to confirm the validity of the network, or to introduce alternative interactions. This is an iterative process that requires the use of computer modeling to run simulations and to generate predictions.
In conclusion, transcriptomic and genetic analyses have provided a wealth of information on meristem function and organ initiation. Discrete, Boolean models have been used to integrate the available data, providing a firm basis for further work. Future efforts should extend these models to more quantitative approaches. In addition, the regulatory networks also have to be studied in space. So far this has been mainly the case for the small regulatory network controlling the CZ of the SAM. Jönsson et al. (2005) used in vivo confocal microscopy to create templates for computational models. Using such templates they developed two spacial models accounting for the organization of the WUS expression domain. One of these models used a reaction-diffusion mechanism in which an activator induced WUS expression. This model was able to organize the WUS expression domain and predicted the reorganization seen in ablation experiments and in mutant backgrounds. Recently, Gordon et al. (2009) used similar spatial, quantified information to construct a model involving both cytokinin signaling and the WUS-CLV feedback loop. Their analysis proposed a scenario that explains how modified cytokinin levels might lead to the formation of a stem cell niche.
Organ Initiation and Positioning: Phyllotaxis as an Emergent Property
The SAM produces organs in very stereotypic arrangements and lateral organs can be positioned in opposite, whorled, or spiraled patterns. To explain the process of organ positioning, also called phyllotaxis, simple models were already proposed in the late 19th century. The still most widely accepted hypothesis is based on a concept of lateral inhibition, where young organs inhibit the formation of new ones in their direct vicinity (for review, see Steeves and Sussex, 1989). By modulating the size of these inhibitory fields, different organ arrangements can be obtained. Already in the 1930s Snow and Snow (1933, 1935, 1952) obtained experimental evidence for such a mechanism, but its precise nature has remained elusive until very recently. The existence of an inhibitory field supposes that the cells interact with their environment via some type of signal. During the last decade, it has become increasingly clear that the plant hormone auxin could be this signal or at least be a major component involved in this signaling process.
It was genetic analysis that provided crucial information on the role of auxin in meristem patterning with the discovery and analysis of the pin-formed1 (pin1) mutant. PIN1 is a member of a family of membrane linked transporters and is required for auxin efflux (e.g. Gälweiler et al., 1998). Importantly, neighboring cells often show coherent PIN localizations, apparently transporting auxin in the same direction. It was therefore suggested that these proteins create fluxes through the tissues, leading to auxin minima and maxima. When PIN1 is impaired at the SAM, auxin transport is greatly reduced and the SAM is no longer able to produce flowers. As a result a naked inflorescence stem is formed. Interestingly, when high concentrations of auxin are applied externally to these naked meristems a primordium is induced. This simple experiment shows that (1) an auxin maximum is apparently required to induce organ formation and (2) that PIN1 is required to create such a maximum. Consequently, it has been proposed that local auxin accumulation at specific places leads to the initiation of new organ primordia (Reinhardt et al., 2003; Heisler et al., 2005; Barbier de Reuille et al., 2006; Jönsson et al., 2006; Smith et al., 2006). This hypothesis was further corroborated by a visual, qualitative characterization of the complex distribution patterns of the PIN1 protein in Arabidopsis. Immunolabeling as well as PIN-GFP fusions, for example, typically showed cells pointing their transporters toward young outgrowing primordia (Reinhardt et al., 2003; Heisler et al., 2005). Together these observations led to a hypothesis where PIN auxin transporters generate auxin maxima at the meristem periphery, causing the initiation of lateral organs. These organs would subsequently act as sinks for the hormone, thus depleting auxin from their direct environment. Depletion would in turn prevent organ formation close to the new primordia. Note that this concept is very close to the original inhibitory field theory discussed above, where the inhibitory activity consists in removing an activator (auxin). Although this hypothesis was very attractive, it was partially based on the simple visual analysis of PIN distributions. In addition, it was not obvious if such a scenario could create stable patterns of phyllotaxis. Two approaches involving computer simulations were used to take these analyses further. First, the properties of the observed PIN distributions were investigated using a combination of image analysis and computer simulations. For this purpose, real images of PIN distribution at the SAM surface were interpreted in the form of connection maps, where neighboring cells were connected with arrows indicating the local direction of auxin transport. Subsequently virtual auxin was injected into these maps to see if the network was able to create auxin maxima and minima (Barbier de Reuille et al., 2006). The simulations showed that virtual auxin was indeed directed to the young primordia, as expected. In addition, they also revealed properties of the PIN1 distribution network that were not readily predicted. In particular, these analyses showed that auxin was transported to the CZ at the meristem summit, suggesting an important role for this part of the SAM in auxin homeostasis.
In parallel to this simulation-based analysis of the observed PIN distributions, several modeling approaches addressed the potential cellular mechanisms behind PIN localization. Both Jönsson et al. (2006) and Smith et al. (2006) tested a scenario where cells would transport auxin to neighbors with higher auxin concentrations, i.e. against the auxin gradient. These models, in the form of virtual tissues, showed that this scenario was indeed able to generate different phyllotactic patterns (see also Merks et al., 2007). They thus confirmed that the initially proposed hypothesis where phyllotaxis depends on auxin transporters generating auxin maxima and auxin depletion zones, was indeed plausible. Although this is important information that can only be obtained via modeling, it is no mathematical proof that the hypothesis is true. Stoma et al. (2008) tested an alternative hypothesis, where cells sense auxin fluxes rather than auxin concentrations. In their model, the cells simply amplify the fluxes of hormone that go through them (so-called canalization; see also Sachs, 1969). Interestingly, this hypothesis was also able to reproduce stable phyllotactic patterns and simulations resulted in realistic predictions regarding PIN localization. A third scenario where both the against the gradient and with the flux were combined also reproduced realistic patterns of PIN localization (Bayer et al., 2009). In this model cells can use either one or the other mechanism, depending on the local auxin concentration. Note that all these models are in line with the general idea of auxin maxima and auxin depletion zones, it is simply the cellular mechanism (sensing fluxes or concentrations), leading to specific PIN localizations that differs. To distinguish between these hypotheses, it is now important to test more specific predictions of the models. For example, the against the gradient model predicts more or less stable auxin maxima at the tip of young primordia, while the with the flux model predicts that initial maxima will be rapidly changed into local minima. In principle, this could be tested by evaluating local auxin concentrations at the SAM, although this will be a challenging task. In parallel, a thorough comparison of the different models in terms of robustness (e.g. sensitivity to changes in parameter values) should also provide further information.
It should be underlined that the virtual tissues mentioned above, like all models, are simplifications of a much more complex reality. They do not, for instance, take into account intercellular spaces, nor do they indicate how auxin fluxes or auxin concentration gradients are sensed. Because of such simplifications, the models are not able to represent all aspects of reality, whether the hypotheses are correct or not. This constitutes an additional problem when we have to evaluate the plausibility of a particular model. The models proposed by Jönsson et al. (2006) and Stoma et al. (2008), for example, did tend to produce unstable phyllotactic patterns. It remains to be seen if this is due to a fundamental problem with the proposed hypotheses or caused by limitations in the modeling methods.
In conclusion, a combination of approaches has led to different plausible and testable hypotheses regarding the establishment of phyllotactic patterns. Without computational modeling, this would not have been possible. All three models are based on the concept of complex systems, where very simple local interactions (e.g. cells transporting against a gradient or with the flux) lead to patterns (in this case specific auxin distributions) at the level of a whole tissue.
Systems Biology and Biophysics: Organ Outgrowth as an Emergent Property
In the previous paragraphs, we have analyzed the gene regulatory networks that control organ initiation and discussed the intercellular auxin transport that is involved in organ positioning. We will now consider morphogenesis, i.e. the generation of organs with a particular shape. Shape, as a geometrical output of gene function, can be defined by two parameters, growth rate and anisotropy (Fig. 2; see also e.g. Coen et al., 2004). From a mechanistic point of view, these two parameters largely depend on the cell wall, which is the major structural component of plant cells. An additional factor is turgor pressure. It is generally thought that this pressure is relatively homogenous over the entire tissue, but this remains to be seen for rapid-growing tissues such as the meristem till what extent the pressure level is modulated. Since little or nothing is known on this issue, we will not consider this factor here.
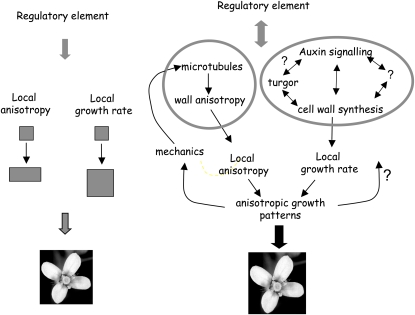
Figure 2.
From gene to shape. Many genes that play a role in the establishment of plant architecture have been identified, but their precise role has never been established. If we want to study gene function in terms of shape, it is important to consider first gene output in terms of geometrical changes during development. The latter can be described with two main parameters, anisotropy and growth rate (e.g. Coen et al., 2004). After the detailed quantification of growth patterns in gene expression domains and in mutants it then becomes possible to attribute specific morphogenetic functions to individual genes. From a more mechanistic point of view, genes have to interfere with the structural components of the cells. Recent findings (Hamant et al., 2008) have shown that anisotropy of growth depends on a feedback loop involving mechanical forces acting on the microtubules. This can be uncoupled from the (auxin) based control of growth rates. The morphogenetic functions of the genes therefore depend on how they influence these two separable processes.
The cell wall is composed of rigid cellulose microfibrils, which are cross-linked to each other by other polysaccharides such as hemicelluloses and pectins. Growth rate partially depends on wall synthesis, causing the cell wall to yield to the internal turgor pressure and the cell to grow. Indeed, interfering with the rate of cellulose synthesis by mutation or drugs substantially modifies growth rates. The degree of cross-linking and remodeling of the matrix also represents an important factor. In addition to its central role in growth rate control, the cell wall controls growth anisotropy. This is because cellulose fibrils are often deposited in highly ordered arrays, favoring cell expansion in specific directions. The initial question as to how the gene regulatory network controls growth rate and anisotropy can therefore be translated into how cell wall synthesis and microfibril orientation are coordinated.
Several groups have started to address the regulation of wall synthesis and anisotropy at the meristem. In a classical study Fleming et al. (1997) showed that the external application of a cell wall-modifying enzyme, expansin, could initiate the outgrowth of organ-like structures on an apex of tomato (Solanum lycopersicum). More recently, Peaucelle et al. (2008) showed that modifications in pectin composition and cross-linking are associated with the rapid outgrowth of young organ primordia. In parallel, Hamant et al. (2008) addressed the regulation of cell anisotropy. It is widely accepted that the orientation of the cellulose microfibrils largely depends on the orientation of the underlying membrane-associated microtubules that apparently serve as tracks along which the cellulose synthase complexes travel. Hamant et al. (2008) therefore analyzed in detail the behavior of these microtubules. It is generally assumed that the meristem is an elastic structure with a relatively rigid surface layer under constant pressure. Based on this postulate, the theoretical force fields at the meristem surface were calculated to show that main stress orientations run in random directions at the top of the meristematic dome, parallel to organ boundaries and perpendicular to the long axis of the stem. Interestingly, it was observed that microtubules follow precisely the same pattern, i.e. they are parallel to the predicted forces. Modifying the forces by changing the shape of the entire meristem or perturbing force patterns using ablations also caused relatively rapid changes in microtubule directions, confirming a causal relationship between physical stresses and cytoskeleton organization. By orienting the microtubules parallel to the main force directions, the cells would also direct their cellulose microfibrils this way, making the cells resist these forces. This process would in particular be important for the formation of sharp physical organ boundaries. Can such a mechanical feedback mechanism where forces feed back on cell anisotropy create the patterns of microtubule orientation and shape changes observed during organ initiation? This question was addressed using models able to express mechanical properties. In one of these models, the surface of the meristem was represented as a network of springs, each spring representing a cell wall. This dome of springs was subsequently put under virtual pressure and the model was asked to calculate the forces sensed by every spring. Every cell in the model was subsequently instructed to stiffen its walls (springs) in function of their angle to the main forces. The simulations were able to reproduce at least qualitatively microtubule behavior. In addition, they could generate simple shapes such as cylinders or outgrowing organs with sharp boundaries. Note that this model is again based on the concept of complex systems: All the cells do in these simulations is to resist locally to forces. At the level of the whole tissue, this subsequently results in specific shapes as an emergent property.
Importantly, this microtubule-based anisotropy can be uncoupled from the auxin-based control of growth rates. If microtubules are depolymerized using the drug oryzalin, cells continue to differentiate at the correct relative positions and relative growth rates seem unperturbed. This suggests that the auxin-based induction of organ outgrowth operates at least partially independently from the control of anisotropy.
CONCLUSION AND PERSPECTIVES: TOWARD AN INTEGRATED VIEW OF ORGAN INITIATION
The results discussed above provide a partially integrated view of the processes that lead to organ formation. Auxin flows generated by cells locally sensing auxin concentrations or auxin fluxes create hormone maxima and minima at the meristematic dome. These auxin concentrations are subsequently directly or indirectly interpreted by the gene regulatory networks in terms of cell wall synthesis rates, causing some cells to grow more quickly than others. The differential growth rates thus generated create tensions and forces in the meristem that finally feed back on microtubule orientation and microfibril texture. Together this leads to the outgrowth of a well-defined organ. Some of the major challenges are now to unravel the pathways between molecular regulatory networks and cell wall modifications, as well as to further determine the precise role of biophysical processes in the feedback between stress and cytoskeleton.
In more general terms, the work illustrates how systems biology is providing a powerful conceptual framework to approach development. Starting from the consideration that living systems are complex systems, we have to study the developing plant at different levels of organization. This requires a range of multidisciplinary methodologies, and one of the main aims is to create a cycle between experimentation and modeling. Since mathematical modeling leads to precise, quantitative predictions, experimentation should, ideally, also be quantitative. Therefore, detailed, quantitative information will be required on a range of parameters such as gene product levels, expression patterns, and growth dynamics.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jan Traas (jan.traas@ens-lyon.fr).
References
- Aida M, Tasaka M (2006) Genetic control of shoot organ boundaries. Curr Opin Plant Biol 9 72–77 [DOI] [PubMed] [Google Scholar]
- Albert R (2007) Network inference, analysis. Mol Syst Biol 19 3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Benítez M, Dávila EB, Chaos A, Espinosa-Soto C, Padilla-Longoria P (2007) Gene regulatory network models for plant development. Curr Opin Plant Biol 10 83–91 [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Bohn-Courseau I, Ljung K, Morin H, Carraro N, Godin C, Traas J (2006) Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc Natl Acad Sci USA 103 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Smith RS, Mandel T, Nakayama N, Sauer M, Prusinkiewicz P, Kuhlemeier C (2009) Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev 23 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971 [DOI] [PubMed] [Google Scholar]
- Coen E, Rolland-Lagan AG, Matthews M, Bangham JA, Prusinkiewicz P (2004) The genetics of geometry. Proc Natl Acad Sci USA 101 4728–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong H (2002) Modeling and simulation of genetic regulatory systems: a literature review. J Comput Biol 9 67–103 [DOI] [PubMed] [Google Scholar]
- Dinneny J, Yadegari R, Fischer R, Yanofsky M, Weigel D (2004) The role of JAGGED in shaping lateral organs. Development 131 1101–1110 [DOI] [PubMed] [Google Scholar]
- Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla E (2004) A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell 16 2923–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A, McQueen-Mason S, Mandel T, Kuhlemeier C (1997) Induction of leaf primordia by the cell wall protein expansin. Science 276 1415–1418 [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gordon S, Chickarmane V, Ohno C, Meyerowitz E (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Heisler M, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz E, et al (2008) Developmental patterning by mechanical signals in Arabidopsis. Science 322 1650–1655 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Long JA, Reddy GV, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hu Y, Poh HM, Chua NH (2006) The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J 47 1–9 [DOI] [PubMed] [Google Scholar]
- Jönsson H, Heisler M, Reddy GV, Agrawal V, Gor V, Shapiro BE, Mjolsness E, Meyerowitz EM (2005) Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics (Suppl 1) 21 i232–i240 [DOI] [PubMed] [Google Scholar]
- Jönsson H, Heisler M, Shapiro BE, Meyerowitz EM, Mjolsness E (2006) An auxin-driven polarized transport model for phyllotaxis. Proc Natl Acad Sci USA 103 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer K, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814 [DOI] [PubMed] [Google Scholar]
- Lohmann J, Hong R, Hobe M, Busch M, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803 [DOI] [PubMed] [Google Scholar]
- Long T, Brady S, Benfey P (2008) Systems approaches to identifying gene regulatory networks in plants. Annu Rev Cell Dev Biol 24 81–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndon RF (1998) The Shoot Apical Meristem. University Press, Cambridge, UK
- Merks RM, Van de Peer Y, Inze D, Beemster GT (2007) Canalization without flux sensors: a traveling-wave hypothesis. Trends Plant Sci 12 384–390 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer R (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G (2008) Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol 18 1943–1948 [DOI] [PubMed] [Google Scholar]
- Rast MI, Simon R (2008) The meristem-to-organ boundary: more than an extremity of anything. Curr Opin Genet Dev 18 287–294 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce E-R, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260 [DOI] [PubMed] [Google Scholar]
- Sablowski R (2007) The dynamic plant stem cell niches. Curr Opin Plant Biol 10 639–644 [DOI] [PubMed] [Google Scholar]
- Sachs T (1969) Polarity and the induction of organized vascular tissues. Ann Bot 33 263–275 [Google Scholar]
- Smith RS, Guyomarc'h S, Mandel T, Reinhardt D, Kuhlemeier C, Prusinkiewicz P (2006) A plausible model of phyllotaxis. Proc Natl Acad Sci USA 103 1301–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow M, Snow R (1933) Experiments on phyllotaxis. II. The effect of displacing a primordium. Philos Trans R Soc Lond B Biol Sci 222 353–400 [Google Scholar]
- Snow M, Snow R (1935) Experiments on phyllotaxis. III. Diagonal splits through decussate apices. Philos Trans R Soc Lond B Biol Sci 225 63–94 [Google Scholar]
- Snow M, Snow R (1952) Minimum areas and leaf determination. Proc R Soc Lond B Biol Sci 139 545–566 [DOI] [PubMed] [Google Scholar]
- Steeves TA, Sussex IA (1989) Patterns in Plant Development, Ed 2. Cambridge University Press, New York
- Stoma S, Lucas M, Chopard J, Schaedel M, Traas J, Godin C (2008) Flux based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLoS Comput Biol 4 1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M (2006) BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J 25 3912–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas J, Doonan J (2001) Cellular basis of shoot apical meristem development. Int Rev Cytol 208 161–206 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV (2009) Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA 106 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]