Abstract
In higher plants, lysophosphatidic acid acyltransferase (LPAAT), located in the cytoplasmic endomembrane compartment, plays an essential role in the synthesis of phosphatidic acid, a key intermediate in the biosynthesis of membrane phospholipids in all tissues and storage lipids in developing seeds. In order to assess the contribution of LPAATs to the synthesis of storage lipids, we have characterized two microsomal LPAAT isozymes, the products of homoeologous genes that are expressed in rapeseed (Brassica napus). DNA sequence homologies, complementation of a bacterial LPAAT-deficient mutant, and enzymatic properties confirmed that each of two cDNAs isolated from a Brassica napus immature embryo library encoded a functional LPAAT possessing the properties of a eukaryotic pathway enzyme. Analyses in planta revealed differences in the expression of the two genes, one of which was detected in all rapeseed tissues and during silique and seed development, whereas the expression of the second gene was restricted predominantly to siliques and developing seeds. Expression of each rapeseed LPAAT isozyme in Arabidopsis (Arabidopsis thaliana) resulted in the production of seeds characterized by a greater lipid content and seed mass. These results support the hypothesis that increasing the expression of glycerolipid acyltransferases in seeds leads to a greater flux of intermediates through the Kennedy pathway and results in enhanced triacylglycerol accumulation.
With increasing environmental challenges and concerns, there is renewed interest in deriving plant-based sustainable alternatives for petroleum products, including carburants, lubricants, and industrial feed stocks. Modifying oilseed crops to produce oils of uniform composition containing fatty acids varying in chain length or possessing reactive functional groups is a primary objective (Jaworski and Cahoon, 2003), as is that of increasing the yield of seed oil (Lardizabal et al., 2008; Zheng et al., 2008). Early success in modifying seed oils to produce the more common fatty acids has been tempered by limited success in the production of high levels of unusual fatty acids (UFAs) in cultivated oilseeds (Thelen and Ohlrogge, 2002; Drexler et al., 2003). Such studies have led to the conclusion that in order to achieve levels of UFAs similar to those present in the oil of native species, enzymatic activities additional to fatty acid modification are necessary to optimize the synthesis (Mekhedov et al., 2001), stability (Eccleston and Ohlrogge, 1998), and channeling (Bafor et al., 1990) of the desired fatty acid into triacylglycerol (TAG).
The synthesis of glycerolipids occurs in the cytoplasm using de novo-synthesized fatty acids exported from the plastid as acyl-CoA thioesters. The fatty acyl groups are incorporated into membrane and storage lipids by the sequential esterification of glycerol-3-phosphate by the action of glycerol-3-phosphate acyltransferase (GPAT; EC 2.3.1.15) at sn-1 to form lysophosphatidic acid followed by lysophosphatidic acid acyltransferase (LPAAT; EC 2.3.1.51) at sn-2 to form phosphatidic acid (PA; Somerville et al., 2000). Dephosphorylation of PA results in the formation of diacylglycerol (DAG), which in developing seeds may be directed into the production of TAG by acyl-CoA-independent reactions or by diacylglycerol acyltransferase (DAGAT; EC 2.3.1.20; Roscoe, 2005). The substrate preferences for acyl-thioesters and the selectivities for the acceptor molecules displayed by the microsomal acyltransferases play a crucial role in establishing the acyl composition of lipids (Frentzen, 1998). The TAG synthesized in most oilseeds of agronomic importance contains fatty acids that are the same as those present in cytoplasmic membrane lipids. In contrast, the seeds of species that synthesize TAGs with exotic fatty acid compositions possess microsomal acyltransferases that facilitate the incorporation of UFAs into storage lipids because of their broad GPAT and/or their selective DAGAT specificities (Wiberg et al., 1994; Frentzen, 1998). Furthermore, oilseeds characterized by TAGs that contain UFAs at sn-2 possess additional seed-specific microsomal LPAATs (Brown et al., 1995; Hanke et al., 1995; Knutzon et al., 1995) that exhibit a wide variation in substrate preference and that serve to ensure the channeling of UFAs to this position, thereby segregating incompatible fatty acids away from membrane lipids.
Cloning of cDNAs from cultivated and exotic plants and the availability of entirely sequenced genomes from plant and algal species have revealed that a minimum of two classes of genes encoding microsomal LPAATs exist (Frentzen, 1998) within a larger, LPAAT-like gene family containing acyltransferases as yet functionally uncharacterized but distinct from GPATs (Roscoe, 2005). The class A microsomal LPAATs defined by Frentzen (1998) possess substrate preferences for C18:1-CoA typical of enzymes involved in membrane lipid synthesis and are ubiquitously expressed in the plant. In contrast, individual members of the class B LPAATs display preferences for distinct, unusual saturated or unsaturated acyl groups and are normally expressed in storage organs. Although class B LPAATs have been exploited to alter the stereochemical composition of rapeseed (Brassica napus) oil to permit the incorporation of modified fatty acids at sn-2 (Lassner et al., 1995; Knutzon et al., 1999), a significant increase in the total amount of UFAs was not accomplished by the expression of the class B LPAATs alone. In contrast, the transformation of rapeseed and Arabidopsis (Arabidopsis thaliana) with a yeast gene encoding a variant LPAAT, SLC1-1, capable of accepting very long chain fatty acyl (VLCFA)-CoA substrates resulted in an increase in the total VLCFAs and, unexpectedly, in total oil content (Zou et al. 1997).
In our efforts to modify the fatty acid composition of oil in rapeseed, in particular to increase the content of VLCFAs, we have addressed the question of optimizing the environment for the correct functioning of LPAATs encoded by transgenes. The above studies using the various LPAAT transgenes indicate that channeling of UFAs into sn-2 of oilseed species remains problematic. The ability to obtain oils with uniform composition strongly depends on the occupancy of sn-2 by UFAs, yet the level of occupancy of sn-2 by fatty acids corresponding to the selectivity of the introduced LPAAT is variable and relatively modest. Occupancy of sn-2 is determined in part by the ability of the LPAAT encoded by the transgene to compete with the endogenous enzyme, a function of the acyl-CoA substrates available to the enzymes and the relative efficiencies of the enzymes to compete for the donor and acceptor substrates. We argued that there is latitude for the reduction of competing activities using an antisense strategy, and although microsomal LPAATs have been cloned from rapeseed, there are no reports of the characterization of the enzyme. Our objectives in this work were to identify and evaluate the potential contribution of LPAAT isozymes to TAG biosynthesis in rapeseed, thereby discerning targets for optimizing efforts to modify oils for industrial purposes. In this study, we catalogue a previously undescribed complexity in microsomal LPAAT diversity and identify a LPAAT isozyme likely to play an important role in TAG synthesis in rapeseed. In contrast to diverged LPAATs of plant origin, we demonstrate a positive effect of the overexpression of microsomal LPAATs on oil content and seed weight.
RESULTS
Sequence Analysis of Rapeseed cDNAs Encoding LPAATs
A cDNA encoding a putative LPAAT isoform was previously isolated in our laboratory via hybridization screening of an immature embryo library of rapeseed with labeled oligonucleotides derived from the maize (Zea mays) endosperm nucleotide sequence (accession no. Z29518). This cDNA sequence was subsequently used to rescreen a second aliquot of the immature embryo library and resulted in the isolation of 13 clones, of which seven contained cDNAs of approximately 1.5 kb. Three clones named BAT1.13, BAT1.12, and BAT1.5 (for Brassica acyl transferase 1) were sequenced to completion and contained cDNAs of 1,466, 1,593, and 1,581 bp, respectively, preceding the poly(A+) tail. The cDNAs BAT1.13 and BAT1.12 shared 89.8% nucleotide identity to each other, and alignment of each of these cDNAs with the National Center for Biotechnology Information (NCBI) nucleotide database using the BLAST2 algorithm revealed the strongest identity (87%) with the rapeseed nucleotide sequence (accession no. Z95637) encoding a putative LPAAT. The BAT1.5 cDNA shared 82.5% nucleotide identity with BAT1.13 and 99% identity with a Brassica oleracea cDNA (accession no. AY616009).
Conceptual translation of the BAT 1.13 and BAT1.12 cDNA sequences revealed the presence of a single open reading frame coding for an identical protein of 390 amino acids, of mass 43.7 kD and pI 9.69. This protein sequence was found to share significant homology with LPAATs of other eukaryotic species in BLAST2 searches of the NCBI database. The highest score, 97% similarity, was obtained with a LPAAT of rapeseed (accession no. Q9XFW4), followed by 96% identity with the Arabidopsis gene product At3g57650, characterized by Kim et al. (2005). The deduced protein sequence encoded by the BAT1.5 cDNA contained 391 amino acids, of mass 43.6 kD and pI 9.76, sharing 95.9% identity and 97.4% similarity to the BAT1.13 protein and 98% identity to a B. oleracea cDNA (accession no. AAT36638). The BAT1.13 and BAT1.5 proteins shared 74% identity with the Limnanthes douglasii LPAAT expressed in seeds and leaves (Brown et al., 1995). The BAT1.13 protein shared 57% similarity to the Saccharomyces cerevisiae LPAAT, SLC1 (Nagiec et al., 1993), and 40% similarity to the Escherichia coli LPAAT encoded by the PlsC gene. No significant homology was evident with the LPAATs that prefer unusual acyl-CoAs as substrates and whose expression is restricted to the seeds in the case of L. douglasii (accession no. Q42870) or the endosperm in the case of coconut (Cocos nucifera; accession no. Q42670).
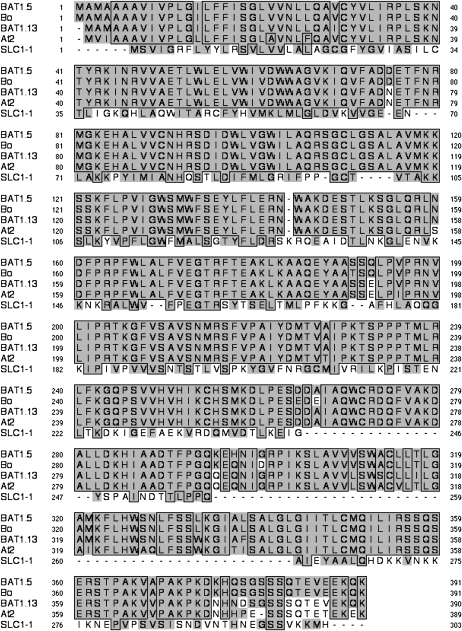
There are six positions at which amino acids differ in a conservative manner and seven nonconservative substitutions between BAT1.13 and BAT1.5, together with additional residues at Ala-7 and Glu-386 present in BAT1.5 and a residue, Asn-375, that is present in BAT1.13 but absent in BAT1.5. Most of the variation between the proteins occurs at the C-terminal region, as shown in Figure 1. Each deduced protein contains a PlsC-like glycerolipid acyltransferase domain (InterProScan, European Bioinformatics Institute; conserved domain, NCBI) located between residues 80 and 200, which includes the highly conserved motif NH(X4)D common to glycerolipid acyltransferases. The deduced rapeseed proteins are predicted to possess at least three transmembrane-spanning regions positioned at residues 6 to 23, 304 to 320, and 337 to 353 (TMPRED, Expasy Switzerland). The most favorable predicted localization of each protein is the endoplasmic reticulum, and a putative dilysine-like endoplasmic reticulum retention signal (EKQK391) is present at the C terminus (pSORT, NIBB Japan).
Figure 1.
Identification of rapeseed LPAAT protein sequences. Comparison of the deduced amino acid sequences of the rapeseed cDNAs BAT1.13 and BAT1.5 with 1-acyl-glycerol-3-phosphate acyltransferases of B. oleracea (accession no. AAT36638), Arabidopsis (accession no. NP567052), and Saccharomyces cerevisiae (accession no. P33333). Outlined boxes indicate identity, shading indicates conservative differences, and unshaded areas indicate nonconservative differences.
Functional Characterization of Rapeseed LPAAT Isoforms
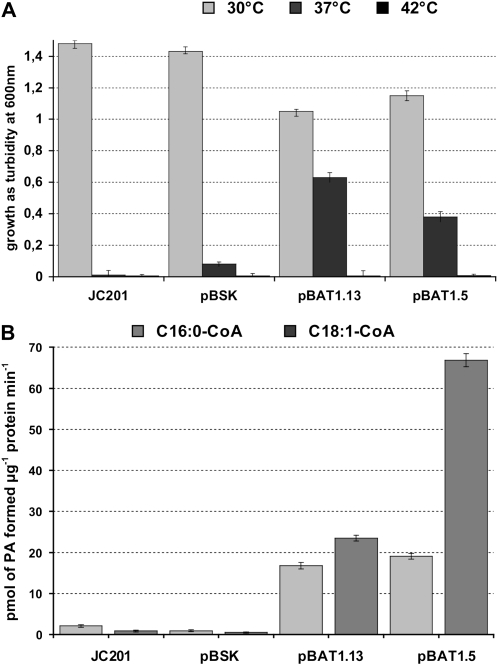
The sequence homologies with LPAATs of yeast and plant origin indicated that BAT1.13 and BAT1.5 cDNAs encoded LPAATs putatively localized at the endoplasmic reticulum and thus likely to be associated with the eukaryotic pathway of lipid synthesis. To verify this hypothesis, we performed complementation and enzymatic assays in the temperature-sensitive E. coli mutant JC201 (Coleman, 1990). This strain is deficient in LPAAT activity when grown at elevated temperatures but grows well at 30°C; thus, the complementation assay is based on the restoration of growth at nonpermissive temperatures. Plasmid DNA containing the BAT1.13 or BAT1.5 cDNAs in frame with the β-galactosidase of the pBluescript SK+ vector was transformed into JC201 cells. Transformed colonies were inoculated and incubated in liquid culture at various temperatures. Growth curves established by monitoring cell densities revealed the BAT1.13 and BAT1.5 cDNAs to be capable of restoring growth to JC201 at 37°C but not at 42°C, as summarized in Figure 2A. The growth of JC201 and vector-only-transformed JC201 was insignificant at 37°C and higher temperatures.
Figure 2.
Functional characterization of rapeseed LPAATs. A, Complementation of an acyltransferase-deficient strain of E. coli. The coding sequences corresponding to the BAT1.13 and BAT1.5 cDNAs were cloned into the pBluescript SK+ vector and transformed into the JC201 plsC− strain. Aliquots from OD600 = 0.5 cultures were grown in the presence of IPTG. The figure shows the OD after 16 h of growth at various temperatures. JC201 indicates strain only; pBSK indicates strain JC201 transformed with vector only; BAT1.13 and BAT1.5 indicate strain JC201 transformed with the vector containing the coding sequence of BAT1.13 or BAT1.5. Data represent means of three independent transformations. B, Rapeseed microsomal LPAAT activities in E. coli membrane preparations. Membranes were isolated from JC201 cells that had been grown at 30°C after induction by IPTG and were assayed in the presence of saturating concentrations of LPA and [1-14C]oleoyl-CoA or [1-14C]palmitoyl-CoA, and the quantity of radioactivity in the PA reaction product was integrated. The genotypes of JC201, pBSK, BAT1.13, and BAT1.5 are as in A. The values represent means of three membrane preparations from independent transformant clones.
In order to confirm that the BAT1.13 and BAT1.5 cDNAs code for a protein possessing LPAAT activity, membranes were isolated from the JC201 strain transformed with plasmids containing the BAT1.13 or the BAT1.5 cDNA and assayed for incorporation of acyl groups into PA using [1-14C]oleoyl-CoA or [1-14C]palmitoyl-CoA (Fig. 2B). LPAAT activity was determined in the presence or absence of 55 μm 1-oleoyl-lysophosphatidic acid (LPA) and varying concentrations of oleoyl-CoA or palmitoyl-CoA. The data presented in Figure 2B were obtained at a saturating concentration of [1-14C]acyl-CoA of 40 μm. The rate of formation of PA from LPA and [1-14C]oleoyl-CoA in the presence of membranes isolated from E. coli JC201 transformed with the vector containing the BAT1.5 cDNA was approximately 140-fold greater compared with membranes isolated from cells transformed with the vector only. For the membranes isolated from E. coli JC201 transformed with the vector containing the BAT1.13 cDNA, the rate of formation of PA from LPA and [1-14C]oleoyl-CoA was approximately 44-fold greater than membranes isolated from the vector-only-transformed cells. Thus, the LPAAT activity with a [1-14C]oleoyl-CoA donor of the BAT1.5-containing membranes was approximately 3-fold greater than the BAT1.13-containing membranes. In contrast, the rates of formation of PA from LPA and [1-14C]palmitoyl-CoA in the presence of membranes isolated from E. coli JC201 transformed with the vector containing the BAT1.5 cDNA or the BAT1.13 cDNA were similar, and 19-fold and 16.5-fold, respectively, greater than membranes isolated from cells transformed with the vector only. Furthermore, the BAT1.5 enzyme exhibits a greater activity with oleoyl-CoA compared with the BAT1.13 enzyme, whereas the activity of each enzyme is similar with respect to palmitoyl-CoA. The LPAAT activity of all membrane fractions was essentially zero in the absence of LPA (data not shown). These results establish a preference of the enzymes encoded by the BAT1.13 and BAT1.5 cDNAs for oleoyl-CoA over palmitoyl-CoA when 1-oleoyl LPA was the acceptor, consistent with a cytoplasm-localized LPAAT associated with the eukaryotic pathway.
Expression of Rapeseed Genes Encoding Microsomal LPAATs in Planta
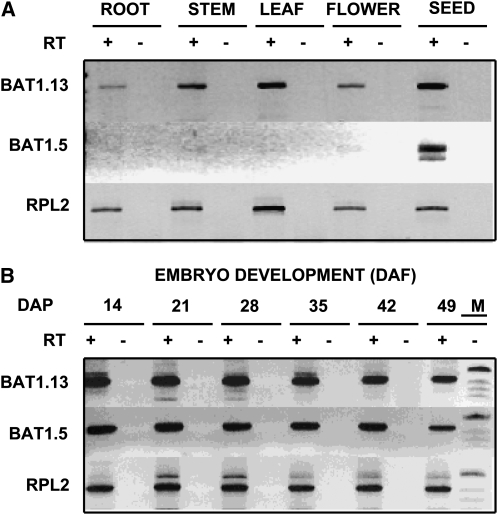
Since we were interested in the contribution of different rapeseed LPAAT isoforms to the synthesis of storage lipids, we established that the BAT1.13 and BAT1.5 genes were both expressed during seed maturation. Reverse transcription (RT)-PCR was performed using oligonucleotides capable of amplifying each transcript in a specific manner. A differential and tissue-specific pattern of expression was evident between the two genes, as shown in Figure 3. BAT1.13 was expressed in root, stem, leaf, flower, and immature seeds of rapeseed, whereas BAT1.5 expression was detected in immature seeds only. In subsequent RT-PCR analysis using RNA isolated from immature embryos of rapeseed, the expression of both BAT1.5 and BAT1.13 was detected at all stages of embryo development at 14 to 49 d after pollination, from the heart stage to the mature embryo entering desiccation. These results indicate that BAT1.13 is expressed in a ubiquitous manner and is thus possibly involved in the production of PA for a constitutive requirement in most tissues, whereas BAT1.5 expression was detected predominantly in the seeds and is thus of potential importance for the provision of PA for both storage and membrane lipid synthesis.
Figure 3.
Expression of rapeseed microsomal LPAATs. Total RNA was purified from various rapeseed tissues (A) and from developing embryos (B) at various days after pollination (DAP), transcribed in the presence (+) of reverse transcriptase (RT), and the resulting cDNA was used as a template for PCR with primers designed to discriminate between BAT1.13 and BAT1.5 transcripts. − indicates the absence of reverse transcriptase. RPL2 represents a control for RNA loading. M corresponds to the DNA size ladder added to the 49 DAP(−) lane. DAF, Days after flowering.
Verification of BAT1.13 and BAT1.5 Promoter Specificities
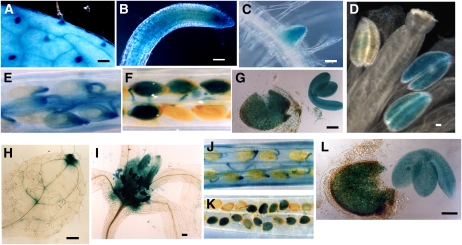
The results obtained by RT-PCR suggested an expression of the two rapeseed LPAAT genes, BAT1.13 and BAT1.5, that differed in a tissue-specific manner. To confirm this apparent difference in the profile of expression, approximately 1 kb of the promoter region of BAT1.13 and BAT1.5 was fused to the GUS reporter gene and incorporated into a binary vector, and Arabidopsis plants were transformed. GUS histochemistry was performed on the various organs of four independent transformants for each of the two promoter∷reporter lines in order to provide information on the precise site of tissue expression (Fig. 4). In the case of the BAT1.13 promoter∷GUS fusions, activity was detected in the cotyledons and in leaves of seedlings, and strong expression was associated with the vascular tissues in these organs and with the basal cell of the trichomes (Fig. 4A). BAT1.13 promoter activity was also detected in the vascular elements of cauline leaves (data not shown). In roots, the BAT1.13 promoter was active at the tip of primary and secondary roots and in mesophyll/cortex tissue behind the elongation zone and in the vascular tissue (Fig. 4, B and C). In flowers, the BAT1.13 promoter was transiently active in the anthers of young, unopened flower buds (Fig. 4D), but the expression disappeared at flower opening as the anthers matured. A weak expression was detected in anthers and/or pollen later after fertilization had occurred. In young siliques at 3 d after flowering, weak BAT1.13 expression was evident in the stigma, in the pistil, and in the elongated carpels after flower opening, together with a stronger BAT1.13 promoter activity in the septum, the funiculus, and the nucellus (Fig. 4E). Between 9 and 12 d after flowering, strong GUS staining was observed in the embryo and endosperm at a stage corresponding to the immature walking stick embryo, which persisted until the mature cotyledonary stage embryo at 18 d after flowering (Fig. 4, F and G).
Figure 4.
Principal sites of BAT1.13 and BAT1.5 gene expression. Histochemical staining is shown for GUS activity in BAT promoter-uidA transgenic Arabidopsis plants. A, Rosette leaf. B, Primary root. C, Emerging secondary root. D, Mature anther. E, Opened silique 3 to 5 d after pollination (DAP). F, Opened silique 9 to 12 DAP. G, Seed coat and embryo 12 to 15 DAP. H, Stained hydratoid of rosette leaf. I, Stipules/emerging leaves. J, Opened silique 3 to 5 DAP. K, Opened silique 9 to 12 DAP. L, Seed coat and embryo 12 to 15 DAP. A to G show the BAT1.13 promoter, and H to L show the BAT1.5 promoter. Bars = 40 μm in A, B, C, H, and I and 80 μm in D, G, and L.
In contrast, evidence of BAT1.5 promoter activity was detected only in the hydathodes of cotyledons and leaves (Fig. 4H) and in the stipules and emerging leaves of seedlings (Fig. 4I). In flowers, expression from the BAT1.5 promoter was similar to that of BAT1.13; that is, transient in the anthers (compare with Fig. 4D), disappearing at flower opening, and weak expression reappearing in the anther after fertilization. Weak BAT1.5 promoter activity was occasionally observed in a few unconnected sections of the vascular tissues of young leaves and in the young roots of some transformant lines. Weak BAT1.5 promoter activity was detected in the carpels, septum, and funiculus in young siliques at 3 d after flowering (Fig. 4J) but decreased thereafter. The BAT1.5 promoter was active in seeds, embryo, and endosperm but not in the integuments, appearing at a stage corresponding to the heart-to-torpedo transition and persisting until the mature cotyledonary embryo (Fig. 4, K and L). The activity of both BAT1.13 and BAT1.5 promoters was uniform in the cotyledons and hypocotyl of the mature embryo. Taken together, these results confirm that the gene encoding the BAT1.13 isozyme is expressed in a ubiquitous, but not a constitutive, manner in most tissues at certain stages of development. In contrast, the gene encoding the BAT1.5 isozyme exhibits a profile of strong expression restricted principally to the developing seed.
Characterization of Arabidopsis Plants Expressing Rapeseed LPAAT Isozymes
In order to evaluate the contributions of seed-expressed LPAAT isoforms to TAG synthesis, binary plant transformation vectors that contained the cDNAs corresponding to the coding sequences of BAT1.13 or BAT1.5 in the sense orientation under the control of the napin 2S2 promoter (Guerche et al., 1990) were constructed and introduced into Agrobacterium tumefaciens, which was subsequently used to transform Arabidopsis plants. The presence of the LPAAT transgene was confirmed in the T1 seedlings by PCR, and a total of 30 BAT1.13 sense plants together with 20 BAT1.5 sense plants containing a single-locus T-DNA insertion were identified. For an analysis of the effect of the LPAAT transgenes on seed lipids, a total of 13 independent T2 homozygous single-copy BAT1.13 plants together with eight BAT1.5 plants were selected at random.
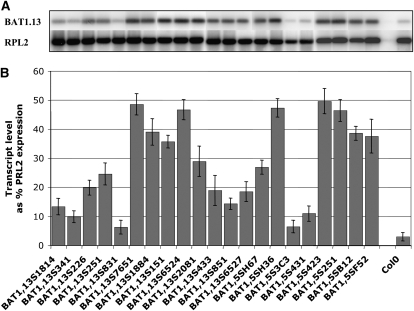
The level of expression of the BAT1.13 and BAT1.5 transgenes in the seeds of the Arabidopsis transformant plants was determined and is documented in Figure 5. We performed RT-PCR with gene-specific primers to discriminate between the expression of the Brassica and Arabidopsis LPAAT genes in the transgenic plants followed by hybridization to a radiolabeled BAT1.13 probe for quantitation. Analysis of RNA isolated from Arabidopsis T3 seeds revealed expression of the Brassica BAT1.13 cDNA or BAT1.5 cDNA in all lines but only a background hybridization signal in the nontransformed plants. Low levels of LPAAT transgene expression were evident for the lines 1.131814, 1.13341, 1.13226, 1.13251, 1.13831, and 1.53C3. The highest levels of transgene expression was detected in the lines 1.13S7651, 1.13S6524, 1.5SH36, and 1.5S423. The BAT1.13 signal after gel-blot analysis of RNA isolated from nontransformed seeds was essentially zero, reflecting background hybridization.
Figure 5.
Expression of BAT1.13 and BAT1.5 cDNAs in transgenic Arabidopsis seeds. Total RNA was purified from mature seeds, reverse transcribed, and the resulting cDNA was used as a template for PCR with primers designed to discriminate the BAT1.13 and BAT1.5 transcripts from that of the endogenous Arabidopsis gene. The resulting reaction mix was migrated on agarose gels and transferred to a nylon membrane. The membrane was hybridized sequentially with a BAT1.13 or a 32P-labeled cDNA followed by a RPL2 32P-labeled probe, and the radioactive signals (A) were integrated using a phosphor imager (B).
Overexpression of Rapeseed LPAAT Isoforms in Arabidopsis Seeds
Our initial characterization of the T2 seeds of eight BAT1.13- and eight BAT1.5-overexpressing lines was conducted by quantifying fatty acids from separate neutral and polar lipid fractions, which revealed that polar lipids represented 14% of the seed lipids of ecotype Columbia (Col-0). Although we did not observe strong differences in the profile of classes of individual polar lipids, we observed that total polar lipids were reduced by approximate means of 10% in BAT1.13 and 15% in BAT1.5 overexpression lines compared with the controls. In contrast, the preliminary analyses of fatty acid content in the TAG fraction extracted from T2 seeds suggested that the expression of each of the microsomal LPAAT isoforms was associated with a greater lipid content in Arabidopsis seeds. Therefore, we extended these analyses to T3 and T4 seeds. Four independent plants of each of the BAT1.13 and BAT1.5 transformant lines were grown together with five individual nontransformed Col-0 plants under the same conditions. There were no discernible morphological or developmental differences between the transformed and nontransformed plants during growth. Flowering time and seed development were similar between transformed plants and nontransformed plants. Siliques were opened between 11 and 13 d after pollination, and numbers of seeds per silique were counted. No significant differences in seed numbers were evident between BAT1.13 and BAT1.5 transformed plants and nontransformed plants. Plants were grown to maturity, and the seeds were collected. The appearance and morphology of the seeds of the BAT1.13 transformants were similar to those of the seeds of nontransformed Col-0 plants. Seed germination was near to 100% and was similar between BAT1.13 and BAT1.5 plants (data not shown).
Whole mature seeds were subjected to direct transesterification, and the resulting fatty methyl esters were quantified via gas chromatography. The total fatty acid content of the seeds of the nontransformed Col-0 plants was 6.07 ± 0.43 μg (n = 25) per seed. These values equate to 33.4% ± 1.88% total lipid content of dry seed weight and are in accordance with the values for Col-0 obtained by Li et al. (2006). The fatty acid composition of the TAG fraction among each of the 13 BAT1.13 and eight BAT1.5 plants did not vary significantly in the proportions of each of the major classes of fatty acids and of the VLCFAs (C20:1 and C22:1) compared with that of the nontransformed control plants grown simultaneously under the same conditions.
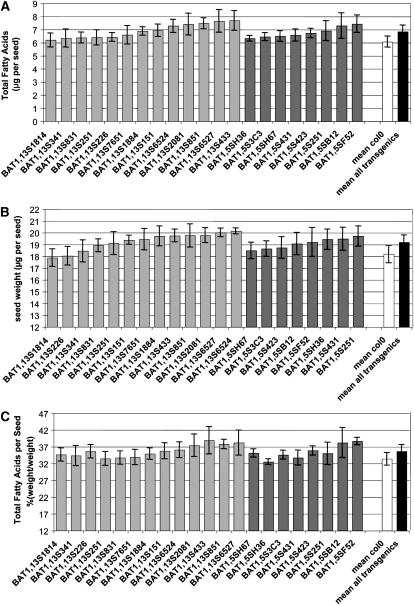
The individual fatty acid methyl esters were quantified from the seeds of the 13 BAT1.13 and eight BAT1.5 lines (Fig. 6A). Fatty acid content was determined on four independent lots of 50 seeds for each transformant line, each lot being a homogeneous mixture of seeds originating from four plants grown under the same conditions. Total fatty acid contents of seeds were determined for the 21 lines on T3 seeds in three independent experiments and repeated on T4 seeds in two independent experiments. The mean value for the total fatty acid content of seeds from the T4 generation, 6.92 ± 0.53 μg, was not significantly different from that of the T3 generation seeds, 6.76 ± 0.51 μg (t test, P < 0.05); therefore, the data obtained from experiments at T3 and T4 generations were considered as five independent biological replicates. The total fatty acid content of the BAT1.13 lines (mean 6.90 ± 0.54 μg) varied in a range between 102% and 127% of that of the nontransformed plants (Fig. 6A). Of the 13 BAT1.13 plants, 10 possessed a greater total fatty acid content per seed (mean 7.08 ± 0.46 μg) that represented a 17% greater content compared with that of seeds of nontransformed plants (t test, P < 0.05). Each of the eight BAT1.5 lines possessed a total fatty acid content in seeds (mean 6.78 ± 0.40 μg) that was greater than that of the nontransformed Col-0 plants grown under the same conditions. The total fatty acid content varied in a range between 104% and 123% of that of the nontransformed plants, with a mean value of 112% (Fig. 6A). Six of the lines possessed a significantly greater total fatty acid content, with a mean value (6.91 ± 0.38) that represented a 14% greater seed lipid content compared with that of the control seeds (t test, P < 0.05). Overall, combining the results for the 13 BAT1.13 and eight BAT1.5 lines, the mean fatty acid content was 6.85 ± 0.49 μg, which represented a 13% greater content than in the Col-0 mean (t test, P < 0.05). Furthermore, the mean total fatty acid content of the 15 lines that possessed significantly greater total fatty acid contents was 7.05 ± 0.43 μg, representing a 16% greater seed lipid content compared with the mean of the combined Col-0 nontransformed plants (t test, P < 0.05).
Figure 6.
Effect of expression of BAT1.13 and BAT1.5 on seed fatty acid content and weight. A, Total fatty acid content in transgenic Arabidopsis seeds. Fatty acid content of seeds was quantified by gas-liquid chromatography of fatty acid methyl esters. Fatty acid content was determined on four independent seed lots of each transgenic line or from seeds of five independent nontransformed Col-0 plants grown under the same conditions. Values represent means of five replicate experiments. “Mean all transgenics” indicates mean of 13 BAT1.13 transformants and eight BAT1.5 transformants. B, Weight of transgenic Arabidopsis seeds. The number of seeds present in an approximately 5-mg aliquot was determined for each lot of seeds of each transformant line together with seeds from five independent nontransformed Col-0 plants grown under the same conditions. Values are averages of two independent aliquots and are expressed as weight per seed. “Mean all transgenics” indicates mean of five replicates of 21 BAT1 transformants and five replicates of five Col-0 plants. C, Total fatty acid content as a proportion of seed weight. The graph is derived from the values used to generate A and B.
The number of mature, dry seeds corresponding to approximately 5 mg was determined and is expressed as per seed weight in Figure 6B. The mean weight for the nontransformed Col-0 seeds (n = 25) was 18.19 ± 0.51 μg per seed. When expressed as a percentage of the weight of the seeds from nontransformed Col-0 plants, individual BAT1.13 transformant lines varied between 98% and 111%. Thus, 10 of the 13 BAT1.13 transformant lines had seed weights that were on average 108% (mean 19.61 ± 0.39 μg; n = 5 for each transgenic line) greater than the control plant seed weights. Similarly, the BAT1.5 transformants had seed weights (mean 19.11 ± 0.44 μg) that varied between 102% and 108% of seeds of the nontransformed plants (Fig. 6B), and the seed weights were on average 7% greater for five of the BAT1.5 (mean 19.39 ± 0.25 μg) transformant lines. The mean seed weights of each of the BAT1.13 and BAT1.5 transformants determined with T3 and T4 seeds were not significantly different; therefore, the mean seed weight for all BAT1.13 and BAT1.5 transformants combined was 19.19 ± 0.63 μg, which was 6% greater than the mean of the nontransformed control seeds (t test, P < 0.05). Taken together, these results suggest that the expression of rapeseed microsomal LPAAT isoforms in Arabidopsis seeds is associated with an increase in TAG content and in seed weight.
DISCUSSION
Our objective in this work was to provide insight into the molecular environment in which PA is synthesized during seed development. Specifically, we wished to identify and evaluate the potential contribution of seed-expressed LPAAT isozymes to TAG biosynthesis in rapeseed. Such information is necessary to optimize the environment for enzymes encoded by transgenes in order to facilitate attempts to modify the fatty acid composition of plant oils. This study revealed the presence of transcripts encoding several microsomal LPAAT isozymes expressed in developing rapeseed that are potentially capable of contributing to TAG biosynthesis and revealed the additional complexity that underlies lipid biosynthesis in a polyploid background. Differences in the profile of expression of the homoeologous LPAAT genes in rapeseed tissues raise the possibility of a specialization of function between the isozymes. In contrast to transgenesis using other LPAATs of plant origin, the expression of each rapeseed LPAAT isozyme in Arabidopsis seeds was associated with an increase in seed mass and total fatty acid content.
Diversity of LPAAT Microsomal Isozymes in Rapeseed
The rapeseed cDNAs BAT1.12, BAT1.13, and BAT1.5 were highly homologous at the nucleotide and deduced protein levels to the L. douglasii LAT1 (Brown et al., 1995) and Arabidopsis LPAT2 (Kim et al., 2005) sequences, which were shown to encode ubiquitous microsomal LPAATs. The BAT1.13 and BAT1.12 cDNAs are independent gene products encoding a protein of identical amino acid composition that is distinct from a third LPAAT isoform (accession no. Q9XFW4). The cDNA BAT1.5 encodes a protein that is more divergent from the other three isoforms but nearly identical to the B. oleracea protein (accession no. AAT36638). The sequence heterogeneity among the four clones originates in the allotetraploid nature of the rapeseed genome and therefore the variation among BAT1.13, BAT1.12, and Q9XFW4, and these genes along with the BAT1.5 gene reflect allelic variation and polymorphism between the parental LPAAT genes present in each donor genome. The near identity with the B. oleracea LPAAT sequence confirms that BAT1.5 is the product of the C genome; consequently, the three other isoforms are likely to be the products of the A genome originating from Brassica rapa. The extent of nucleotide variation among the BAT1.13, BAT1.12, and Q9XFW4 cDNAs suggests that these are derived from genes that were either multiplied by triplication and originally present in the ancient progenitor genomes of the parental diploid genomes of rapeseed or possibly the result of subsequent and more recent gene rearrangements (Parkin et al., 2003). Thus, in contrast to the orthologous LPAAT in Arabidopsis encoded by the gene At3g57650, there exist a minimum of four microsomal LPAAT isoforms expressed in rapeseed that are likely to be associated with the Kennedy pathway. The apparent redundancy of cytoplasmic LPAATs in rapeseed may mask an alteration, or an acquisition of novel functions or modes of regulation, that may contribute to phenotypic diversity (Lukens et al., 2004). Furthermore, this diversity among rapeseed LPAATs may be an important aspect to be considered in understanding the partitioning of fatty acids between membrane and storage lipids in seeds and has implications for attempts to modify seed oil composition. We have shown that LPAAT activity is additive in developing rapeseed; therefore, the ability to selectively suppress a seed-specific isozyme with high specific activity and selectivity toward oleoyl-CoA presents a potential route to obtaining uniform oil composition without compromising membrane lipid synthesis.
The Homoeologous Genes BAT1.13 and BAT1.5 Encode Functional LPAAT Isozymes
We selected the BAT1.13 and BAT1.5 clones for functional characterization as representative of the extent of sequence variation among the cDNAs encoding microsomal LPAATs isolated from the rapeseed embryo library. The complementation of the bacterial LPAAT-deficient strain indicated that both BAT1.13 and BAT1.5 proteins possess LPAAT activity in vivo, and this was confirmed by enzymatic assays that further revealed a preference for oleoyl-CoA over palmitoyl-CoA as an acyl group donor in the conversion of LPA to PA. Preliminary selectivity assays using reciprocal combinations of labeled and unlabeled oleoyl- and palmitoyl-CoA substrates indicated that BAT1.13 is more strict in its preference for C18:1 acyl groups than is BAT1.5, which seems to be more relaxed with respect to C16:0 acyl groups (Maisonneuve, 2004). We did not detect activity with lysophospholipids other than LPA in membrane extracts of E. coli expressing the Bn-LPAATs BAT1 (plastidial), BAT1.13, and BAT1.5 in the presence of [14C]palmitoyl-CoA or [14C]oleoyl-CoA with either LPC or lysophosphatidylethanolamine (T.J. Roscoe and J.-J. Bessoule, unpublished data). This would suggest that while the Arabidopsis lysophosphatidylethanolamine acyltransferases encoded by At1g80950 and At2g45670 exhibit a degree of promiscuity with respect to their lysophospholipid preferences (Stålberg et al., 2009), the microsomal LPAATs represented by At3g57650, GUO45434 (BAT1.13), and GUO45435 (BAT1.5) are authentic LPAATs using only LPA acceptors. The sequence divergence among members of the Arabidopsis LPAAT-like gene family (Roscoe, 2005), represented by the AtLPAAT2 clade containing At3g57650 and the AtLPAAT5/AtLPAAT8 clade containing At1g80950 and At2g45670, reflects this selectivity for phospholipid acceptors, as does the AtLPAAT9 clade containing At1g78690, which contains an N-acetylphosphatidylethanolamine synthase (Faure et al., 2009).
The BAT1.13 and BAT1.5 proteins, therefore, possess the enzymatic characteristics typical of the microsomal enzyme involved in both membrane and storage lipid synthesis in most cultivated oilseeds (Frentzen, 1998), including rapeseed (Sun et al., 1988). Although each LPAAT isozyme exhibited a preference for oleoyl-CoA, BAT1.5 was characterized by a consistent 2- to 3-fold greater specific activity in comparison with BAT1.13 when measured in bacterial membrane extracts. The apparent difference in catalytic properties between the two isozymes should be determined by the relatively small number of variant residues between the two proteins; in this context, the charge variation at residue D75N between the proteins may be of significance, since it is in the proximity of the conserved motif NH(X4)D important for catalytic activity in glycerolipid acyltransferases (Heath and Rock, 1998) and for activity of the plastidial LPAAT (Maisonneuve et al., 2000). Since we show that both LPAAT isozymes are functionally active in vitro, it may be assumed that these proteins act additively in planta in the tissues where they are coexpressed; thus, each isozyme is capable of contributing to the PA pool destined for membrane and storage lipid synthesis in seeds.
Differential Expression of Rapeseed Microsomal LPAAT Genes
The genes encoding the two rapeseed microsomal LPAAT isozymes show strong differences in their profiles of expression in planta. The expression of BAT1.13 was ubiquitous in tissues of all organs examined, including seeds, suggesting that the BAT1.13 isoform is associated with the Kennedy pathway. This inference is confirmed by the close homology of protein sequences between BAT1.13 and the microsomal LPAAT encoded by the gene At3g57650 characterized by Kim et al. (2005) and deduced to be the unique LPAAT associated with the eukaryotic pathway of lipid synthesis in Arabidopsis. In contrast, the expression of BAT1.5, although detected in some tissues of root and leaf at certain stages, was predominant throughout seed development and, taken together with the higher rates of activity over the ubiquitous isozyme, albeit determined in vitro, indicated the potential for a significant contribution to storage lipid biosynthesis in rapeseed. In rapeseed, a polyploid oilseed selected for production of high quantities of TAG, it is conceivable that a specialization of overlapping function may exist between the LPAAT isozymes whereby the enhanced rate of catalysis and seed expression of the BAT1.5 enzyme serve to augment the rate of synthesis of storage lipids during seed development. Such genetic versatility may have arisen as a consequence of the gene duplication that occurred prior to or during formation of the allotetraploid rapeseed (Parkin et al., 2003). To avoid problems of altered gene dosage, polyploid cells suppress or reduce the expression of copies of certain genes, and this may occur via loss of function or by specialization of function mediated by partitioning tissue or cell specificity (Lynch and Force, 2000). Evidence for such subfunctionalization has been shown in Gossypium tetraploids, where a high frequency of homoeologous gene pairs, copies from each parental genome donor, show dissimilar levels of expression; furthermore, a large proportion of gene pairs show differential expression among organs (Adams et al., 2003). The mechanisms by which changes in gene expression occur are diverse and poorly understood (Chen and Ni, 2006) but probably initially occur via epigenetic mechanisms (Wolfe and Matzke, 1999). That a regulation of gene dosage of LPAATs is necessary and occurs is supported by microsynteny analysis of the duplicated chromosomal regions in Arabidopsis, which has revealed that four members of the LPAAT-like gene family, including the microsomal LPAAT encoded by At3g57650, are localized in chromosomal regions that were duplicated; however, in each case, one of the two duplicated copies has been eliminated (R. Guyot and T. Roscoe, unpublished data).
Enhanced Oil Content and Seed Weight by Overexpression of Microsomal LPAATs
To evaluate the relative contribution of seed-expressed LPAAT isozymes to TAG biosynthesis in rapeseed, we expressed the coding sequences of the BAT1.13 and BAT1.5 cDNAs in Arabidopsis seeds under the control of the napin 2S2 promoter (Guerche et al., 1990), which is expressed throughtout the embryo during seed maturation from the globular stage embryo. We anticipated that the use of a seed-specific promoter would circumvent potential problems related to perturbation of membrane lipid metabolism in vegetative tissues associated with altered dosage of LPAAT. The fatty acid composition of the TAG in seeds of the transgenic lines was not significantly different from that of the nontransformed control plants, which is consistent with the substrate preference of the two rapeseed LPAATs and that of the endogenous Arabidopsis enzyme (Kim et al., 2005). The average seed weight and the accumulation of seed storage lipids were enhanced by the expression of either of the rapeseed LPAAT isozymes in the majority of the transgenic lines examined (Fig. 6). Since the substrate preference of each isozyme is similar, the results for each of the individual BAT1.13 and BAT1.5 lines may be summarized together, showing that the overexpression of a microsomal LPAAT results in an increase of 6% in the average seed weight and a mean increase of 13% in total fatty acid content over the nontransformed control seeds.
The enhancement of seed TAG content by a microsomal LPAAT homologous to the endogenous enzyme is remarkable because such an effect has been observed only when the yeast SLC1-1 gene, encoding a variant LPAAT (Nagiec et al., 1993), was used to transform rapeseed and Arabidopsis (Zou et al., 1997). In this latter study, the variant yeast LPAAT, because of its broad substrate preference, resulted in the production of TAG that was greatly enriched in VLCFAs at sn-2, the position from which these unusual acyl groups are excluded in crucifers. Unexpectedly, expression of the SLC1-1 also led to substantial increases in seed oil content that were accompanied by increases in seed weight. In contrast, although the expression of a Limnanthes LPAAT in rapeseed under the control of a napin promoter resulted in the incorporation of VLCFAs into the TAG at sn-2 (Lassner et al., 1995), there was no increase in seed oil content observed. In the case of SLC1-1, the substantial increases in TAG content that were not accounted for by the increases in VLCFA indicated that the expression of the yeast LPAAT had increased the flux of fatty acids through the entire Kennedy pathway to TAG. The authors suggested, therefore, that the synthesis of PA catalyzed by LPAAT may constitute a regulatory step in the synthesis of TAG in oilseeds and that the positive effect on oil is a consequence of a less stringent regulation of the yeast enzyme, thereby allowing the creation of a new sink for lipid deposition. Alternatively, it was proposed that the relaxed acyl acceptor selectivity of the enzyme may be the decisive factor (Frentzen, 1998) that may allow scavenging of phospholipid intermediates toward TAG.
The positive effect of the rapeseed LPAAT isozymes on the accumulation of storage lipids in Arabidopsis seeds documented in this work is similar to the effect of the yeast LPAAT but cannot be explained in terms of altered regulation or substrate preference as proposed for SLC1-1. Our results are consistent with the idea that flux through the entire pathway from fatty acid synthesis to utilization in TAG was increased by the expression of the rapeseed LPAAT in addition to that of the endogenous enzyme. These findings support the idea that conversion of LPA to PA is a potentially rate-limiting step in the provision of intermediates for membrane or TAG synthesis but that whereas membrane homeostasis is likely to be strongly regulated (Shank et al., 2001; Wallis and Browse, 2002), the increased pool of intermediates favors channeling into a more neutral and flexible TAG sink. Alignment of the yeast SLC1-1 LPAAT reveals a significant degree of homology with both the ubiquitous (class A) LPAATs (Fig. 1) and to a lesser extent the plant seed-specific (class B) LPAATs, but the latter proteins are not homologous to the ubiquitous plant LPAATs. Predictions of transmembrane-spanning domains (TOPRED; Expasy) suggest that the yeast protein may be less constrained by membrane topology than proteins belonging to either of the two classes of plant LPAATs. That the Limnanthes and coconut LPAATs do not stimulate oil accumulation may reflect the extent of sequence divergence between these specialized prokaryote-like enzymes and the ubiquitous plant enzyme that may prevent efficient interactions with other protein components of the Kennedy pathway.
In the case of BAT1.13- and BAT1.5-overexpressing lines, there was an overall positive correlation between the TAG content and average seed weight (r2 = 0.49), and certain lines possessing higher TAG contents (BAT1.13S6527 and BAT1.5S251) also had higher seed weights, although other lines with low lipid contents (BAT1.5SH36) had high seed weights. The expression of total fatty acid content on a seed weight basis reveals that half of the transgenic lines examined possess a significantly greater proportion of seed oil, in a range of approximately 35.6% to 38.9% dry seed weight (mean 37.2% ± 1.29%), representing an average of 11% greater seed oil content compared with Col-0 (Fig. 6C). Thus, in some lines, the greater seed weight may be in part attributable to a greater TAG content; however, other seed components including oleosins could also contribute to the increase in mass, whereas in other lines, the increase in TAG content may be at the expense of other seed constituents such as storage proteins or dry matter content. Overall, the total fatty acid content and the relative level of BAT1.13 and BAT1.5 expression was poorly correlated, although a degree of correlation exists (r2 = 0.24) between low levels of LPAAT transgene expression in lines that differ nonsignificantly in total fatty acid content from the nontransformed control. The correlation between the transformant lines characterized by a significantly greater fatty acid content than the controls and the expression of the transgenes is weak (r2 = 0.16), possibly indicating a minimal threshold of gene expression to influence lipid synthesis (25%–30% RPL2; Fig. 5B). This lack of a strong correlation between the quantitative composition of seeds and the expression of lipid-related transgenes or transgene enzymatic activities (Zou et al., 1997; Jako et al., 2001; Xu et al., 2008) is not surprising, since variation in additional posttranscriptional parameters may influence the relative contribution to seed metabolism; our primary objective was to verify the expression of the transgene in each of the transformation events. In our initial studies, we produced 16 transgenic lines expressing the BAT1.13 and BAT1.5 cDNAs in an antisense orientation under the control of the napin promoter that were characterized by substantially reduced seed total fatty acid contents compared with the Col-0 controls. These observations, taken together with the data in Figure 5, indicate a broad correlation between LPAAT expression levels and oil content.
The enhancement of storage lipid accumulation associated with the expression of the yeast (Zou et al., 1997) and the Brassica LPAAT isozymes described in this work resembles the increase of seed oil content and seed weight obtained by the overexpression of DAGAT reported by Jako et al. (2001). These authors argue that flexibility exists in TAG deposition in developing seeds, which may be related to mechanisms that signal the sink size; for example, enhanced acyltransferase activity may deplete the acyl-CoA pool, signaling a need to increase fatty acid synthesis. The inference that altering the expression of glycerolipid acyltransferases in seeds will modify the flux through the entire pathway to TAG synthesis is further supported by the report that overexpression of either native or endomembrane-targeted, plastidial, and bacterial GPATs also seems to enhance seed oil content (Jain et al., 2000). Thus, control of TAG synthesis in seeds may be shared among the three acyltransferases, consistent with the recent finding that genes encoding Kennedy pathway enzymes are up-regulated in seeds possessing enhanced TAG content that express a DAGAT transgene (Sharma et al., 2008). Taken together, increasing the quantity of any of the Kennedy pathway acyltransferases results in elevated TAG content of seeds consistent with an augmentation of the pool of LPA, PA, and DAG intermediates and a feed-forward enhancement of storage lipid sink size. Apparently, the supply of precursor fatty acyl groups and glycerol-3-phosphate responds via feedback signaling to meet this increased demand. The modest increases in TAG content attained by increasing the supply of fatty acids, for example, via the engineering of an increased expression of acetyl-CoA carboxylase in the plastid (Roesler et al., 1997), compared with the substantial augmentation of TAG accumulation achieved by overexpression of Kennedy pathway acyltransferases may indicate that fatty acid utilization represents the more important limitation on storage lipid accumulation, at least in crucifer species. This inference is supported by the studies on metabolic control analysis conducted in rapeseed, which suggested that a greater level of control was exerted at the level of TAG assembly than at fatty acid synthesis (Ramli et al., 2002). In this work, we have not addressed the question of yield; although we observed that the greater TAG content did not affect the number of seeds per silique, we did not attempt to determine the number of siliques per plant. However, an increased TAG content resulting from overexpression of the Arabidopsis DAGAT did not negatively affect the number of seeds per plant (Jako et al., 2001).
In conclusion, we have catalogued previously undescribed diversity in microsomal LPAAT isozymes of rapeseed and identified the BAT1.5 isozyme as being of particular interest in TAG synthesis due to its greater rate of activity and because of its preferential expression in developing seeds. We have shown that the overexpression of genes encoding the class A-type microsomal LPAATs, like the yeast SLC1-1 gene, leads to an increase in seed weight and a greater accumulation of lipid in seeds, characteristics that were not conferred by transgenes encoding plant prokaryote-like LPAATs.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana Col-0) nontransformed control plants and transformed plants were grown simultaneously in a sterilized soil/vermiculite mix in a growth chamber under continuous illumination of 100 μE m−2 s−1 provided by a mix of tungsten and photosynthetically enhanced fluorescent lighting at 22°C and controlled humidity. The plants were moved to new positions each day in order to minimize any effects of environmental variations in growth room conditions. Vernalized rapeseed (Brassica napus ‘Gaspard’) seedlings were grown in a greenhouse under natural lighting in springtime, and siliques were harvested at 1-week intervals after flowering.
Isolation of Rapeseed cDNAs
A cDNA library derived from immature rapeseed embryos (Barret et al., 1998) was initially screened using an oligonucleotide (5′-AATCATCGGAGTGATATTGATTGG-3′) derived from the maize (Zea mays) sequence (accession no. Z29518) labeled with fluorescein at the 3′ end (GC Healthcare). Library screening and recovery of recombinant pBluescript SK+ plasmids by in vivo excision were carried out according to the protocols of the supplier (Stratagene). A rapeseed partial cDNA was isolated that shared 64% identity with the maize sequence and was subsequently radiolabeled and used to rescreen the embryo library. The sequences of the inserts of the positive clones were determined using DyeDeoxy Terminator cycle sequencing (Applied Biosystems) on double-stranded DNA templates with an ABI 377A sequencer. The cDNA sequences were identified by comparison with the NCBI database, and deduced proteins were aligned using ClustalW.
Characterization of Rapeseed LPAATs by Expression in Escherichia coli
The thermosensitive strain E. coli JC201 was used in the complementation experiments (Coleman, 1990). The rapeseed cDNAs were cloned in the reading frame for expression as a β-galactosidase fusion protein. JC201 was transformed via heat shock and was selected on ampicillin plates containing 1 mm isopropylthio-β-galactoside (IPTG) at 38°C. Complementing colonies were inoculated into starter cultures and grown to an optical density at 600 nm of 0.5 at 30°C. Aliquots were grown in the presence of IPTG at 30°C, 37°C, and 42°C, and growth curves were constructed using data obtained from three individual complementation experiments.
The 1-acyl-glycerol-3-phosphate acyltransferase activity of E. coli membrane fractions expressing the rapeseed LPAATs was determined as described by Bourgis et al. (1999). JC201 cells were transformed with the coding sequences corresponding to the BAT1.13 or BAT1.5 cDNA cloned into the pBluescript SK+ vector (Stratagene). Membranes were isolated from cells that had been grown at 30°C for 3 h after induction by 1 mm IPTG. The standard assay comprised 50 mm Tris-HCl, pH 8, 1 mm MgCl2, the presence or absence of 55 μm oleoyl-LPA, 10 μm [1-14C]acyl CoA, and 50 μg of membrane fraction and was incubated for 15 min at 30°C. The products of the reaction were separated by thin-layer chromatography, and the quantity of radioactivity in the PA band was integrated using a phosphor imager.
RNA Isolation and RT-PCR
Total RNA was extracted from rapeseed tissues and embryos dissected from seeds by the protocol described by Kay et al. (1987). RT-PCR was performed using the Pro-Star kit (Stratagene) using 100 ng of DNase-treated RNA as template. Primers (Eurogentec) were derived from the BAT1.13 and BAT1.5 cDNA sequences and were designed to detect gene-specific transcripts: BAT1.131 (5′-CACAAGAGTACGCTGCCTCCTCTGAG-3′), BAT1.132 (5′-GCAAGAACTTCATTGCTCCAAGAAT-3′), BAT1.51 (5′-ACAAGAGTACGCAGCCTCCTCTCAG-3′), and BAT1.52 (5′-GTAAGAACTTCATTGCTCCAAGAGT-3′). RNA loading was controlled by PCR amplification of a fragment of the RPL2 gene using the primers RPL25 (5′-GGTGATCGTGGTGTCCTCGCTAGAGC-3′) and RPL23 (5′-GTCTGCCTTGGCAGCTGAAGCAGC-3′).
LPAAT Promoter-Reporter Constructs
The promoter regions corresponding to the sequence upstream of the predicted translation start sites of BAT1.13 and BAT1.5 were isolated from rapeseed genomic DNA by a PCR walking technique (Devic et al., 1997). Two successive walks from the coding sequence into the promoter region for BAT1.5 were performed using the primer pairs BAT153 (5′-GGATTTTGAGCTTAGAATCATATGTGC-3′) and BAT154 (5′-CAGCAACGGAATCTAAGAGCAGAAGAGGG-3′), followed by BAT155 (5′-CCCCTTATATCTTATCGTTGATCAGATCGACCC-3′) and BAT156 (5′-CACACAGCTAGAACTGATCTCTTCTACCCACG3′). A single PCR walk was performed for BAT1.13 using the primer pair TJR-BAT1131 (5′-GTTCTTTGCCCATTCGATTGAAGGTTTCATT-3′) and TJR-BAT1132 (5′-TAGACAGAGGTCGAATAAGAACATAACAAAT-3′).
After cloning and sequencing of the PCR walk fragments, primers were then designed to amplify a 903-bp sequence upstream of the initiation codon of BAT1.13 from rapeseed genomic DNA using the primers BP113X (5′-TACCGCTCGAGCGAAAGACTTAACAGTTAACATGCATTCACG-3′) containing a XhoI restriction site and BP113N (5′-CCAACGCCATGGAATCCAACCAACAC-3′) containing a NcoI site. Similarly, a 1.096-kb sequence upstream of the initiation codon of the BAT1.5 sequence was amplified using the primers BP15X (5′-TACCGCTCGAGCGTATTTTCTTAATCCAAGTGCAAAAAGC-3′) and BP15N (5′-CCATCGCCATGGAACCTAAGGAATCC-3′). The promoter fragments were ligated into the vector pRTL2 (Restrepo et al., 1990) containing the uidA (GUS) gene after digestion with XhoI and NcoI. A cassette containing each LPAAT promoter fused to the GUS gene was recovered by digestion with PstI and XbaI and finally cloned into the pBIB-Hygro binary vector (Becker, 1990). The authenticity of all constructs was verified by sequencing prior to transformation of Agrobacterium tumefaciens.
Construction of LPAAT Expression Vectors for Plant Transformation
Binary plant transformation vectors that contained the cDNAs corresponding to the coding sequences of BAT1.13 and BAT1.5 in the sense orientation under the control of a seed-specific promoter were constructed. PCR was performed to introduce XhoI restriction sites at each extremity of the coding sequence using the cloned BAT1.5 and BAT1.13 cDNAs as templates. The primer pairs used were BAT1135E (5′-GTACGCTCGAGCATGGCGATGGCAGCAGCTGTG-3′), BAT1133E (5′-CTAGGCTCGAGTTTACTTCTGCTTCTCTACTTCTG-3′), BAT155E (5′-CTACGCTCGAGCATGGCGCATGGCAGCAGCAGCAGTG-3′), and BAT153E (5′-CTAGGCTCGAGTTTACTTCTGCTTCTCCTCCACTTC-3′). The amplimers were ligated into a XhoI-digested pBSK vector, subsequently released by XhoI digestion, and purified. The coding sequences of BAT1.13 and BAT1.5 were then ligated in the sense orientation into a XhoI-digested pMM5 vector containing the At2S2 promoter (Guerche et al., 1990). The cassette containing the At2S2 promoter, the LPAAT sequence, and the nos terminator was released by NotI digestion and ligated into NotI-digested pEC2 (Guerche et al., 1990), a vector containing a kanamycin resistance gene for bacterial selection and the Basta (glufosinate) resistance gene for plant selection and used to transform A. tumefaciens.
Transformation of A. tumefaciens and Arabidopsis
Competent A. tumefaciens cells (strain C58C1) were transformed with the pBIB-Hygro or pEC constuct via heat shock at 37°C for 5 min. The transformed cells were plated on Luria-Bertani agar containing the appropriate antibiotics and grown at 28°C. Arabidopsis plants were transformed by floral dip (Clough and Bent, 1998) followed by vacuum infiltration (Bechtold et al., 1993).
Selection of Transformed Arabidopsis Plants
The seeds were selected on plates containing Murashige and Skoog germination medium supplemented with 30 mg mL−1 hygromycin for the LPAAT promoter∷reporter lines or 100 mg mL−1 Basta (Dr. Ehrenstorfer GmbH) for the LPAAT sense lines. After 10 d, green-leafed, antibiotic-resistant, or herbicide-resistant seedlings were scored and/or were transferred to soil. The presence of the LPAAT transgene was confirmed by PCR with genomic DNA isolated from leaves of the T1 seedlings using the primers GUS1 (5′-CACGGGTTGGGGTTTCTA-3′) with BAT1.131 (5′-CACAAGAGTACGCTGCCTCCTCTGAG-3′) or BAT1.51 (5′-ACAAGAGTACGCAGCCTCCTCTCAG-3′) for sense constructs. Segregation analyses were performed on the T2 seedlings grown in the presence of Basta to determine the number of T-DNA insertion loci. Plants homozygous for the transgene were identified by screening T3 seeds for 100% resistance to Basta.
Determination of LPAAT Transgene Copy Number
Genomic DNA was isolated from 0.5 g of young leaf tissue of transformed lines using the cetyl-trimethyl-ammonium bromide protocol of Doyle and Doyle (1990). The DNA was digested with EcoRI and electrophoresed on a 0.8% agarose gel. The DNA fragments were transferred to a nylon membrane (Hybond N+; GE Healthcare), and a BAT1.13 DNA fragment was amplified using primers BAT1.131 and BAT1.132 and radiolabeled with [32P]dCTP via random priming. The probe was hybridized, and the membrane was washed to high stringency and exposed using a phosphor imager, allowing the discrimination of single or multiple LPAAT copies
GUS Histochemistry
Seedlings and various plant organs of the LPAAT promoter∷reporter transformed lines were incubated in a solution of 100 mm potassium phosphate, pH 7, 10 mm EDTA, 0.1% Triton X-100, 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide for 16 h at 37°C. The tissues were decolorized with 70% ethanol. Tissues were observed and photographed with a Zeiss Axioplan microscope. Seeds were treated and examined in Hoyer's solution.
Quantitation of BAT1.13 Expression in Transgenic Seeds
Total RNA was purified from mature Arabidopsis seeds at 15 to 18 d after fertilization using the Oligotex mRNA kit (Qiagen), 2.5 μg of RNA was reverse transcribed using the first-strand synthesis kit (Stratagene), and the resulting cDNA was used as a template for PCR with primers BAT1.131 and BAT1.132 designed to discriminate the BAT1.13 transcript from that of the endogenous Arabidopsis gene. PCR was limited to 25 cycles at 51°C. The resulting reaction mix was migrated on agarose gels and transferred to a Hybond N+ membrane. The membrane was subsequently hybridized with BAT1.13 and RPL2 32P-labeled probes and washed to high stringency, and the radioactive signals were integrated using a phosphor imager after 1- and 3-h exposures.
Determination of Seed Weight
The number of seeds (cleaned of debris and counted twice with a dissecting microscope) present in two independent aliquots of approximately 5 mg of seed was determined. Each aliquot of seed was taken from a mix of equal proportions of four independent plants grown under the same conditions in order to minimize plant and environmental variations. The seeds were dried in open tubes in a desiccator for 24 h prior to weighing and counting (Li et al., 2006).
Lipid Extraction and Quantitative Fatty Acid Analysis
Four aliquots of 50 Arabidopsis seeds of each line obtained as described above were subjected to direct transmethylation by the addition of 1 mL of methanol containing 2.5% H2SO4 (v/v) and heated at 90°C for 90 min essentially as described by Li et al. (2006), except that a heptadecanoic acid internal standard at a final concentration of 5 ng μL−1 was included for quantification. After cooling, 1.5 mL of deionized water and 0.75 mL of hexane were added. Fatty acid methyl esters were extracted into the hexane phase by vigorous shaking followed by centrifugation at 1,500g for 5 min. A 1-μL sample was taken from the organic phase for gas chromatography (Hewlett-Packard 5890 series II) on a 15-m × 0.53-mm Carbowax column (Altech) with flame ionization detection and integration. The oven temperature was programmed for 1 min at 160°C, followed by a 20°C min−1 ramp to 190°C and a second ramp of 5°C min−1 to 210°C, and maintained at this temperature for a further 6 min. The retention times of fatty acid methyl esters were determined by comparison with standards, and they were quantified using the heptadecanoic acid methyl ester standard.
The nucleotide sequences corresponding to the BAT1.13, BAT1.5, and BAT1.12 cDNAs have been entered in the GenBank database under the accession numbers GUO45434, GUO45435, and GUO45436, respectively.
Acknowledgments
We thank Martine Devic for critical and incisive comments on this paper. T.J.R. thanks Jack Coleman for the gift of JC201. Michele Laudie and Cristel Berger are thanked for DNA sequencing. The support of Etienne Poirat and Françoise Labelette in project administration is gratefully acknowledged.
This work was supported by the French Agence de l'Environnement et de la Maitrise de l'Energie and the Organisation Nationale Interprofessionelle des Oléagineux (grant no. BOU0049).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Thomas J. Roscoe (thomas.roscoe@ird.fr).
References
- Adams KL, Cronn R, Percifeld R, Wendel JF (2003) Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ specific-reciprocal silencing. Proc Natl Acad Sci USA 100 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafor M, Jonsson L, Stobart AK, Stymne S (1990) Regulation of triacylglycerol biosynthesis in embryos and microsomal preparations from the developing seeds of Cuphea lanceolata. Biochem J 272 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret P, Delourme R, Renard M, Domergue F, Lessire R, Delseny M, Roscoe TJ (1998) A rapeseed FAE1 gene is linked to the E1 locus associated with variation in the content of erucic acid. Theor Appl Genet 96 177–186 [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris 316 1194–1199 [Google Scholar]
- Becker D (1990) Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res 18 203–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Kader JC, Barret P, Renard M, Robinson D, Robinson C, Delseny M, Roscoe TJ (1999) A plastidial lysophosphatidic acid acyltransferase from oilseed rape. Plant Physiol 120 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AP, Brough CL, Kroon JTM, Slabas AR (1995) Identification of a cDNA that encodes a 1-acyl-sn-glycerol-3-phosphate acyltransferase from Limnanthes douglasii. Plant Mol Biol 29 267–278 [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Ni Z (2006) Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays 28 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coleman J (1990) Characterisation of Escherichia coli cells deficient in 1-acyl sn-glycerol-3-phosphate acyltransferase activity. J Biol Chem 265 17215–17221 [PubMed] [Google Scholar]
- Devic M, Albert S, Delseny M, Roscoe TJ (1997) Efficient PCR walking on plant genomic DNA. Plant Physiol Biochem 35 331–339 [Google Scholar]
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissues. Focus 12 13–15 [Google Scholar]
- Drexler H, Spiekermann P, Meyer A, Domergue F, Zank T, Sperling P, Abbadi A, Heinz E (2003) Metabolic engineering of fatty acids for breeding of new oilseed crops: strategies, problems and first results. J Plant Physiol 160 779–802 [DOI] [PubMed] [Google Scholar]
- Eccleston V, Ohlrogge JB (1998) Expression of lauroyl-ACP carrier protein thioesterase in Brassica napus seeds induces pathways for both fatty acid oxidation and biosynthesis and implies a set point for triacylglycerol accumulation. Plant Cell 10 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure L, Coulon D, Laroche-Traineau J, Le Guedard M, Schmitter JM, Testet E, Lessire R, Bessoule JJ (2009) Discovery and characterization of an Arabidopsis thaliana N-acylphosphatidylethanolamine synthase. J Biol Chem 284 18734–18741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzen M (1998) Acyltransferases from basic science to modified seed oils. Fett/Lipid 100: 161–166 [Google Scholar]
- Guerche P, Tire C, De Sa FG, De Clercq A, Van Montagu M, Krebbers E (1990) Differential expression of the Arabidopsis 2S albumin genes and the effects of increasing gene family size. Plant Cell 2 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke C, Peterek G, Wolter FP, Frentzen M (1995) A plant acyltransferase involved in triacylglycerol biosynthesis complements an Escherichia coli sn-1 acyl-glycerol-3-phosphate acyltransferase mutant. Eur J Biochem 232 806–810 [PubMed] [Google Scholar]
- Heath RJ, Rock CO (1998) A conserved histidine is essential for glycerolipid acyltransferase catalysis. J Bacteriol 180 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, Coffey K, Kumar A, MacKensie SL (2000) Enhancement of seed oil content by expression of glycerol-3-phosphate acyltransferase genes. Biochem Soc Trans 28 958–961 [PubMed] [Google Scholar]
- Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Cahoon EB (2003) Industrial oils from transgenic plants. Curr Opin Plant Biol 6 178–184 [DOI] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236 1299–1302 [DOI] [PubMed] [Google Scholar]
- Kim UK, Li Y, Huang AHC (2005) Ubiquitous and endoplasmic reticulum located lysophosphatidic acid acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17 1073–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutzon DS, Hayes TR, Wyrick A, Xiong H, Davies HM, Voelker TA (1999) Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric acid rapeseed oil and can increase total laurate levels. Plant Physiol 120 739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutzon DS, Lardizabal KD, Nelsen JS, Bleibaum JL, Davis HM, Metz JG (1995) Cloning of a coconut endosperm cDNA encoding a 1-acyl-sn-1-acyl glycerol-3-phosphate acyltransferase that accepts medium chain length substrates. Plant Physiol 109 999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardizabal K, Effertz R, Levering C, Mai J, Pedroso MC, Jury T, Aasen E, Gruys K, Bennett K (2008) Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol 148 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassner MW, Levering CK, Davies HM, Knutzon DS (1995) Lysophosphatidic acid acyltransferase from meadowfoam mediates insertion of erucic acid at the sn-2 position of triacylglycerol in transgenic rapeseed oil. Plant Physiol 109 1389–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to plant variation. Phytochemistry 67 904–915 [DOI] [PubMed] [Google Scholar]
- Lukens LW, Quijada PA, Udall J, Pires JC, Schranz ME, Osborn TC (2004) Genome redundancy and plasticity within ancient and recent Brassica crop species. Biol J Linn Soc Lond 82 665–674 [Google Scholar]
- Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve S (2004) Contribution à l′étude des 1-acyl glycerol-3-phosphate acyltransférases microsomales de Brassica napus. PhD thesis. Université de Perpignan, Perpignan, France
- Maisonneuve S, Bessoule JJ, Lessire R, Delseny M, Roscoe TJ (2000) Mutagenesis of a plastidial lysophosphatidic acid acyltransferase. Biochem Soc Trans 28 961–969 [PubMed] [Google Scholar]
- Mekhedov S, Cahoon EB, Ohlrogge J (2001) An unusual seed-specific 3 ketoacyl-ACP synthase associated with the biosynthesis of petroselinic acid in coriander. Plant Mol Biol 47 507–518 [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Wells GB, Lester RL, Dickson RC (1993) A suppressor gene that enables Saccharomyces cerevisiae to grow without making phospholipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem 268 22156–22163 [PubMed] [Google Scholar]
- Parkin IA, Sharpe AG, Lydiate DJ (2003) Patterns of genome duplication within the Brassica napus genome. Genome 46 291–303 [DOI] [PubMed] [Google Scholar]
- Ramli US, Baker DS, Quant PA, Harwood JL (2002) Control analysis of lipid biosynthesis in tissue cultures from oil crops shows that flux control is shared between fatty acid synthesis and lipid assembly. Biochem J 364 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of potyviral proteins. Plant Cell 2 967–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler K, Shintani D, Savage L, Bodupalli S, Ohlrogge J (1997) Targetting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol 113 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe TJ (2005) Identification of acyltransferases controlling triacylglycerol biosynthesis in oilseeds using a genomics-based approach. Eur J Lipid Sci Technol 107 256–262 [Google Scholar]
- Shank KJ, Su P, Brglez I, Boss WF, Dewey RE, Boston RS (2001) Induction of lipid metabolic enzymes during the endoplasmic reticulum stress response in plants. Plant Physiol 126 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Anderson M, Kumar A, Zhang Y, Giblin EM, Abrams SR, Zaharia LI, Taylor DC, Fobert PR (2008) Transgenic increases in seed oil content are associated with the differential expression of novel Brassica-specific transcripts. BMC Genomics 9 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Browse J, Jaworski JG, Ohlrogge JB (2000) Lipids. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 456–527
- Stålberg K, Ståhl U, Stymne S, Ohlrogge J (2009) Characterization of two Arabidopsis thaliana acyltransferases with preference for lysophosphatidylethanolamine. BMC Plant Biol 9 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Cao YZ, Huang AHC (1988) Acyl coenzyme A preference of the glycerol phosphate pathway in the microsomes from maturing seeds of palm, maize and rapeseed. Plant Physiol 94 1194–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4 12–21 [DOI] [PubMed] [Google Scholar]
- Wallis JG, Browse J (2002) Mutants of Arabidopsis reveal many roles for membrane lipids. Prog Lipid Res 41 254–278 [DOI] [PubMed] [Google Scholar]
- Wiberg E, Tillberg E, Stymne S (1994) Substrates of diacylglycerol acyltransferase in microsomes from developing oil seeds. Phytochemistry 36 573–577 [Google Scholar]
- Wolfe AP, Matzke MA (1999) Epigenetics: regulation through repression. Science 286 481–486 [DOI] [PubMed] [Google Scholar]
- Xu J, Francis T, Mietkiewska E, Giblin EM, Barton DL, Zhang Y, Zhang M, Taylor DC (2008) Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol J 6 799–818 [DOI] [PubMed] [Google Scholar]
- Zheng P, Allen WB, Roesler K, Williams ME, Zhang S, Li J, Glassman K, Ranch J, Nubel D, Solawetz W, et al (2008) A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet 40 367–372 [DOI] [PubMed] [Google Scholar]
- Zou J, Katavic V, Giblin EM, Barton DL, MacKenzie SL, Keller WA, Hu X, Taylor DC (1997) Modification of seed oil content and acyl composition in the Brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 9 909–923 [DOI] [PMC free article] [PubMed] [Google Scholar]