Abstract
Lipoxygenases (LOXs) are crucial for lipid peroxidation processes during plant defense responses to pathogen infection. A pepper (Capsicum annuum) 9-LOX gene, CaLOX1, which encodes a 9-specific lipoxygenase, was isolated from pepper leaves. Recombinant CaLOX1 protein expressed in Escherichia coli catalyzed the hydroperoxidation of linoleic acid, with a Km value of 113. 9 μm. Expression of CaLOX1 was differentially induced in pepper leaves not only during Xanthomonas campestris pv vesicatoria (Xcv) infection but also after exposure to abiotic elicitors. Transient expression of CaLOX1 in pepper leaves induced the cell death phenotype and defense responses. CaLOX1-silenced pepper plants were more susceptible to Xcv and Colletotrichum coccodes infection, which was accompanied by reduced expression of defense-related genes, lowered lipid peroxidation, as well as decreased reactive oxygen species and lowered salicylic acid accumulation. Infection with Xcv, especially in an incompatible interaction, rapidly stimulated LOX activity in unsilenced, but not CaLOX1-silenced, pepper leaves. Furthermore, overexpression of CaLOX1 in Arabidopsis (Arabidopsis thaliana) conferred enhanced resistance to Pseudomonas syringae pv tomato, Hyaloperonospora arabidopsidis, and Alternaria brassicicola. In contrast, mutation of the Arabidopsis CaLOX1 ortholog AtLOX1 significantly increased susceptibility to these three pathogens. Together, these results suggest that CaLOX1 and AtLOX1 positively regulate defense and cell death responses to microbial pathogens.
To effectively combat invasion by microbial pathogens, plants activate distinct defense responses that are specifically effective. Despite the presence of plant immune systems, many pathogens can evade or suppress host defense mechanisms. Lipoxygenase (LOX) pathways are crucial for lipid peroxidation processes during plant defense responses to pathogen infection (Casey and Hughes, 2004). Plant LOXs are key enzymes involved in the generation of fatty acid derivatives in oxylipin metabolism.
LOXs comprise a family of non-heme-iron-containing fatty acid dioxygenases, which are ubiquitous in plants and animals (Brash, 1999). LOXs catalyze the conversion of polyunsaturated fatty acids such as linoleic acid into hydroperoxides that are in turn converted to oxylipins. These primary products, which may cause oxidative damage to plant membranes during the hypersensitive response (HR; Slusarenko, 1996), are enzymatically metabolized into traumatin, jasmonic acid (JA), and methyl jasmonate (MeJA). These latter compounds are involved in diverse physiological functions in plant growth and development, senescence, and stress responses. Plant LOXs can be classified as 9-LOXs or 13-LOXs according to the position at which oxygen is incorporated into linoleic acid or linolenic acid, the most important substrates for LOX catalysis in plants (Feussner and Wasternack, 2002). LOX enzymatic activity initiates the different biosynthetic pathways that result in the accumulation of distinct oxylipins. The most understood functional aspects of oxylipin pathways have come mainly from studies of JA produced through the action of 13-LOXs but not 9-LOXs. The metabolism of 13-LOX has been described in tobacco (Nicotiana tabacum) leaves infected by an avirulent strain of Pseudomonas syringae pv phaseolicola (Kenton et al., 1999). During bacterial infection, JA accumulates in tobacco leaves prior to cell death (Kenton et al., 1999). The level of LOX activity and gene expression also increases in tobacco plants during infection with Phytophthora parasitica var nicotianae (Christophe et al., 1996; Rancé et al., 1998). However, the defense-related functions of 9-LOXs are not fully understood. Both 9-LOXs and oxidative processes are proposed to be involved in the HR of tobacco induced by the avirulent pathogen Pseudomonas syringae pv syringae (Montillet et al., 2005). The production of free fatty acid hydroperoxides via the 9-LOX pathway in tobacco is crucial for hypersensitive cell death induced by cryptogein, a purified protein from Phytophthora cryptogea (Rusterucci et al., 1999). The function of LOXs in defense against pathogens is likely to be related to the synthesis of fatty acid hydroperoxides and of volatile products with signaling functions (Rusterucci et al., 1999) and antimicrobial activity (Croft et al., 1993; Weber et al., 1999). Gao et al. (2007) recently suggested that oxylipin metabolism mediated by a specific 9-LOX, ZmLOX3, may be involved in fungal pathogenesis in maize (Zea mays). ZmLOX3 loss-of-function mutants are susceptible to Aspergillus flavus and Aspergillus nidulans infection (Gao et al., 2009).
LOX activity may initiate the synthesis of signal molecules or induce structural and metabolic changes in the cell, ultimately leading to cell death that has been termed the HR (Maccarrone et al., 2001). Plant cell death occurs during various phases of development, senescence, and responses to abiotic and biotic stresses, and in particular, in response to pathogen invasion (Morel and Dangl, 1997). Activation of LOXs in plants may be involved in cell death induced by pathogens (Buonaurio and Servili, 1999; Rusterucci et al., 1999). The induction of HR-like cell death by the activation of the 9-LOX-encoding gene GhLOX1 was shown in cotton (Gossypium hirsutum) plants during Xanthomonas campestris pv malvacearum infection (Marmey et al., 2007). LOX activity increases in parallel with the induction of HR symptoms in tobacco; however, in compatible interactions, LOX activity is delayed and reaches much lower levels (Montillet et al., 2002). In cotton, high LOX activity supports cell death during X. campestris pv malvacearum infection (Sayegh-Alhamdia et al., 2008). The HR, an important defense reaction of plants to pathogen infection, is accompanied by lipid peroxidation processes. In particular, 9-LOX-dependent lipid peroxidation operates during cryptogein-induced HR in tobacco leaves (Rusterucci et al., 1999). In potato (Solanum tuberosum), lipid peroxidation occurs as a controlled and directed process that is facilitated by the action of a specific 9-LOX during the HR (Göbel et al., 2003; Montillet et al., 2005). GhLOX1 is associated with salicylic acid (SA) accumulation during the HR of cotton to X. campestris pv malvacearum (Marmey et al., 2007).
The bacterial plant pathogen Xanthomonas campestris pv vesicatoria (Xcv) is the causative agent of bacterial spot disease on pepper (Capsicum annuum) and tomato (Solanum lycopersicum) plants. To identify genes involved in the HR-based innate immune response in pepper, we have isolated and functionally characterized defense-related genes encoding PR1 (for pathogenesis-related protein 1; Kim and Hwang, 2000; Hong and Hwang, 2005), chitinase (Hong et al., 2000), chitin-binding protein (Lee et al., 2001), thionin (Lee et al., 2000), SAR 8.2 (Lee and Hwang, 2003), peroxidase (Choi et al., 2007), and menthone reductase (Choi et al., 2008) from pepper leaves infected with the Xcv avirulent strain Bv5-4a. In this study, we used a cDNA macroarray method (Jung and Hwang, 2000) to isolate a novel pepper gene, CaLOX1, which encodes a 9-LOX and is specifically induced by avirulent Xcv infection of pepper leaves. The purified CaLOX1 protein was expressed in Escherichia coli and investigated for LOX activity. Virus-induced gene silencing (VIGS) is a widely used, powerful technique for reverse genetics. VIGS vectors derived from the Tobacco rattle virus (TRV) are the most popular for VIGS. Recently, a VIGS method was established for the functional characterization of defense-related genes in pepper (Baulcombe, 1999; Burch-Smith et al., 2006; Choi et al., 2007; Chung et al., 2007). Here, we analyzed the effect of CaLOX1 loss of function during pathogen infection using TRV-based VIGS of the CaLOX1 gene. Arabidopsis (Arabidopsis thaliana) plants that constitutively overexpressed CaLOX1 were also examined to determine the gain-of-function phenotype of CaLOX1 in plant defense. We further functionally characterized the Arabidopsis mutants lox1-1 and lox1-2, which have T-DNA insertions in AtLOX1, a putative CaLOX1 ortholog. Analysis of the function of CaLOX1 in pepper and Arabidopsis plants provided insight into the role of CaLOX1 expression in defense responses and the hypersensitive cell death of plants following pathogen invasion.
RESULTS
Isolation, Sequence Analysis, and LOX Activity of CaLOX1
We isolated pathogen-induced pepper genes using a pepper cDNA library prepared from pepper leaves inoculated with the Xcv avirulent strain Bv5-4a (Jung and Hwang, 2000). Among the Xcv-induced pepper cDNA clones, the pepper LOX gene CaLOX1 was identified. CaLOX1 contains 76 bp at the 5′ untranslated region and 184 bp at the 3′ untranslated region. Translation of the unique open reading frame present in CaLOX1 indicated that it codes for an 861-amino acid protein with an estimated molecular mass of 97.9 kD and an estimated pI of 5.42 (Supplemental Fig. S1). Sequence alignment of the predicted amino acid sequence of CaLOX1 showed that it shares identities greater than 60% with other plant LOXs from potato, tobacco, tomato, and Arabidopsis (Supplemental Fig. S2). In particular, CaLOX1 showed the greatest match (92% identity) to potato LOX (Kolomiets et al., 1996). Analysis of the deduced amino acid sequence of CaLOX1 identified two conserved LOX domains for plant LOX-related proteins: the PLAT_LH (for polycystein-1, LOX, α-toxin) or LH2 (for lipoxygenase homology 2) domain and the LOX domain. The deduced CaLOX1 protein contains conserved His residues (positions 547, 556, and 715) that are also observed in other plant LOXs and that have been implicated in iron binding and enzyme catalytic activity (Prigge et al., 1996).
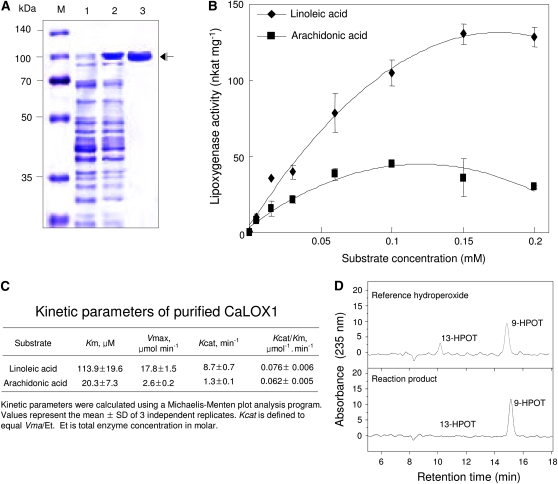
The enzyme activity of CaLOX1 was tested by expressing full-length CaLOX1 in E. coli as a fusion protein with an N-terminal His tag. Purified recombinant CaLOX1 expressed in E. coli was detected on Coomassie Brilliant Blue-stained SDS-PAGE gels as an apparent single polypeptide (Fig. 1A). To assay enzyme activity, affinity-purified recombinant CaLOX1 was incubated with linoleic acid or arachidonic acid as a substrate at various concentrations (Fig. 1B). We first determined the effects of pH and substrate concentration on CaLOX1 activity. The optimal pH for purified CaLOX1 activity ranged from 6 to 6.5 (Supplemental Fig. S3A). Enzyme activity with arachidonic acid as a substrate was lower, whereas linoleic acid was a good substrate (Supplemental Fig. S3B). Optimal conditions for CaLOX1 enzyme activity were pH 6.0 to 6.5 and 0.1 to 1.5 mm with linoleic acid as a substrate at 25°C (Supplemental Fig. S3C).
Figure 1.
Purification, enzyme activity, and positional specificity of CaLOX1. A, SDS-PAGE of the recombinant His-tagged CaLOX1 fusion protein. Total protein extracts and purified protein were analyzed by 10% SDS-PAGE and stained with Coomassie Brilliant Blue. Lane M, Molecular markers (kDa); lane 1, uninduced soluble fraction of E. coli BL21 cell extracts expressing His-tagged CaLOX1 protein; lane 2, soluble fraction of E. coli BL21 cell extracts expressing His-tagged CaLOX1 protein after isopropyl β-d-thiogalactopyranoside induction; lane 3, Purified His-tagged CaLOX1 fusion protein. B, LOX activity of CaLOX1 as determined with various concentrations of linoleic acid as a substrate. Values are presented as means ± sd. C, Kinetic parameters of purified CaLOX1. Kinetic parameters were calculated using a Michaelis-Menten plot analysis program. Values represent means ± sd of three independent replicates. D, Determination of positional specificity of CaLOX1. Reaction products were separated by reverse-phase HPLC to resolve the hydroxy isomers of linoleic acid. [See online article for color version of this figure.]
We then calculated the kinetic parameters for the substrates linoleic acid and arachidonic acid under standard reaction conditions using a Michaelis-Menten plot analysis program (Fig. 1C). The Km values of CaLOX1 for linoleic acid and arachidonic acid were 113.9 and 20.3 μm, respectively, and the kcat values for the two substrates were 8.7 and 1.3 min−1, respectively. The Km, Vmax, and kcat values for linoleic acid were much higher than those for arachidonic acid, suggesting that linoleic acid has a significantly higher affinity for purified CaLOX1 than does arachidonic acid. Estimates of kcat/Km of the purified CaLOX1 for linoleic acid and arachidonic acid were 0.076 and 0.062 μm−1min−1, respectively, suggesting that the catalytic efficiencies of both substrates for CaLOX1 are different. This indicates that linoleic acid is an in planta substrate suitable for CaLOX1
To determine the positional specificity of CaLOX1, we further identified the reaction products from the CaLOX1 enzyme assay following the HPLC analysis. As shown in Figure 1D, 9-hydroperoxylinolenic (9-HPOT) acid, but not 13-HPOT acid, was mainly produced by the reaction of purified CaLOX1 with linoleic acid as a substrate, indicating that CaLOX1 is indeed a 9-specific LOX.
Expression of CaLOX1 in Response to Biotic and Abiotic Stresses
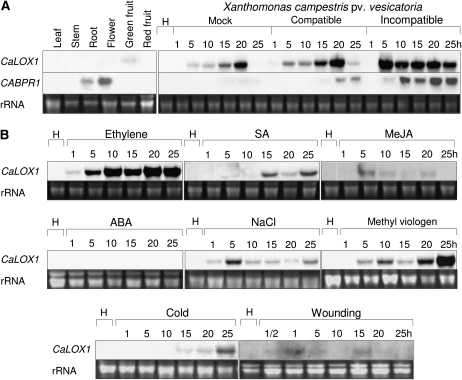
To determine the organ-specific expression of CaLOX1 in pepper plants, we performed RNA gel-blot analysis of expression of CaLOX1 in leaf, root, stem, flower, green fruit, and red fruit (Fig. 2A). The CaBPR1 gene was used as a comparable control (Kim and Hwang, 2000). CaLOX1 transcripts were slightly detected in green fruit but not in other pepper organs.
Figure 2.
RNA gel-blot analysis of pepper CaLOX1 and CaBPR1 expression genes in pepper leaves. The blots were hybridized with biotin probes for the CaLOX1 3′ untranslated region and full-length CaBPR1 cDNA. A, Expression of CaLOX1 and CaBPR1 in various organs of pepper plants and in pepper leaves at different time points after inoculation with the virulent strain Ds1 and the avirulent strain Bv5-4a of Xcv. B, Expression of CaLOX1 in pepper leaves at various time points after treatment with ethylene (10 mL L−1), SA (5 mm), MeJA (100 mm), ABA (100 mm), NaCl (200 mm), methyl viologen (100 mm), cold (4°C), and wounding. H, Healthy leaves.
LOXs and their metabolites have been demonstrated to be involved in plant defense responses to diverse pathogens and stresses (Siedow, 1991; Porta and Rocha-Sosa, 2002; Liavonchanka and Feussner, 2006). Expression profiles of CaLOX1 in response to the virulent Ds1 and the avirulent Bv5-4a strains of Xcv were examined in infected pepper leaves during both compatible and incompatible interactions (Fig. 2A). Northern analysis of total RNA extracted from infected leaves revealed that the CaLOX1 transcript accumulated in pepper leaves infected with virulent or avirulent Xcv strains. Expression of CaLOX1 was induced 1 h after infection by the virulent strain Ds1, peaked at 20 h, and decreased thereafter. In contrast, induction of CaLOX1 by the avirulent strain Bv5-4a was rapid, peaking at 5 h and strongly maintained 5 to 25 h after inoculation. Expression of CaLOX1 was also significantly increased by mock inoculation. These data indicated that the induction of CaLOX1 transcript in mock-inoculated or virulent Xcv-inoculated leaves may be due, in part, to the wounding effect.
Ethylene, SA, abscisic acid (ABA), methyl viologen, and MeJA are well-known modulators of defense responses in plants. Pepper plants at the six-leaf stage were used to study the CaLOX1 function in stress-induced responses. Treatment with ethylene, SA, NaCl, and methyl viologen significantly induced CaLOX1 in pepper leaves (Fig. 2B). However, CaLOX1 transcripts were not or were weakly detected in leaves treated with ABA, MeJA, cold, and wounding. In particular, treatments with ethylene and methyl viologen were very effective in triggering CaLOX1 expression.
Induction of the Cell Death Response by Transient Expression of CaLOX1 in Pepper Leaves
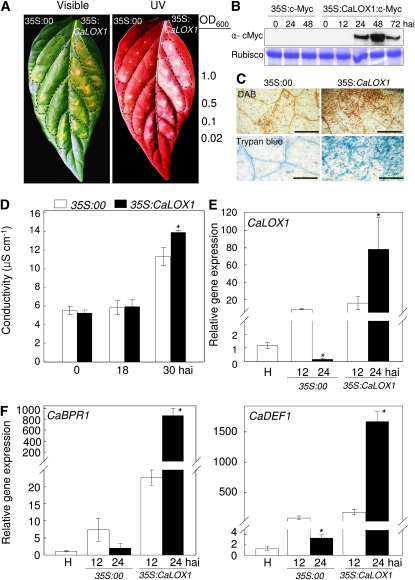
To determine whether the CaLOX1 gene is involved in cell death, we transiently expressed CaLOX1 in pepper leaves infiltrated with Agrobacterium tumefaciens carrying 35S:00 (empty vector) or 35S:CaLOX1 construct. The empty vector control did not induce cell death or a necrotic response (Fig. 3A), whereas a necrotic phenotype was distinctly observed in pepper leaves infiltrated with different concentrations of Agrobacterium (35S:CaLOX1). Cell death lesions in plants can be visualized by fluorescence under UV light, indicating the cell death-related production of phenolic compounds (Dixon and Paiva, 1995). UV illumination of pepper leaves revealed that such compounds were synthesized and accumulated in regions surrounding the Agrobacterium (35S:CaLOX1)-infiltrated area (Fig. 3A). We confirmed the Agrobacterium-mediated transient expression of CaLOX1 in pepper leaves using an immunoblotting assay. Expression of c-Myc-tagged CaLOX1 was specifically detected in leaf tissues 24, 48, and 72 h after inoculation; however, the protein expressing 35S:c-Myc was not detected in pepper leaves (Fig. 3B). Cell death and hydrogen peroxide (H2O2) production in leaves transiently expressing CaLOX1 was visualized by trypan blue and 3,3′-diaminobenzidine (DAB) staining, respectively, but was not visualized at all, or only weakly, in leaves transiently expressing the empty vector (Fig. 3C). We employed an ion leakage test to analyze the severity of cell necrosis caused by plasma membrane damage. Pepper leaves that transiently expressed CaLOX1 exhibited more ion leakage 30 h after agroinfiltration than did empty vector control leaves (Fig. 3D). Changes in defense-related gene expression in leaves transiently expressing CaLOX1 and empty vector control were examined using real-time reverse transcription (RT)-PCR. Expression of the CaLOX1, CaBPR1 (for basic pathogenesis-related gene), and CaDEF1 (for defensin) genes was rapidly and strongly induced by transient expression of CaLOX1 (Fig. 3, E and F). The transcript levels of CaBPR1 and CaDEF1 increased continuously during the transient expression of CaLOX1, indicating that the CaLOX1-mediated expression of defense-related genes is required for hypersensitive cell death in pepper leaves.
Figure 3.
Transient expression of CaLOX1 and cell death in pepper leaves infiltrated with Agrobacterium GV3101 carrying the 35S:00 or 35S:CaLOX1 construct. A, Visible and UV light-illuminated phenotypes of the transient expression in leaves 3 d after agroinfiltration with different bacterial concentrations (OD600). B, Immunoblotting assay of the Agrobacterium-mediated transient expression of CaLOX1 in pepper leaves. C, Staining with trypan blue and DAB of transient expression in leaf tissues 1 d after agroinfiltration. Bars = 0.2 mm. D, Ion leakage from transiently expressing leaves at different time points after agroinfiltration. E and F, Induction of CaLOX1, CaBPR1, and CaDEF1 as measured by real-time RT-PCR. The data of the relative transcript levels are normalized to the expression of the 18S rRNA gene as an internal control. Values are presented as means ± sd. Asterisks indicate significant differences between the 35S:CaLOX1 and 35S:00 constructs (Student's t test, P < 0.05). H, Healthy leaves; hai, hours after infiltration.
Enhanced Susceptibility of CaLOX1-Silenced Pepper to Bacterial and Fungal Infection
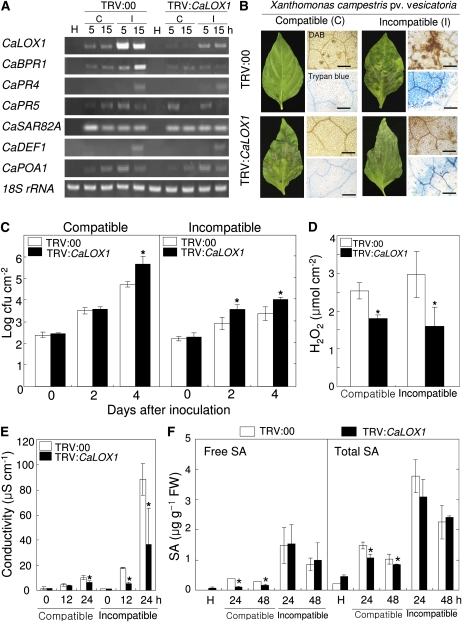
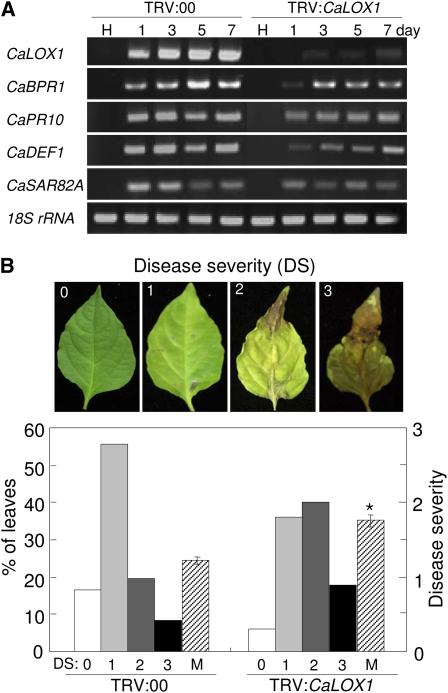
To investigate the CaLOX1 loss-of-function phenotype in pepper, we used the VIGS technique to knock down the expression level of CaLOX1 with TRV vectors containing 621 bp of CaLOX1 cDNA (TRV:CaLOX1) or lacking the CaLOX1 insert (TRV:00). Three to 4 weeks after VIGS, empty vector control (TRV:00) and silenced (TRV:CaLOX1) plants were challenged with the virulent strain Ds1 (compatible) and the avirulent strain Bv5-4a (incompatible) of Xcv. Five and 15 h after inoculation with Xcv (106 colony-forming units [cfu] mL−1), CaLOX1 transcript levels were analyzed by RT-PCR in empty vector and silenced plants. CaLOX1 expression was compromised in CaLOX1-silenced plants compared with empty vector control plants. We further evaluated whether CaLOX1 silencing affects the resistance response to Xcv infection. Expression levels of pepper defense-related genes in empty vector control and silenced plants were analyzed by RT-PCR. As shown in Figure 4A, avirulent Xcv infection strongly induced the CaBPR1 gene in empty vector control plants; however, CaBPR1 expression was weakly or not at all induced in CaLOX1-silenced plants infected with avirulent Xcv. The expression of CaPR4 (putative antifungal protein), CaPR5 (osmotin-like protein), CaSAR82A (SAR8.2), CaDEF1, and CaPOA1 (ascorbate peroxidase) in CaLOX1-silenced leaves was similar to that in empty vector control leaves during both virulent and avirulent Xcv infections. CaLOX1-silenced plants exhibited enhanced susceptibility to the Xcv virulent strain Ds1 and the avirulent strain Bv5-4a (Fig. 4, B and C). The Ds1 and Bv5-4a strains (5 × 104 cfu mL−1) grew significantly less in empty vector control leaves than in CaLOX1-silenced leaves during infection, especially 4 d after inoculation. Five days after inoculation, more symptoms of susceptibility were observed in CaLOX1-silenced plants. We also stained Xcv-infected empty vector control and silenced leaves with trypan blue and DAB. Strongly polymerized DAB (indicative of H2O2 formation) and HR were detected in empty vector control leaves 24 h after inoculation with Bv5-4a, whereas HR and H2O2 accumulation were distinctly reduced in CaLOX1-silenced leaves. In compatible interactions with Ds1, both empty vector control and CaLOX1-silenced leaves did not exhibit H2O2 formation or hypersensitive cell death. In addition, we also measured H2O2 by xylenol orange assay, in which H2O2 is reduced by ferrous ions in an acidic solution that forms a ferric product-xylenol orange complex (Fig. 4D). Low levels of H2O2 were detected during the Xcv infection in CaLOX1-silenced leaves compared with empty vector control leaves. As shown in Figure 4E, cell death, which is assessed by monitoring cellular ion leakage as a measure of membrane damage, significantly declined in CaLOX1-silenced leaves during the Xcv infection. SA data obtained by HPLC analysis show that lower free and total SA (free SA plus Glc-conjugated SA) levels accumulated in CaLOX1-silenced leaves infected by the virulent (compatible) Xcv strain compared with empty vector control leaves (Fig. 4F). However, no significant differences in SA levels were detected between empty vector control and silenced leaves during the avirulent Xcv infection. The role of CaLOX1 in JA signaling was also determined by comparing JA levels in empty vector control and CaLOX1-silenced plants. The JA levels were drastically induced during the Xcv infection. However, no significant differences in JA levels were found between empty vector control and CaLOX1-silenced plants during infection, except the 18-h early stage of infection by the virulent Xcv (Supplemental Fig. S4A). Collectively, these results indicate that silencing of the CaLOX1 gene compromises the basal defense response of pepper to Xcv infection.
Figure 4.
Enhanced susceptibility of CaLOX1-silenced plants to infection by the virulent strain Ds1 (compatible [C]) and the avirulent strain Bv5-4a (incompatible [I]) of Xcv (106 cfu mL−1). A, RT-PCR analysis of the expression of CaLOX1 and pepper defense-related genes in empty vector control (TRV:00) and CaLOX1-silenced plants (TRV:CaLOX1). The experiments were performed three times with 28 PCR cycles. H, Healthy leaves; CaBPR1, basic pathogenesis-related protein 1; CaPR4, putative antifungal protein; CaPR5, osmotin-like protein; CaSAR8.2, SAR8.2; CaDEF1, defensin 1; CaPOA1, ascorbate peroxidase 1; 18S rRNA, control. B, Disease symptoms on leaves of empty vector control and silenced plants inoculated with Xcv Ds1 and Bv5-4a. Five days after inoculation, empty vector control plants showed only mild symptoms, whereas CaLOX1-silenced plants showed extensive lesions (left). Infected leaves were stained with trypan blue and DAB 1 d after Xcv inoculation (right). Bars = 0.2 mm. C, Bacterial growth in leaves of the empty vector control (TRV:00) and CaLOX1-silenced plants 0, 2, and 4 d after inoculation (5 × 104 cfu mL−1). D, Quantification of H2O2 in leaves of empty vector control plants and CaLOX1-silenced plants 15 h after inoculation with 107 cfu mL−1 Xcv using the xylenol orange assay. E, Ion leakage from the leaf tissues at different time points after inoculation with Xcv. F, Levels of SA in the empty vector control and CaLOX1-silenced leaves infected by Xcv. FW, Fresh weight. Values are presented as means ± sd. Asterisks indicate significant differences from wild-type plants (Student's t test, P < 0.05). [See online article for color version of this figure.]
We then challenged control and CaLOX1-silenced pepper plants with the fungal pathogen Colletotrichum coccodes. Leaves of CaLOX1-silenced plants infected with C. coccodes exhibited more chlorotic lesions than did empty vector control plants (Fig. 5). High levels of expression of CaLOX1 were induced in C. coccodes-infected empty vector control plants at the six-leaf stage, whereas silencing of CaLOX1 compromised CaLOX1 expression in pepper leaves. Pepper defense-related genes, such as CaBPR1, CaPR10, CaSAR82A, and CaDEF1, were weakly induced in CaLOX1-silenced plants compared with empty vector control plants. To evaluate disease development in the inoculated plants, disease severity was rated based on a 0 to 3 scale, where 0 = no visible symptoms, 1 = small circular or irregular gray-brown spots on leaves, 2 = dark-brown lesions with mild chlorosis, and 3 = enlarged dark-brown lesions with severe chlorosis (Fig. 5B). The disease severity of CaLOX1-silenced plants infected with C. coccodes was greater than that of empty vector control plants (Fig. 5B). Seven days after inoculation, more susceptible chlorotic symptoms were observed in leaves of CaLOX1-silenced plants than in leaves of empty vector control plants. Together, these results indicate that the expression of CaLOX1 is involved in the resistance of pepper plants to Xcv and C. coccodes.
Figure 5.
Enhanced susceptibility of CaLOX1-silenced pepper plants to C. coccodes infection. A, Expression of defense-related genes in leaves of empty vector control and silenced plants. H, Healthy leaves. The experiments were performed three times, and 28 PCR cycles were carried out. B, Disease symptoms and disease severity in leaves of empty vector control and silenced pepper plants 5 d after inoculation. Asterisks indicate significant differences between the means as determined by Student's t test (P < 0.05). M, Means of disease severities. [See online article for color version of this figure.]
LOX Activity in Pepper Leaves
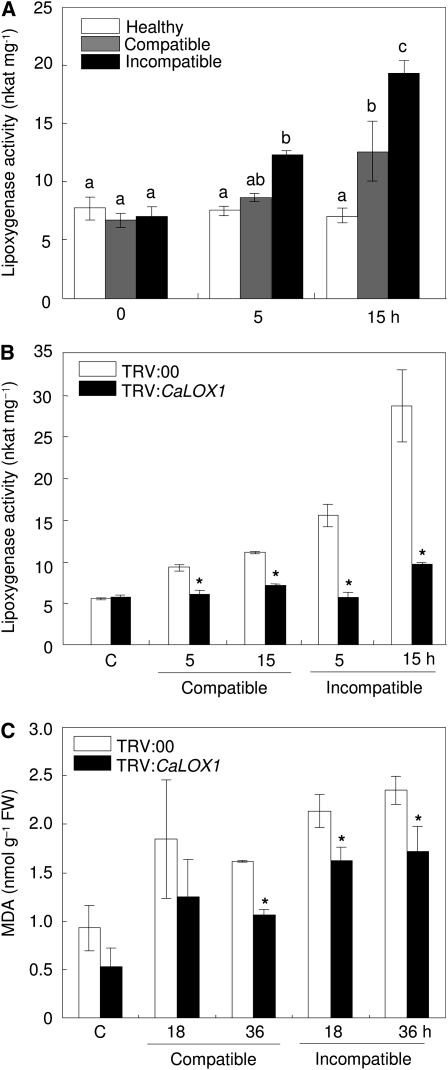
To determine whether CaLOX1 activity is induced by pathogen infection, we tested for LOX activity in protein extracts of pepper leaves infected with the virulent Ds1 and avirulent Bv5-4a strains of Xcv (Fig. 6). Profiles of LOX activity in extracts from healthy and infected leaves are shown in Figure 6A. Infection with Xcv Bv5-4a induced significantly higher LOX activity in pepper leaves than did infection with Xcv Ds1; however, LOX activity remained at low levels in healthy leaves. We then examined LOX activity in unsilenced (TRV:00) and silenced (TRV:CaLOX1) pepper plants during Xcv infection (Fig. 6B). Empty vector control plants (TRV:00) displayed higher levels of LOX activity during infection with avirulent Xcv compared with those infected with the virulent strain. However, LOX activity was not significantly enhanced in CaLOX1-silenced plants by Xcv infection, indicating that silencing of CaLOX1 compromised LOX activity. These data suggest that pepper plants defend themselves against pathogen infection by activating the LOX pathway.
Figure 6.
Enhanced activity of LOX in pepper leaves infected with Xcv. A, LOX activity in pepper leaves inoculated with the virulent (compatible) Ds1 and avirulent (incompatible) Bv5-4a strains of Xcv. B, LOX activity in leaves of unsilenced (TRV:00) and silenced (TRV:CaLOX1) pepper plants infected with the Ds1 and Bv5-4a strains. Linoleic acid (0.1 mm) was used as a substrate. All experiments were performed three times with similar results. C, Quantification of lipid peroxidation (MDA concentration) in leaves of empty vector control (TRV:00) and silenced (TRV:CaLOX1) pepper plants infected with Xcv. FW, Fresh weight. Values are presented as means ± sd. Different letters indicate significant differences as determined by the lsd test (P < 0.05). Asterisks indicate significant differences between wild-type and inoculated plants (Student's t test, P < 0.05).
Induction of lipid peroxidation was also assessed by determining the accumulation of thiobarbituric acid-reactive substances in the pepper leaves infected by Xcv (Fig. 6C). The lipid peroxidation was significantly induced in pepper leaves by Xcv infection. Lower levels of malondialdehyde (MDA), which is a decomposition product formed by peroxidation of polyunsaturated fatty acids in the membranes, were detected in CaLOX1-silenced leaves than in empty vector control plants (TRV:00) during the Xcv infection. This indicates that the CaLOX1 gene functions in lipid peroxidation of leaf tissues of pepper.
Enhanced Resistance of CaLOX1-OX Transgenic Arabidopsis to Bacterial Infection
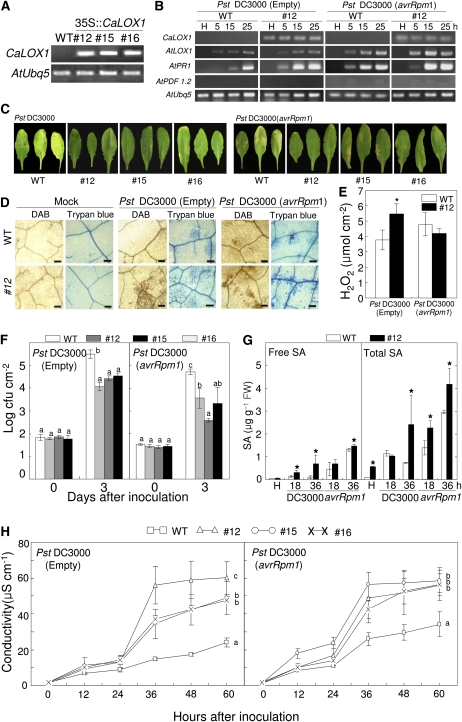
To determine the gain-of-function phenotype of the pepper CaLOX1 gene in planta, transgenic Arabidopsis lines that express a cauliflower mosaic virus 35S promoter-driven CaLOX1 construct were generated using an Agrobacterium-mediated gene transfer system (Clough and Bent, 1998). At least 10 independent transgenic lines expressed CaLOX1, whereas CaLOX1 transcripts were not observed in wild-type (ecotype Columbia [Col-0]) plants (Fig. 7A). Three CaLOX1-OX transgenic lines, 12, 15, and 16, were selected for further study. The CaLOX1 transgene was constitutively expressed in the representative Arabidopsis transgenic line 12 during Pseudomonas syringae pv tomato (Pst) infection (Fig. 7B). We investigated time course expression of some defense-related genes such as AtPR1 and AtPDF1.2, which serve as markers in the SA and JA pathways, during infection with Pst DC3000 and Pst DC3000 (avrRpm1) in CaLOX1-OX transgenic plants. Induction of AtPR1, but not AtLOX1 and AtPDF1.2, greatly and rapidly increased in the transgenic plants during Pst infection compared with wild-type plants. The CaLOX1-OX transgenic plants, in which LOX is activated, may respond differentially to pathogen infection. We examined their responses to Pst DC3000 and to Pst DC3000 (avrRpm1). Pst DC3000 is an important bacterial pathogen and a valuable model organism to study plant-pathogen interactions (Quirino and Bent, 2003). Arabidopsis plants were inoculated with 106 cfu mL−1 Pst. As shown in Figure 7C, wild-type plants inoculated with Pst DC3000 showed disease symptoms, whereas a limited necrotic response was observed in the infected areas of leaves of transgenic lines 3 d after inoculation. Wild-type plants inoculated with Pst DC3000 (avrRpm1) showed a typical HR. Similarly, CaLOX1 transgenic lines also showed enhanced resistance to Pst DC3000 (avrRpm1) compared with wild-type plants. We further quantified the expression of several defense-related genes in CaLOX1-OX Arabidopsis transgenic plants. AtLOX1, 9-position specific LOX, was strongly expressed in Arabidopsis leaves during Pst infection, especially in the incompatible interaction with CaLOX1-OX Arabidopsis, which is consistent with the recent finding of Bannenberg et al. (2009). AtPR1, which serves as a marker for induction of the SA pathway, was rapidly and strongly induced in CaLOX1-OX transgenic plants; however, AtPDF1.2, a marker gene for the JA pathway, was not at all expressed during Pst infection. To characterize the observed necrotizing process at the microscopic level, 5-week-old wild-type and transgenic plant leaves were infiltrated with Pst DC3000 and Pst DC3000 (avrRpm1). Cell death and H2O2 production were detected by staining with trypan blue (dark blue) and DAB (dark brown), respectively (Fig. 7D). A significant macroscopic HR was observed in transgenic plants 24 h after inoculation with Pst DC3000 but not in either wild-type or transgenic plants infected by Pst DC3000 (avrRpm1) (data not shown). In plants inoculated with Pst DC3000 (avrRpm1), the HR was more strongly induced in transgenic plants than in wild-type plants (Fig. 7D). The visual observation of DAB staining was quantified by measurements of H2O2 levels using xylenol orange assay (Fig. 7E). Levels of H2O2 significantly increased in CaLOX1-OX transgenic leaves during the virulent Pst DC3000 infection. CaLOX1-OX transgenic plants generated H2O2 and underwent significant cell death when inoculated with either the virulent or avirulent strain at 106 cfu mL−1. Treatment with MgCl2 solution, as a negative control, did not lead to H2O2 production or to cell death in wild-type or transgenic lines. In this study, all three transgenic lines gave similar results.
Figure 7.
Enhanced resistance of CaLOX1-OX Arabidopsis transgenic lines to Pst infection. A, RT-PCR analysis of the expression of the CaLOX1 transgene in 35S:CaLOX1 lines. B, RT-PCR analysis of the expression of Arabidopsis defense-related genes in wild-type (WT) and CaLOX1-OX lines of Arabidopsis plants after inoculation with 106 cfu mL−1 Pst DC3000 or Pst DC3000 (avrRpm1). The experiments were performed three times with 28 PCR cycles. H, Healthy leaves; AtUbQ5, control. C, Disease symptoms on leaves infected with Pst DC3000 or Pst DC3000 (avrRpm1). Leaves of 4-week-old Arabidopsis plants were infiltrated with a suspension (106 cfu mL−1) of Pst DC3000 or Pst DC3000 (avrRpm1), and photographs were taken 5 d later. D, DAB and trypan blue staining of ROS accumulation and cell death, respectively, in leaves 24 h after inoculation with Pst DC3000 or Pst DC3000 (avrRpm1). Bars = 0.1 mm. E, Quantification of H2O2 in leaves of wild-type and CaLOX1-OX lines 12 h after inoculation with 106 cfu mL−1 Pst DC3000 or Pst DC3000 (avrRpm1) using the xylenol orange assay. F, Bacterial growth in leaves inoculated with Pst DC3000 or Pst DC3000 (avrRpm1). G, Levels of SA in the empty vector control and CaLOX1-silenced leaves 18 and 36 h after inoculation with Xcv. FW, Fresh weight. H, Ion leakage from the leaf tissues inoculated with Pst DC3000 or Pst DC3000 (avrRpm1). Values are presented as means ± sd. Different letters indicate significant differences as determined by the lsd test (P < 0.05) in three independent experiments.
The CaLOX1 function was further studied by monitoring bacterial growth in CaLOX1-OX transgenic lines. We tested the growth of virulent Pst DC3000 and avirulent Pst DC3000 (avrRpm1) in wild-type and transgenic plants 0 and 3 d after inoculation with 106 cfu mL−1 (Fig. 7F). Bacterial growth immediately after inoculation (day 0) indicated that an equal quantity of bacterial inoculum was infiltrated into leaves of both wild-type and transgenic plants. Higher growth of Pst DC3000 and Pst DC3000 (avrRpm1) was observed in wild-type plants 3 d after inoculation compared with transgenic plants (Fig. 7F). To determine whether CaLOX1 overexpression affects the SA pathway, we also quantified SA levels in wild-type and CaLOX1-OX transgenic leaves. Free and total SA (free SA plus Glc-conjugated SA) levels accumulated faster and greater in CaLOX1-OX transgenic leaves than in wild-type leaves during Pst DC3000 infection (Fig. 7G). These support the idea that the elevated SA levels may be due to the induction of AtPR1 expression conferred by ectopic CaLOX1 overexpression. In contrast, there were no significant differences in JA levels between wild-type and CaLOX1-OX transgenic Arabidopsis plants infected with Pst DC3000 and Pst DC3000 (avrRpm1) (Supplemental Fig. S4B). These results suggest that CaLOX1 overexpression confers defense responses to Pst infection via the SA-dependent signaling pathway. The Pst DC3000- and Pst DC3000 (avrRpm1)-dependent HR in CaLOX1-OX transgenic plants was examined using an electrolyte leakage assay (Fig. 7H). Cell death associated with the HR causes the release of electrolytes, which is measured as a change in the conductance of a bath solution (Orlandi et al., 1992). Transgenic plants infected by Pst DC3000 and Pst DC3000 (avrRpm1) exhibited significantly higher levels of ion leakage compared with wild-type plants. These data indicate that ectopic expression of the CaLOX1 transgene in Arabidopsis enhanced basal resistance to bacterial pathogen infection, which was accompanied by a HR response.
Enhanced Susceptibility of AtLOX1∷T-DNA Plants to Bacterial Infection
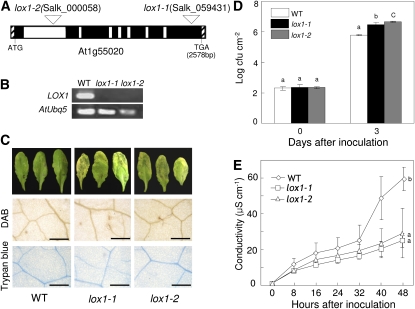
To investigate the loss-of-function phenotype of the CaLOX1 ortholog AtLOX1 in Arabidopsis, we isolated two putative AtLOX1 insertion lines, SALK_000058 and SALK_059431, from the Salk Institute T-DNA insertion library database (http://signal.salk.edu/cgi-bin/tdnaexpress) by a BLAST search. The predicted amino acid sequence of AtLOX1 was 68% identical to that of CaLOX1 (Supplemental Fig. S2), and they both contain the LOX-specific PLAT_LH and LOX domains. Homozygous T-DNA insertion lox1 mutants were obtained by screening each ortholog line by PCR with specific primer pairs. Figure 8A shows a schematic of lox1 genomic structures indicating the positions of the T-DNA insertions. The loss-of-function mutants were named lox1-1 and lox1-2. RT-PCR analysis revealed that AtLOX1 transcripts were not detected in lox1-1 and lox1-2 plants inoculated with Pst DC3000 (avrRpm1) (Fig. 8B). Three days after inoculation with the virulent strain Pst DC3000, severe chlorotic symptoms developed in lox1 plants, whereas typical susceptible symptoms did not occur in wild-type plants (Fig. 8C). Dead cells or cells with damaged membranes were stained with DAB and trypan blue, which indicated that H2O2 formation and cell death did not occur in wild-type and lox1 leaves. Significantly, Pst DC3000 grew well in lox1-1 and lox1-2 mutants (Fig. 8D). The growth of Pst DC3000 was 5-fold greater in lox1-1 and lox1-2 mutants than in wild-type plants. Electrolyte leakage of the two mutant lines, lox1-1 and lox1-2, was significantly lower than that of wild-type plants during virulent Pst infection (Fig. 8E). Together, these data indicate that the AtLox1 mutants exhibited enhanced susceptibility to virulent Pst infection.
Figure 8.
Enhanced susceptibility of Arabidopsis ortholog mutants, lox1-1 and lox1-2, of CaLOX1 to Pst infection. A, Schematic representation of T-DNA insertion sites in lox11 (SALK_059431) and lox1-2 (SALK_000058) and genomic structure of the Arabidopsis LOX1 gene. Exons and introns are represented as black and white boxes, respectively. B, RT-PCR analysis of expression of the lox1-1 and lox12 genes. WT, Wild type. C, ROS accumulation, cell death phenotype, and disease symptoms on leaves 24 h (DAB and trypan blue staining) and 3 d after inoculation with Pst DC3000 (106 cfu mL−1), respectively. Bars = 0.2 mm. D, Bacterial growth in leaves inoculated with Pst DC3000. E, Ion leakage from leaf tissues inoculated with Pst DC3000. Values are presented as means ± sd. Different letters indicate significant differences as determined by the lsd test (P < 0.05) in three independent experiments. [See online article for color version of this figure.]
LOX Activity in CaLOX1-OX and Arabidopsis lox1 Mutant Plants
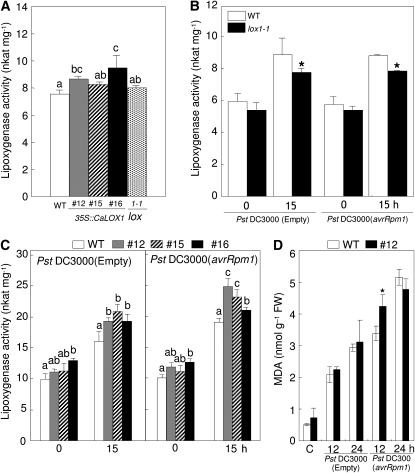
We speculated whether the enhanced resistance of CaLOX1-OX plants is due to increased LOX activity; hence, we analyzed LOX activity in wild-type and CaLOX1-OX Arabidopsis (Fig. 9). The levels of LOX activity were higher in the CaLOX1-OX transgenic lines 12, 15, and 16 relative to that of wild-type plants (Fig. 9A), suggesting that CaLOX1 is enzymatically active in heterologous transgenic Arabidopsis. We also measured LOX activity in the Arabidopsis mutant lox1-1 (Fig. 9B) and found that it was similar to that in wild-type plants. However, the enhanced LOX activity in lox1-1 leaves was lower than that in wild-type leaves 15 h after infection with Pst DC3000 and Pst DC3000 (avrRpm1). These data suggest that mutation of Arabidopsis AtLOX1 may compromise the level of LOX activity.
Figure 9.
Altered LOX activity in Arabidopsis leaves infected with Pst DC3000 and Pst DC3000 (avrRpm1). A, LOX activity in leaf extracts from wild-type (Col-0 [WT]), CaLOX1-OX, and lox1-1 mutant plants. B, LOX activity in Arabidopsis wild-type and lox1-1 leaves infected with Pst. C, LOX activity in Arabidopsis wild-type and CaLOX1-OX leaves. Linoleic acid (0.1 mm) was used as a substrate. All experiments were performed three times with similar results. D, Quantification of lipid peroxidation (MDA concentration) in Arabidopsis wild-type and CaLOX1-OX leaves infected with Pst. FW, Fresh weight. Values are presented as means ± sd. Different letters indicate significant differences as determined by the lsd test (P < 0.05) in three independent experiments. Asterisks indicate significant differences between wild-type and inoculated plants (Student's t test, P < 0.05).
To further substantiate the effect of CaLOX1 overexpression on LOX activity in plants, we analyzed wild-type and CaLOX1-OX Arabidopsis plants during infection with Pst DC3000 and Pst DC3000 (avrRpm1) (Fig. 9C). In CaLOX1-OX plants, the level of LOX activity was somewhat higher in healthy leaves than in wild-type plants. As expected, infection with virulent Pst DC3000 or avirulent Pst DC3000 (avrRpm1) also induced significantly higher LOX activities in CaLOX1-OX Arabidopsis compared with wild-type plants. Importantly, either virulent or avirulent Pst DC3000 infection drastically stimulated lipid peroxidation in wild-type and CaLOX1-OX Arabidopsis leaves (Fig. 9D). However, there were no significant differences in MDA levels between wild-type and CaLOX1-OX plants during Pst infection, except for a high level of lipid peroxidation in CaLOX1-OX leaves 12 h after inoculation with avirulent Pst DC3000 (avrRpm1).
Distinct Responses of Arabidopsis CaLOX1-OX and lox1 Plants to Hyaloperonospora arabidopsidis Infection
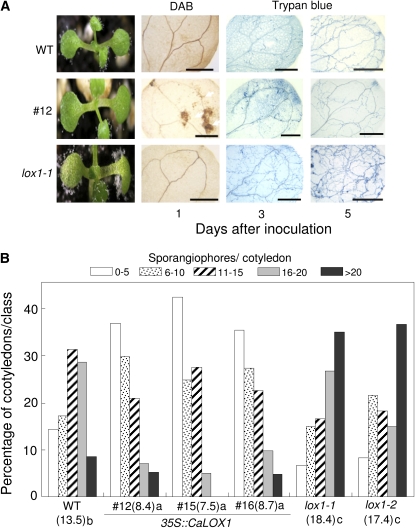
To determine whether CaLOX1-OX transgenic and lox1 plants are resistant to the biotrophic oomycete H. arabidopsidis, we inoculated over 100 seedlings of each line with a suspension of an asexual inoculum of H. arabidopsidis isolate Noco2 (5 × 104 conidiosporangia mL−1), which is virulent to Arabidopsis Col-0. As shown in Figure 10A, H. arabidopsidis isolate Noco2 was highly virulent to wild-type cotyledons, which exhibited high levels of mycelial growth, sporulation, and sporangiophores. In contrast, CaLOX1-OX transgenic plants were significantly resistant to H. arabidopsidis isolate Noco2, with less sporangiophore formation in cotyledons compared with wild-type plants. Three and 5 d after inoculation, H2O2 production and restricted hyphal growth were observed in transgenic plants, as observed by DAB and trypan blue staining. We also tested lox1 mutants for resistance to H. arabidopsidis and found that they were more susceptible to H. arabidopsidis isolate Noco2 than were wild-type plants. DAB staining of infection sites was not observed in these cotyledons. Hyphal growth, and early and heavy asexual sporulation, were distinctly enhanced in lox1 mutants relative to wild-type and CaLOX1-OX transgenic plants.
Figure 10.
Distinct responses of wild-type (WT), CaLOX1 transgenic, and T-DNA insertion mutant (lox1-1 and lox1-2) plants to H. arabidopsidis isolate Noco2. A, Disease reactions on 7-d-old cotyledons inoculated with an asexual spore suspension of H. arabidopsidis. Diseased cotyledons were photographed 7 d after inoculation and stained with DAB (1 d after inoculation) and trypan blue (3 and 5 d after inoculation). Bars = 0.5 mm. B, Quantification of sporangiophores produced on at least 50 cotyledons inoculated with H. arabidopsidis isolate Noco2. Different letters indicate significant differences as determined by the lsd test (P < 0.05) in three independent experiments. [See online article for color version of this figure.]
Effects of the gain or loss of function of CaLOX1 and AtLOX1 on plant resistance to H. arabidopsidis infection were evaluated by measuring asexual sporulation on Arabidopsis cotyledons (Fig. 10B). These levels varied for each line, ranging from heavy to low. The percentage of sporangiophore formation and the average number of sporangiophores were lower in CaLOX1-OX transgenic plants than in wild-type plants. In contrast, lox1 mutants exhibited higher asexual sporulation than did wild-type plants. Together, these results indicate that the CaLOX1 and AtLOX1 genes play a crucial role in basal resistance to the biotrophic oomycete H. arabidopsidis.
Distinct Responses of Arabidopsis CaLOX1-OX and lox1 Plants to Alternaria brassicicola Infection
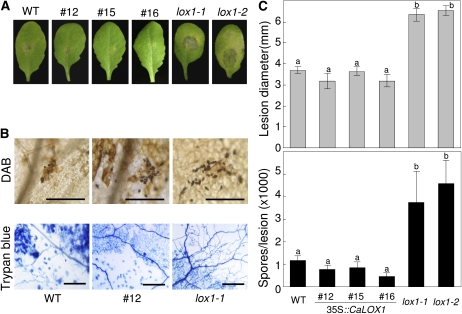
Previous studies reported that LOX expression is regulated by JA during infection with fungal pathogens (Melan et al., 1993). We examined whether CaLOX1 is involved in plant resistance to the fungal pathogen A. brassicicola. Because wild-type (Col-0) plants are known to be incompatible with A. brassicicola (Moreno et al., 2005), we used the Arabidopsis ecotype Col-0 as a comparable control for resistance to A. brassicicola. The HR was induced in wild-type and CaLOX1-OX transgenic plants by A. brassicicola infection, which results in the development of brown necrotic lesions on leaves (Narusaka et al., 2003). The restricted hyphal growth, cell death, and production of H2O2 during infection were similar in wild-type and CaLOX1-OX transgenic plants. In contrast, infection of lox1 mutants by A. brassicicola resulted in spreading lesions and abundant sporulation (Fig. 11, A and C). As shown in Figure 11B, restricted fungal spread and high levels of H2O2 were detected in wild-type and CaLOX1-OX plants by trypan blue and DAB staining. However, in lox1 mutants inoculated with A. brassicicola, the infection hyphae spread profusely from the site of inoculation. The number of spores produced at the infection site was determined using the spore count assay (Fig. 11C). Spores were abundantly produced on leaves of lox1 mutants compared with wild-type and CaLOX1-OX plants. The lox1 mutants were clearly more susceptible to A. brassicicola infection than were wild-type and CaLOX1-OX transgenic plants. Together, these results indicated that CaLOX1 is necessary for plant defense responses to the fungal pathogen A. brassicicola.
Figure 11.
Disease development on Arabidopsis wild-type (WT), CaLOX1-OX transgenic, and lox1 leaves inoculated with A. brassicicola. Leaves of 4-week-old plants were challenged with 10-μL droplets containing 5 × 104 mL−1 fungal spores. A, Disease symptoms. B, Diseased leaves stained with DAB and trypan blue. Microscopic images show stained fungal structures and damaged plant cells. Bars = 0.05 mm (DAB) and 0.15 mm (trypan blue). C, Quantification of disease development. Top, average diameter of lesions caused by A. brassicicola. The lesion sizes are averages of 30 lesions per line. Lesion diameter is presented ± sd. Bottom, average numbers of newly formed spores per lesion. Spores on 10 inoculated leaves per line were counted and are presented as means ± sd. Different letters indicate significant differences as determined by the lsd test (P < 0.05) in three independent experiments. [See online article for color version of this figure.]
DISCUSSION
Here, we report the cloning of the pepper 9-LOX gene CaLOX1, which is involved in plant defense and cell death responses to microbial pathogens. Activation of CaLOX1 was crucial for cell death and defense responses in pepper leaves, which were accompanied by reactive oxygen species (ROS) accumulation, lipid peroxidation, SA accumulation, and defense-related gene expression. The strong activity of CaLOX1 also was shown to induce plant defense and cell death in response to pathogen infection. Our molecular genetic study using CaLOX1-silenced pepper plants, CaLOX1-OX Arabidopsis, and Arabidopsis lox1 mutants revealed that the enhanced expression of CaLOX1 and defense-related genes such as the SA-responsive CaBPR1 is required for defense and cell death in plants. In contrast to the CaLOX1 overexpression phenotype, CaLOX1-silenced and lox1 mutants exhibited enhanced susceptibility to pathogen invasion. Overexpression of CaLOX1 in Arabidopsis conferred broad-spectrum resistance to infection by H. arabidopsidis and A. brassicicola as well as Pst. The data presented here suggest that CaLOX1 functions as a positive regulator of broad-spectrum resistance and cell death responses during pathogen infection.
Isolation, Enzyme Activity, and Position Specificity of the Pepper 9-LOX Gene and Protein
LOX genes have been widely used to study the role of JA and oxylipins in defense responses during abiotic and biotic stresses, but their biological functions are not fully understood (Porta and Rocha-Sosa, 2002). Our results here provide evidence of a role for the pepper 9-LOX gene, CaLOX1, in regulating disease resistance. Sequence alignment analysis revealed that CaLOX1 shares high homology with LOXs from potato, tobacco, tomato, and Arabidopsis. The His residues that act as ligands to the active site iron are highly conserved in LOXs (Prigge et al., 1996). The pepper CaLOX1 protein contains conserved His residues involved in catalysis, as observed in other LOXs (Hornung et al., 1999). The presence of these conserved His residues predicted that CaLOX1 was a LOX.
Linolenic and linoleic acids are the most common plant substrates for LOXs (Siedow, 1991), and linoleic acid is particularly important as a substrate (Lorenzi et al., 2006). A comparison of the substrate specificity of the six Arabidopsis LOXs revealed that the 9-LOXs AtLOX1 and AtLOX5 oxygenate linoleic acid and linolenic acid rather than arachidonic acid (Bannenberg et al., 2009). The catalytic efficiency of CaLOX1 (kcat/Km) was higher with linoleic acid than with arachidonic acid, which suggests that linoleic acid is an efficient substrate for the formation of the enzyme-substrate complex. The LOX pathway has several branches and produces many signaling molecules that respond to a variety of pathogens or stress factors. That is, LOXs are the entry point to pathways that provide a variety of oxylipin molecules involved in signaling for stress responses. LOX enzymes oxygenate polyunsaturated fatty acids in a position-specific manner. The biosynthesis of plant oxylipins is initiated by primary fatty acid oxygenases, including the 9- and 13-LOXs (Bannenberg et al., 2009). Specifically, only 13-hydroperoxylinolenic acid produced by 13-LOX leads to the biosynthesis of JA (Feussner and Wasternack, 2002). However, JA is not synthesized through 9-hydroperoxylinolenic acid by 9-LOX. The positional specificity of the LOX enzyme is a crucial factor to determine whether the LOX is involved in the biosynthesis of JA. HPLC analysis of the reaction products for the positional specificity of CaLOX1 revealed that 9-hydroperoxylinolenic acid is mainly produced by the reaction of purified CaLOX1 with linoleic acid as a substrate, which suggests that CaLOX1 is a 9-LOX.
CaLOX1 Is Required for the Cell Death Response in Pepper
CaLOX1 was highly expressed during the HR of pepper leaves during the incompatible interaction with Xcv Bv5-4a. This suggests an essential role for LOX in establishing the HR. HR cell death may be due, in part, to lipid peroxidation initiated by LOX activity. This hypothesis is supported by the findings of Jalloul et al. (2002) and Montillet et al. (2002) that lipid peroxidation and LOX activity are stimulated in the HR of plants to pathogenic bacteria. Importantly, LOX activity also increased rapidly and to a higher level in HR cell death during Xcv infection. The accumulation of LOXs is specifically associated with leaf cell death in Arabidopsis (Montillet et al., 2002). LOX activation may be associated with the HR and H2O2 oxidative stress to induce programmed cell death (Maccarrone et al., 2001).
CaLOX1-dependent cell death was observed in parallel with enhanced CaLOX1-transient expression in pepper plants infiltrated with Agrobacterium (35S:CaLOX1). Intriguingly, CaLOX1 transient expression strongly induced the SA-responsive gene CaBPR1 and the JA-responsive gene CaDEF1 in pepper leaves. Simultaneously, UV-visible fluorescence and ROS accumulation were also detected in leaves transiently expressing CaLOX1, suggesting an accumulation of ROS and phenolic compounds associated with cell death. These results raise the possibility that CaLOX1 acts as a trigger of cell death responses in pepper plants during pathogen infection. Metabolic products in the LOX pathway are likely to be required for the induction of cell death responses that limit pathogen growth at the infection site. For example, 9- and 13-HPOT cause cell death in lentil (Lens culinaris) root protoplasts (Maccarrone et al., 2000). Signal transduction pathways that are dependent on SA or JA may be activated during the cell death response in pepper transiently expressing CaLOX1, leading to biosynthesis or the release of potential antimicrobial effector molecules (Morel and Dangl, 1997). HR cell death is closely related to the generation of lipid peroxides and ROS (Porta and Rocha-Sosa, 2002). Accumulation of hydrogen peroxides in the cell death region in CaLOX1-transiently expressing pepper leaves is consistent with the findings of Maccarrone et al. (2000) that early production of H2O2 during the HR in lentil root protoplasts induces cell death as well as an increase in LOX activity.
Silencing of CaLOX1 Suppresses Defense Responses to Microbial Pathogens in Pepper
To investigate the loss of function of CaLOX1 in pepper plants, we used the VIGS technique with the TRV vector system, a fast and highly effective tool for gene down-regulation (Liu et al., 2002; Hein et al., 2005; Sarowar et al., 2007). CaLOX1-silenced pepper plants showed not only enhanced susceptibility to the virulent fungus C. coccodes and the bacterial pathogen Xcv Ds1 but also a delayed HR to the avirulent Xcv Bv5-4a. These observations suggest that CaLOX1 is required for basal defense and HR-mediated resistance to C. coccodes and Xcv infection. Interestingly, reduced induction of CaBPR1 (a SA marker gene) in the silenced plants was intimately associated with the inhibition of SA accumulation during the virulent Xcv infection. The suppression of CaLOX1 in CaLOX1-silenced plants also seems to be crucial for the inhibition of HR-associated cell death. Importantly, levels of CaLOX1 transcripts in unsilenced and CaLOX1-silenced plants paralleled LOX activities. CaLOX1 induction and LOX activity were low in CaLOX1-silenced plants. These results are consistent with the earlier demonstration that LOX transcript accumulation patterns and LOX activities are coincident in potato (Kolomiets et al., 2000). In our study, VIGS of the CaLOX1 gene also inhibited localized cell death in pepper leaves infected with pathogens, which was accompanied by the suppression of H2O2 accumulation, lipid peroxidation, SA accumulation, and cell death-related genes. These results suggest that lipid peroxidation and ROS and SA accumulation in pepper are crucial for the execution of hypersensitive cell death and defense responses induced by CaLOX1 expression.
Expression of CaLOX1 and the CaLOX1 Ortholog AtLOX1 Confers Basal Resistance to a Broad Range of Pathogens in Arabidopsis
CaLOX1 overexpression in Arabidopsis plants conferred enhanced resistance to Pst DC3000, Pst DC3000 (avrRpm1), H. arabidopsidis, and A. brassicicola. Infection with Pst DC3000 (avrRpm1) induced a HR and H2O2 accumulation in CaLOX1-OX Arabidopsis. During Pst DC3000 infection, CaLOX1-OX plants exhibited small HR-like necrotic lesions at the infection site and inhibition of pathogen growth. Infection of CaLOX1-OX leaves by pathogens resulted in oxidative stress, which led to the irreversible damage of cellular membranes and the appearance of necrotic disease symptoms. Moreover, CaLOX1-OX Arabidopsis plants exhibited an increase in LOX activity compared with wild-type plants. Upon infection of CaLOX1-OX plants with Pst DC3000 and Pst DC3000 (avrRpm1), LOX activity was significantly enhanced, suggesting that CaLOX1 is critically involved in the defense response in heterologous Arabidopsis. In contrast, plants homozygous for T-DNA insertions of the Arabidopsis CaLOX1 ortholog (the lox1-1 and lox1-2 mutants) were susceptible to infection by Pst DC3000, H. arabidopsidis, and A. brassicicola. As expected, we observed decreased LOX activity in the Arabidopsis lox1 mutant relative to wild-type plants during Pst infection. Interestingly, this suggests that AtLOX1 may function in the Arabidopsis defense response to Pst infection in a manner similar to that in CaLOX1-OX Arabidopsis lines. CaLOX1 overexpression in transgenic Arabidopsis plants enhanced AtPR1 expression during Pst DC3000 and Pst DC3000 (avrRpm1) infection. The increased level of SA-responsive AtPR1 expression in CaLOX1-OX Arabidopsis plants supports the notion that CaLOX1 may be involved in SA-mediated signaling pathways during pathogen infection. In contrast, JA-mediated signaling may not affect disease resistance in Arabidopsis, because JA levels were similar in wild-type and CaLOX1-OX plants.
More specifically, H2O2 production, lipid peroxidation, and SA accumulation in CaLOX1-OX Arabidopsis plants may be involved in the execution of cell death and defense response to Pst infection, as suggested in tobacco by Montillet et al. (2002, 2005). Plant LOXs have been proposed to play a role in responses to wounding and JA, which triggers gene activation during wound response and necrotrophic fungal pathogen infection (Bleé, 2002). One such pathogen, A. brassicicola, kills host cells at early stages of infection, causing tissue damage (Glazebrook, 2005). Infection with A. brassicicola resulted in a resistance response in wild-type and CaLOX1-OX transgenic leaves. These resistance symptoms are consistent with the HR in Arabidopsis Col-0 plants infected by A. brassicicola (Narusaka et al., 2003). In contrast, lox1 mutants were highly susceptible to A. brassicicola. The remarkable involvement of CaLOX1 and AtLOX1 in mediating resistance to pathogen attack suggests that these LOX genes are highly conserved for disease resistance in plants. Analyses of CaLOX1-OX and lox1 mutant plants revealed that plant disease resistance is conferred by LOX gene expression during pathogen infection.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Pepper (Capsicum annuum ‘Nockwang’) plants were raised in a plastic tray (55 × 35 × 15 cm) after seed germination. At the two-leaf stage, plants were transplanted and grown in pots containing mixed soil (peat moss:vermiculite:perlite, 3:1:1, v/v/v) at 28°C with a 16-h-light/8-h-dark regime at a light intensity of 70 μmol photons m−2 s−1. 35S:CaLOX1, lox1-1, lox1-2, and wild-type plants in the Arabidopsis (Arabidopsis thaliana) Col-0 background were used in this study. Arabidopsis plants were grown in pots containing peat moss, perlite, and vermiculite (2:1:2, v/v/v) at 24°C with a 14-h-light/10-h-dark regime at a light intensity of 130 μmol photons m−2 s−1. Prior to sowing on soil, seeds were vernalized at 4°C for 2 d to overcome dormancy.
cDNA Library Screening and DNA Sequencing
A pepper cDNA library was constructed from pepper leaves 18 h after inoculation with Xanthomonas campestris pv vesicatoria. The cDNAs were cloned into the vector λ ZAPII using the λ ZAPII-cDNA library synthesis kit (Stratagene). Pathogen-induced cDNAs were identified using cDNA probes from uninoculated pepper leaves or leaves inoculated with the Xcv avirulent strain Bv5-4a (Jung and Hwang, 2000). DNA sequencing was performed with an ABI 310 DNA sequencer (PE Biosystems) using PRISM BigDYE Terminator sequencing ready reaction kits. Nucleotide sequences were assembled and analyzed using DNA Strider 1.2.1. Database searches were run with the BLASTx program (http://www.ncbi.nlm.nih.gov/BLAST/) and the ExPASy Proteomics Server (http://www.expasy.org). The CaLOX1 cDNA was found among sequenced clones.
Production of Recombinant CaLOX1 in Escherichia coli
The CaLOX1 gene was cloned into the vector pET28a to generate pET28a∷CaLOX1, which encodes CaLOX1 with a His tag fused to the N terminus. The CaLOX1 gene was amplified by PCR with the forward primer 5′-GGATCCATGTTACTGGAAAAGATTG-3′ with a BamHI site and the reverse primer 5′-GAATTCTATATCGACACACTGTTGGGTA-3′ with an EcoRI site, based on the CaLOX1 full sequence. For CaLOX1 expression, pET28a∷CaLOX1 was transformed into E. coli BL21 (DE3). Transformant bacteria were incubated in a flask containing 50 mL of Luria-Bertani medium supplemented with kanamycin (50 μg mL−1) and cultured at 37°C, until optical density at 600 nm (OD600) reached approximately 0.5. CaLOX1 protein expression was induced in the bacterial culture for 10 h at 18°C by the addition of isopropyl β-d-thiogalactopyranoside (IPTG) to 0.5 mm. pET28a∷CaLOX1 was purified with 1.5 mL of nickel-nitrilotriacetate agarose resin (Invitrogen) in a 10-mL purification column, from which the target protein was eluted with an imidazole solution (250 mm). Purified CaLOX1 was subjected to 10% SDS-PAGE and stained with Coomassie Brilliant Blue to confirm purity.
CaLOX1 Enzyme Assay
Protein extracts were prepared from healthy and bacteria-infected pepper and Arabidopsis leaves, which were homogenized with 1 mL of 0.1 m sodium phosphate buffer (pH 6.5) using a pestle and mortar. The homogenate was centrifuged at 15,000g for 30 min at 4°C, and the resulting supernatant was used as the enzyme source. Protein concentrations of crude fractions were determined with a dye-binding protein assay kit (Bio-Rad) following the manufacturer's instructions with bovine serum albumin as a standard.
LOX activity in the soluble fractions was spectrophotometrically measured by monitoring the increase of the conjugated diene hydroperoxide at A234 at 25°C for 10 min (Richard et al., 1992; Rancé et al., 1998). The assay mixture included 0.1 m sodium phosphate buffer (pH 6.5), 0.1 mm linoleic acid (Sigma), and 5 to 10 μL of resuspended enzyme in a total volume of 1.2 mL. The enzyme reaction was initiated by adding purified CaLOX1 from E. coli or protein extracts from plants, and the change in absorbance was recorded. LOX activity is expressed in nkat mg−1 protein using a molar extinction coefficient of 25,000 m−1 cm−1.
Kinetic parameters of hydroperoxidation of linoleic acid and arachidonic acid for purified CaLOX1 were analyzed, and the linear parts of the kinetic progress curves were evaluated as described (Seo et al., 2001). Various concentrations (0–200 μm) of substrates were tested to determine the kinetic parameters. Km, Vmax, and kcat for each substrate were calculated using Prism 5.0 (GraphPad Software). Kinetic parameters were determined by analyzing Michaelis-Menten plots from three independent experiments.
Positional Specificity of CaLOX1
Positional specificity of CaLOX1 protein was analyzed by silica gel HPLC using a Zorbax SIL column (4.6 × 150 mm; Agilent). Purified recombinant CaLOX1 protein was incubated for 5 min in 0.1 m sodium phosphate buffer (pH 6.5) containing 0.1 mm linoleic acid (Sigma) at 25°C. The reaction was stopped by the addition of 0.1 m HCl. Reaction products were extracted in 10:1 (v/v) hexane:ethanol. After shaking for 1 min, the organic layer was applied to the HPLC column using a solvent system of hexane:ethyl ether:acetic acid (70:30:1, v/v/v) and a flow rate of 1 mL min−1. 9-HPOT and 13-HPOT standards were purchased from Cayman Chemical (Mizuno et al., 2003).
Pathogen Inoculation and Disease Rating
Pepper plants were inoculated with the virulent strain Ds1 and avirulent strain Bv5-4a of Xcv (Jung and Hwang, 2000). Xcv was grown at 28°C on yeast nutrient broth (5 g of yeast extract, 8 g of nutrient broth, and 1 L of water). Pepper plants at the six-leaf stage were inoculated by infiltrating bacterial suspension (5 × 108 cfu mL−1; Chung et al., 2007; Kim et al., 2007). Inoculated plants were incubated for 18 h in a moist chamber at 28°C and then in a growth room as described previously (Jung and Hwang, 2000). Infected leaves were sampled at various time points after inoculation.
Pseudomonas syringae pv tomato strain DC3000 and Pst DC3000 (avrRpm1) were used. The bacteria were grown at 28°C in yeast nutrient broth containing rifampicin (50 mg L−1). To study bacterial growth, bacterial suspensions (106 cfu mL−1) were infiltrated into Arabidopsis leaves using a syringe. At appropriate time points, three independent leaves infiltrated with Pst were harvested.
Colletotrichum coccodes isolate 2-25 was used in this study. The fungus was grown on oatmeal agar plates for 5 to 7 d at 28°C in the dark. Conidia concentration was adjusted to 5 × 105 mL−1 with sterile tap water using a hemacytometer. Pepper plants at the six-leaf stage were sprayed with the conidial suspension in 0.05% Tween 20, incubated in a moist chamber in the dark for 48 h at 28°C, and then raised in the growth room.
Hyaloperonospora arabidopsidis isolate Noco2 was maintained on Arabidopsis Col-0 plants by weekly subculturing. To produce large quantities of inoculum, 7- to 10-d-old seedlings were inoculated with H. arabidopsidis and the spores produced on cotyledons were collected in water. Seven-day-old seedlings were sprayed with a suspension of asexual inoculum (5 × 104 conidiosporangia mL−1). The inoculated seedlings were covered with a transparent dome to maintain high humidity (80%–100%) and grown for 7 d at 17°C (Lee et al., 2008). Seven days after inoculation, sporangiophore production was monitored using a dissection microscope. Asexual sporulation of H. arabidopsidis was visually assessed by counting the number of sporangiophores on both sides of the cotyledons. Disease ratings consisted of five classes based on the number of sporangiophores: zero to five, six to 10, 11 to 15, 16 to 20, and over 20 sporangiophores per cotyledon.
Alternaria brassicicola was cultured on potato dextrose agar medium at 24°C for 10 d. Spore concentrations were determined using a hemacytometer and adjusted to 5 × 104 conidia mL−1. A. brassicicola was inoculated by placing 10-μL droplets of suspension on leaves of 5-week-old plants. For mock treatment, 10-μL droplets of water were placed onto the leaves. Inoculated plants were kept at 100% relative humidity at 24°C. Four days after inoculation, lesion diameters and the numbers of newly formed spores were measured (Oh et al., 2005).
Stress Treatment
SA (5 mm), methyl viologen (10 mm), ABA (100 μm), and MeJA (100 μm) were applied onto pepper leaves at the six-leaf stage. Plants treated with MeJA were tightly sealed in a plastic bag. For ethylene treatment, the plants were sealed in a flask and aliquots of pure ethylene gas were injected with a syringe into the flask to 10 μL L−1. For salt treatment, pepper plants were removed from the soil and soaked in 200 mm NaCl. For cold treatment, pepper plants were exposed to low temperature (4°C). To induce drought stress, plants were exposed to dehydration by withholding water. For wounding experiments, leaves were pricked with a needle. The treated pepper plants were sampled at various time points after treatment, frozen in liquid nitrogen, and stored at −70°C until used for RNA isolation.
RNA Gel-Blot and RT-PCR Analysis
Total RNA was isolated from leaf tissues of pepper and Arabidopsis plants using TRIzol (Invitrogen) according to the manufacturer's instructions. For RNA gel-blot analysis, total RNA (10 μg) was separated on 1.2% formaldehyde agarose gels and transferred onto a nylon membrane (Hybond+; Amersham Pharmacia Biosciences), followed by UV cross-linking. Probes were prepared from the corresponding pepper and Arabidopsis cDNAs using 14-dCTP-biotin (Invitrogen). The membrane was prehybridized for 1 h and hybridized for 16 h in dextran solution (1 mm EDTA, 7% [w/v] SDS, 0.25 m disodium phosphate, pH 7.2, and 5% dextran sulfate) with a labeled probe at 65°C. Bands were detected using x-ray film. For RT-PCR analysis, first-strand cDNA was synthesized using 1 μg of total RNA, oligo(dT) primer, and avian myeloblastosis virus reverse transcriptase (Bio Basic) following the manufacturer's instructions. Aliquots of RT reaction products were used for RT-PCR using ExTaq DNA polymerase (Takara) with the following gene-specific primers: CaLOX1 forward (5′-TGCAGGTTACCTCCCAAATCGCCCA-3′) and CaLOX1 reverse (5′-CTATATCGACACACTGTTGGGTATTCCTT-3′); CaBPR1 forward (5′-CAGGATGCAACACTCTGGTGG-3′) and CaBPR1 reverse (5′-ATCAAAGGCCGGTTGGTC-3′) for CaBPR1 (accession no. AF053343); and CaDEF1 forward (5′-CAAGGGAGTATGTGCTAGTGAGAC-3′) and CaDEF1 reverse (5′-TGCACAGCACTATCATTGCATAC-3′) for CaDEF1 (accession no. AF442388).
TRV-Based VIGS of the CaLOX1 Gene in Pepper
pTRV1 and pTRV2 were used for VIGS of the CaLOX1 gene (Liu et al., 2002). To specifically silence CaLOX1, a 621-bp fragment of the CaLOX1 cDNA was amplified by PCR and cloned into pTRV2 to generate pTRV2:CaLOX1. Agrobacterium tumefaciens GV3101 containing pTRV1 or pTRV2:CaLOX1 was coinfiltrated into fully expanded pepper cotyledons using a syringe (OD600 = 0.4). As a positive control, cotyledons were infiltrated with a VIGS clone containing the Phytoene Desaturase gene. As a negative control, cotyledons were infiltrated with an empty vector clone (TRV:00). Plants were placed in a growth room at 24°C with a 16-h-light/8-h-dark photoperiod cycle for growth and viral spread. The virulent Ds1 and avirulent Bv5-4a strains of Xcv were infiltrated into unsilenced (TRV:00) and silenced (TRV:CaLOX1) pepper leaves to examine gene expression (106 cfu mL−1) and to assay bacterial growth (5 × 104 cfu mL−1).
Transgene Construction and Plant Transformation
To generate the 35S:CaLOX1 construct, a full-length CaLOX1 fragment was amplified by PCR and subcloned into the vector pCR2.1-TOPO (Invitrogen). CaLOX1 was digested with XbaI and BamHI and inserted into the vector pBIN 35S. Constructs were verified by sequencing. Transgenic Arabidopsis plants expressing CaLOX1 were generated by the floral dip method using Agrobacterium GV3101 (Clough and Bent, 1998). Transformed seeds were selected on Murashige and Skoog (Duchefa) plates containing 50 μg mL−1 kanamycin. Independent kanamycin-resistant transgenic plants were identified by PCR amplification of an cDNA insert of the expected size.
Transient Expression Assay of CaLOX1
The CaLOX1 construct was cloned in pBIN 35S and expressed in Agrobacterium GV3101, which was cultured to OD600 = 1.0 in induction medium (10 mm ethanesulfonic acid, pH 5.7, 10 mm MgCl2, and 200 μm acetosyringone) and diluted to OD600 = 0.02. Bacterial suspensions expressing CaLOX1 were injected at different concentrations into pepper leaves using a syringe.
Protein Extraction and Immunoblotting
Agrobacterium-mediated transient expression of c-Myc-tagged 35S:CaLOX1 and c-Myc-tagged empty vector in pepper leaves was analyzed by immunoblotting using an anti-c-Myc antibody (Sigma). For protein extraction, pepper leaves were harvested at different time points after infiltration with Agrobacterium GV3101 harboring these vectors. Total proteins were obtained from 1 g of leaf tissue homogenized in 1 mL of extraction buffer (50 mm HEPES, pH 7.4, 50 mm NaCl, 10 mm EDTA, 0.2% Triton X-100, and 1× proteinase inhibitor cocktail [Roche]). Crude protein extracts were immunoprecipitated on an anti-c-Myc agarose affinity gel (Sigma) overnight at 4°C. The immunoprecipitates were rinsed four times with 1× phosphate-buffered saline buffer, resuspended in SDS sample loading buffer, subjected to SDS-PAGE, and transferred onto Hybond-P membranes (GE Healthcare Bioscience) by wet electroblotting. Fusion proteins were detected using a mouse anti-c-Myc antibody (Sigma) at 1:2,000 dilution.
Identification of T-DNA Insertion Lines
Homozygous insertion mutant seeds were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/abrc/) at Ohio State University. The lox1-1 (SALK_059431) and lox1-2 (SALK_000058) mutants were isolated from the SALK T-DNA lines. The seeds were planted on Murashige and Skoog agar plates containing kanamycin (50 μg mL−1), and kanamycin-resistant plants were transferred to soil. Mutant plants were confirmed by PCR using T-DNA and gene-specific primer sets, as described on the T-DNA Express homepage (http://signal.salk.edu/tdnaprimers.html). Two sets of PCR were carried out using the three primers LP (left gene-specific primer), RP (right gene-specific primer), and LB (left border primer of the T-DNA insertion) for each SALK line: lox1-1 LP primer, 5′-AGCTCCTTGAACCTCACTTCC-3′; lox1-1 RP primer, 5′-GAGACGCTATTTGGAATTCCC-3′; lox1-2 LP primer, 5′-TTTTCTTTTTGCGTTCTCGAG-3′; lox1-2 RP primer, 5′-CCACTGATAAAAAGAATATTTTGCC.-3′; and LBa1 primer, 5′-TGGTTCACGTAGTGGGCCATCG-3′.
Measurement of Ion Leakage
Leaves of 5-week-old Arabidopsis plants were infiltrated with 106 cfu mL−1 Pst D3000 and Pst DC3000 (avrRpm1) to determine ion leakage. Leaf discs (1 cm in diameter) were removed immediately following infiltration, washed in 30 mL of double distilled water for 30 min, and transferred into 20 mL of double distilled water. Conductance was then measured with a conductivity meter (model sensION7; Hach; Mackey et al., 2003).
Quantification of Lipid Peroxidation
Lipid peroxidation was estimated by determining the MDA contents in pepper and Arabidopsis plants as described previously (Heath and Packer, 1968). Leaf samples (200 mg) were homogenized in 0.5% TCA. The homogenates were centrifuged at 10,000g for 15 min at 4°C. Extracts was mixed with 0.5% thiobarbituric acid prepared in 20% TCA and incubated at 95°C for 30 min. The reaction was stopped on ice, and samples were centrifuged at 12,000g for 15 min. The absorbance of the resultant supernatant was measured at 532 and 600 nm. The nonspecific A600 was subtracted from the A532. The MDA concentration was determined using the extinction coefficient 155 mm−1 cm−1.
Staining with DAB, Trypan Blue, and Aniline Blue
To monitor plant cell death and fungal growth, control leaves or leaves inoculated with pathogens were stained with 1 mg mL−1 DAB (Sigma), lactophenol-trypan blue (10 mL of lactic acid, 10 mL of glycerol, 10 g of phenol, and 10 mg of trypan blue, dissolved in 10 mL of distilled water), and 0.01% aniline blue in 150 mm K2HPO4, pH 9.5. After DAB staining overnight, the leaves were cleared by boiling for 10 min in ethanol and destained overnight in ethanol. For trypan blue staining, infected plants were boiled for 5 min in the staining solution and destained overnight in chloral hydrate (2.5 g mL−1). For aniline blue staining, infected plants were cleared of chlorophyll using alcoholic lactophenol and rinsed in 50% ethanol and water. The samples were then stained for 30 min with 0.01% aniline blue (Rigano et al., 2007) and mounted in 70% glycerol for microscopic observation.
Quantification of H2O2 by Xylenol Orange Assay
H2O2 production in plants was spectrophotometrically measured using xylenol orange assay, which forms a complex with the Fe3+ produced by the hydroperoxide-based oxidation of Fe2+ (Bindschedler et al., 2001). One milliliter of assay reagent [25 mm FeSO4 and 25 mm (NH4)2SO4, dissolved in 2.5 m H2SO4] was added to 100 mL of 125 μm xylenol orange and 100 mm sorbitol. To measure the H2O2 amount, leaf discs excised using a cork borer were withdrawn and centrifuged at 5,000g for 10 min, and 100 μL of the supernatant was added to 1 mL of xylenol orange reagent. After 30 min of incubation, the peroxide-mediated oxidation of Fe2+ to Fe3+ was determined by measuring the A560 of the Fe3+-xylenol orange complex.
Measurement of SA
SA and SA glycoside were extracted and quantified according to the method described by Verberne et al. (2002). Leaf tissue samples (0.5 g) were frozen in liquid nitrogen, ground to a fine power, and sequentially extracted with 90% and 100% methanol. As an internal standard for SA, 3-hydroxybenzoic acid (Sigma) was added at a mass ratio of 50 mg g−1 leaf fresh weight. SA was determined by fluorescence (excitation 305 nm, emission 405 nm) after separation on a C18 reverse-phase HPLC column (Waters).
Extraction and Measurement of JA
For analysis of JA, leaf samples were extracted with ice-cold 100% methanol and centrifuged at 10,000g for 10 min. Supernatants were decanted and pellet reextracted with 100% methanol. Dihydrojasmonic acid was added as an internal standard for quantification of JA. Extraction of JA was continued by rotating the samples at 28°C for 2 h. Extracted samples was adjusted to 70% methanol with ice-cold water, and pH was lowered to 3 with HCl. The samples were passed through a Sep-Pak C18 cartridge (Waters), which has been prewashed with 70% methanol. Cartridges were washed with 70% methanol, and eluates were combined. Samples were partitioned with CHCl3, dried over anhydrous MgSO4, and methylated with methylation reagent followed by hexane:tert-butyl methyl ether (1:1, v/v). The supernatants were decanted and stored at −20°C. JA was analyzed by gas chromatography-mass spectrometry (Agilent 6890) with a DB-5MS column (30 m × 0.25 mm, 0.25 μm; J&W Scientific). The temperature gradient was 60°C for 1 min, 60°C to 190°C at 15°C min−1, 190°C to 220°C at 5°C min−1, 220°C to 290°C at 25°C min−1, and 290°C for 1 min.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ377808 (CaLOX1), AF053343 (CaBPR1), AF442388 (CaDEF1), AF442387 (CaPOA1), AF313766 (CaSAR82), AF244122 (CaPR4), AF244121 (CaPR10), At1g55020 (AtLOX1), and At3g62250 (AtUBQ5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide and derived amino acid sequences of CaLOX1 cDNA encoding pepper LOX.
Supplemental Figure S2. Alignment of the deduced amino acid sequence of CaLOX1 with other plant LOXs.
Supplemental Figure S3. LOX activity of recombinant His tag-CaLOX1 expressed in E. coli BL21 (DE3).
Supplemental Figure S4. Levels of JA in pepper and Arabidopsis.
Supplementary Material
Acknowledgments
We thank Dr. S.P. Dinesh-Kumar (Yale University) for the pTRV1 and pTRV2 vectors and Dr. U. Bonas (Martin-Luther-Universitaet) for the A. tumefaciens strain GV3101.
This work was supported by the Crop Functional Genomics Center of the 21st Century, Frontier Research Program (grant no. CG1133), funded by the Ministry of Education, Science, and Technology, Korea, and by the BioGreen21 Program (grant no. 20070401034028), Rural Development Administration, Korea.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Byung Kook Hwang (bkhwang@korea.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bannenberg G, Martínez M, Hamberg M, Castresana C (2009) Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 44 85–95 [DOI] [PubMed] [Google Scholar]
- Baulcombe DC (1999) Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol 2 109–113 [DOI] [PubMed] [Google Scholar]
- Bindschedler LV, Minibayeva F, Gardber SL, Gerrish C, Davies DR, Bolwell GP (2001) Early signalling events in the apoplastic oxidative burst in suspension cultured French bean cells involve cAMP and Ca2+. New Phytol 151 185–194 [DOI] [PubMed] [Google Scholar]
- Bleé E (2002) Impact of phyto-oxylipins in plant defense. Trends Plant Sci 7 315–322 [DOI] [PubMed] [Google Scholar]
- Brash AR (1999) Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J Biol Chem 274 23679–23682 [DOI] [PubMed] [Google Scholar]
- Buonaurio R, Servili M (1999) Involvement of lipoxygenase, lipoxygenase pathway volatiles, and lipid peroxidation during the hypersensitive reaction of pepper leaves to Xanthomonas campestris pv. vesicatoria. Physiol Mol Plant Pathol 54 155–169 [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP (2006) Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey R, Hughes RK (2004) Recombinant lipoxygenases and oxylipin metabolism in relation to food quality. Food Biotechnol 18 133–170 [Google Scholar]
- Choi HW, Kim YJ, Lee SC, Hong JK, Hwang BK (2007) Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol 145 890–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HW, Lee BG, Kim NH, Park Y, Lim CW, Song HK, Hwang BK (2008) A role for a menthone reductase in resistance against microbial pathogens in plants. Plant Physiol 148 383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe V, Martina R, Joelle F, Marie-Laure P, Marie-Thérèse ET (1996) Lipoxygenase gene expression in the tobacco-Phytophthora parasitica nicotianae interaction. Plant Physiol 112 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E, Oh SK, Park JM, Choi D (2007) Expression and promoter analyses of pepper CaCDPK4 (Capsicum annuum calcium dependent protein kinase 4) during plant defense response to incompatible pathogen. Plant Pathol J 145 890–904 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Croft KPC, Juttner F, Slusarenko AJ (1993) Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv. phaseolicola. Plant Physiol 101 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53 275–297 [DOI] [PubMed] [Google Scholar]
- Gao X, Brodhagen M, Isakeit T, Brown SH, Göbel C, Betran J, Feussner I, Keller NP, Kolomiets M (2009) Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol Plant Microbe Interact 22 222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Shim WB, Göbel C, Kunze S, Feussner I, Meeley R, Balint-Kurti P, Kolomiets M (2007) Disruption of a maize 9-lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol Plant Microbe Interact 20 922–933 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43 205–227 [DOI] [PubMed] [Google Scholar]
- Göbel C, Feussner I, Rosahl S (2003) Lipid peroxidation during the hypersensitive response in potato in the absence of 9-lipoxygenases. J Biol Chem 278 52834–52840 [DOI] [PubMed] [Google Scholar]
- Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125 189–198 [DOI] [PubMed] [Google Scholar]
- Hein I, Pacak MB, Hrubikova K, Williamson S, Dinesen M, Soenderby IE, Sundar S, Jarmolowski A, Shirasu K, Lacomme C (2005) Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol 138 2155–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JK, Hwang BK (2005) Induction of enhanced disease resistance and oxidative stress tolerance by overexpression of pepper basic PR-1 gene in Arabidopsis. Physiol Plant 124 267–277 [Google Scholar]
- Hong JK, Jung HW, Kim YJ, Hwang BK (2000) Pepper gene encoding a basic class II chitinase is inducible by pathogen and ethephon. Plant Sci 159 39–49 [DOI] [PubMed] [Google Scholar]
- Hornung E, Walter M, Kühn H, Feussner I (1999) Conversion of cucumber linoleate 13-lipoxygenase to a 9-lipoxygenating species by site-directed mutagenesis. Proc Natl Acad Sci USA 96 4192–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalloul A, Montillet JL, Assigbetse K, Agnel JP, Delannoy E, Triantaphylides C, Daniel JF, Marmey P, Geiger JP, Nicole M (2002) Lipid peroxidation in cotton:Xanthomonas interactions and the role of lipoxygenases during the hypersensitive reaction. Plant J 32 1–12 [DOI] [PubMed] [Google Scholar]
- Jung HW, Hwang BK (2000) Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 13 136–142 [DOI] [PubMed] [Google Scholar]
- Kenton P, Mur LAJ, Atzorn R, Wasternack C, Draper J (1999) Jasmonic acid accumulation in tobacco hypersensitive response lesions. Mol Plant Microbe Interact 12 74–78 [Google Scholar]
- Kim BS, Kim YC, Shin KS, Kim JH (2007) Near-isogenic lines for genes conferring hypersensitive resistance to bacterial spot in chili pepper. Plant Pathol J 23 155–160 [Google Scholar]
- Kim YJ, Hwang BK (2000) Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol Plant 108 51–60 [Google Scholar]
- Kolomiets MV, Chen H, Gladon RJ, Braun EJ, Hannapel DJ (2000) A leaf lipoxygenase of potato induced specifically by pathogen infection. Plant Physiol 124 1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomiets MV, Hannapel DJ, Gladon RJ (1996) Nucleotide sequence of a cDNA clone for a lipoxygenase from abscisic acid-treated potato leaves (accession no. U60202) (PGR 96–069). Plant Physiol 112 44512226401 [Google Scholar]
- Lee SC, Hong JK, Kim YJ, Hwang BK (2000) Pepper gene encoding thionin is differentially induced by pathogens, ethylene and methyl jasmonate. Physiol Mol Plant Pathol 56 207–216 [Google Scholar]
- Lee SC, Hwang BK (2003) Identification of the pepper SAR8.2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta 216 387–396 [DOI] [PubMed] [Google Scholar]
- Lee SC, Hwang IS, Choi HW, Hwang BK (2008) Involvement of the pepper antimicrobial protein CaAMP1 gene in broad spectrum disease resistance. Plant Physiol 148 1004–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Kim YJ, Hwang BK (2001) A pathogen-induced chitin binding protein gene from pepper: its isolation and differential expression in pepper tissues treated with pathogens, ethephon, methyl jasmonate or wounding. Plant Cell Physiol 42 1321–1330 [DOI] [PubMed] [Google Scholar]
- Liavonchanka A, Feussner I (2006) Lipoxygenases: occurrence, functions and catalysis. J Plant Physiol 163 348–357 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar S (2002) Virus-induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- Lorenzi V, Maury J, Casanova J, Berti L (2006) Purification, product characterization and kinetic properties of lipoxygenase from olive fruit (Olea europaea L.). Plant Physiol Biochem 44 450–454 [DOI] [PubMed] [Google Scholar]
- Maccarrone M, Melino G, Finazzi-Agro A (2001) Lipoxygenases and their involvement in programmed cell death. Cell Death Differ 8 776–784 [DOI] [PubMed] [Google Scholar]
- Maccarrone M, Van Zadelhoff G, Veldink GA, Vliegenthart JFG, Finazzi-Agro A (2000) Early activation of lipoxygenase in lentil (Lens culinaris) root protoplasts by oxidative stress induces programmed cell death. Eur J Biochem 267 5078–5084 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector avrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389 [DOI] [PubMed] [Google Scholar]
- Marmey P, Jalloul A, Alhamdia M, Assigbetse K, Cacas JL, Voloudakis AE, Champion A, Clerivet A, Montillet JL, Nicole M (2007) The 9-lipoxygenase GhLOX1 gene is associated with the hypersensitive reaction of cotton Gossypium hirsutum to Xanthomonas campestris pv. malvacearum. Plant Physiol Biochem 45 596–606 [DOI] [PubMed] [Google Scholar]
- Melan MA, Dong X, Endara ME, Davis KR, Ausubel FM, Peterman TK (1993) An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol 101 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Iida T, Takano A, Yokoyama M, Fujimura T (2003) A new 9-lipoxygenase cDNA from developing rice seeds. Plant Cell Physiol 44 1168–1175 [DOI] [PubMed] [Google Scholar]
- Montillet JL, Agne JP, Ponchet M, Vailleau F, Roby D, Triantaphylidès C (2002) Lipoxygenase-mediated production of fatty acid hydroperoxides is a specific signature of the hypersensitive reaction in plants. Plant Physiol Biochem 40 633–639 [Google Scholar]
- Montillet JL, Chamnongpol S, Rusterucci C, Dat J, van de Cotte B, Agnel JP, Battesti C, Inze D, Van Breusegem F, Triantaphylides C (2005) Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Dangl JL (1997) The hypersensitive response and the induction of cell death in plants. Cell Death Differ 4 671–683 [DOI] [PubMed] [Google Scholar]
- Moreno JI, Martin R, Castresana C (2005) Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J 41 451–463 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Narusaka M, Seki M, Ishida J, Nakashima M, Kamiya A, Enju A, Sakurai T, Satoh M, Kobayashi M, et al (2003) The cDNA microarray analysis using an Arabidopsis pad3 mutant reveals the expression profiles and classification of genes induced by Alternaria brassicicola attack. Plant Cell Physiol 44 377–387 [DOI] [PubMed] [Google Scholar]
- Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee SM, Cheong HS, Park OM (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi EW, Hutcheson SW, Baker CJ (1992) Early physiological responses associated with race-specific recognition in soybean leaf tissue and cell suspensions treated with Pseudomonas syringae pv. glycinea. Physiol Mol Plant Pathol 40 173–180 [Google Scholar]
- Porta H, Rocha-Sosa M (2002) Plant lipoxygenases: physiological and molecular features. Plant Physiol 130 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge ST, Boyington JC, Gaffney BJ, Amzel LM (1996) Structure conservation in lipoxygenases: structural analysis of soybean lipoxygenase-1 and modeling of human lipoxygenases. Proteins 24 275–291 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Bent AF (2003) Deciphering host resistance and pathogen virulence: the Arabidopsis/Pseudomonas interaction as a model. Mol Plant Pathol 4 517–530 [DOI] [PubMed] [Google Scholar]
- Rancé I, Fournier J, Esquerré-Tugayé MT (1998) The incompatible interaction between Phytophthora parasitica var. nicotianae race 0 and tobacco is suppressed in transgenic plants expressing antisense lipoxygenase sequences. Proc Natl Acad Sci USA 95 6554–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard MB, Yamamoto H, Choi DI, Ricker KE, Bernard LW (1992) Rapid stimulation of 5-lipoxygenase activity in potato by the fungal elicitor arachidonic acid. Plant Physiol 100 1448–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigano LA, Payette C, Brouillard G, Marano MR, Abramowicz L, Torres PS, Yun M, Castagnaro AP, Oirdi ME, Dufour V, et al (2007) Bacterial cyclic β-(1,2)-glucan acts in systemic suppression of plant immune responses. Plant Cell 19 2077–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusterucci C, Montillet JL, Agnel JP, Battesti C, Alonso B, Knoll A, Bessoule JJ, Etienne P, Suty L, Blein JP, et al (1999) Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J Biol Chem 274 36446–36455 [DOI] [PubMed] [Google Scholar]
- Sarowar S, Oh HW, Cho HS, Baek KH, Seong ES, Joung YH, Choi GJ, Lee S, Choi D (2007) Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J 51 792–802 [DOI] [PubMed] [Google Scholar]
- Sayegh-Alhamdia M, Marmey P, Jalloul A, Champion A, Petitot S, Clerivet A, Nicole M (2008) Association of lipoxygenase response with resistance of various cotton genotypes to the bacterial blight disease. J Phytopathol 156 542–549 [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow JN (1991) Plant lipoxygenase: structure and function. Annu Rev Plant Physiol Plant Mol Biol 42 145–188 [Google Scholar]
- Slusarenko AJ (1996) The role of lipoxygenase in plant resistance to infection. In GJ Piazza, ed, Lipoxygenase and Lipoxygenase Pathway Enzymes. AOCS Press, Champaign, IL, pp 176–197
- Verberne MC, Brouwer N, Delbianco F, Bol JF, Linthorst HJM, Verpoorte R (2002) Method for the extraction of the volatile compound salicylic acid from tobacco leaf material. Phytochem Anal 13 45–50 [DOI] [PubMed] [Google Scholar]
- Weber H, Chételat A, Caldelari D, Farmer EE (1999) Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell 11 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.