Phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31) is an enzyme playing a crucial role in photosynthesis of C4 plants. Here, we identify anionic phospholipids as novel regulators that inhibit C4 PEPC activity and provide evidence that the enzyme partially localizes to membranes.
PEPC catalyzes the β-carboxylation of phosphoenolpyruvate (PEP) in a reaction that yields oxaloacetate and inorganic phosphate. In C4 plants, it notably performs the initial fixation of atmospheric CO2 in photosynthesis, while it also has an anaplerotic function in coordinating carbon and nitrogen metabolism in all plants (Chollet et al., 1996; Vidal and Chollet, 1997). The PEPC protein is subject to distinct but interrelated mechanisms of posttranslational regulation by allosteric positive (e.g. Glc-6-P, triose-P) or negative (e.g. l-malate, Asp) effectors, as well as phosphorylation of the protein at its N-terminal domain (Nimmo, 2003).
Previously, we have identified C3 PEPC isoforms as phosphatidic acid (PA)-binding proteins from tomato (Solanum lycopersicum) and Arabidopsis (Arabidopsis thaliana) suspension-cultured cells in a proteomics screen (Testerink et al., 2004). Phospholipids can affect both localization and activity of a diverse range of proteins, including protein kinases and phosphatases (Testerink and Munnik, 2005; Hurley, 2006; Lemmon, 2008; Munnik and Testerink, 2009; Stahelin, 2009), but also directly regulate metabolic enzymes, such as Escherichia coli pyruvate oxidase (Neumann et al., 2008), mammalian CTP:phosphocholine cytidyltransferase (Johnson et al., 2003; Cornell and Taneva, 2006; Taneva et al., 2008), and wheat (Triticum aestivum) phosphoethanolamine N-methyltransferase (Jost et al., 2009).
Using PA-affinity beads, we show here that C4-type PEPC also binds PA. Moreover, we found that C4 PEPC activity, unlike its C3 counterpart, was inhibited by addition of PA. Other anionic phospholipids, but not neutral, zwitterionic, or positively charged lipids, were also able to block PEPC activity. Western analysis of crude biochemical fractions of sorghum (Sorghum bicolor) leaf extracts revealed that although most PEPC was present in the soluble fraction, a subpool was membrane associated. A possible physiological role for PEPC recruitment to membranes is discussed.
C4 PEPC BINDS PA AND ITS ACTIVITY IS INHIBITED BY ANIONIC PHOSPHOLIPIDS
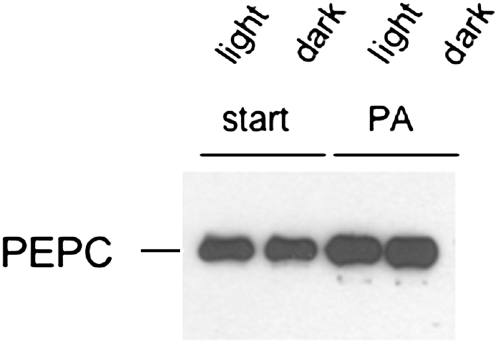
Arabidopsis and tomato C3 PEPC isoforms from suspension cells were previously identified as PA-binding proteins (Testerink et al., 2004). As we are interested in regulation of PEPC from C4 plants, we tested whether C4 PEPC can bind PA. C4 PEPC indeed showed affinity for PA coupled to Sepharose beads (Fig. 1). Similar binding was observed for both illuminated leaves or leaves kept in the dark.
Figure 1.
Sorghum PEPC has affinity for PA. Protein extracts from light- or dark-treated sorghum leaves were analyzed for PA binding using PA-affinity beads described before (Anthony et al., 2004; Testerink et al., 2007). Start material (start; 15% of total input loaded; lanes 1 and 2) and bound fraction (PA; lanes 3 and 4) were analyzed using SDS-PAGE followed by western blotting.
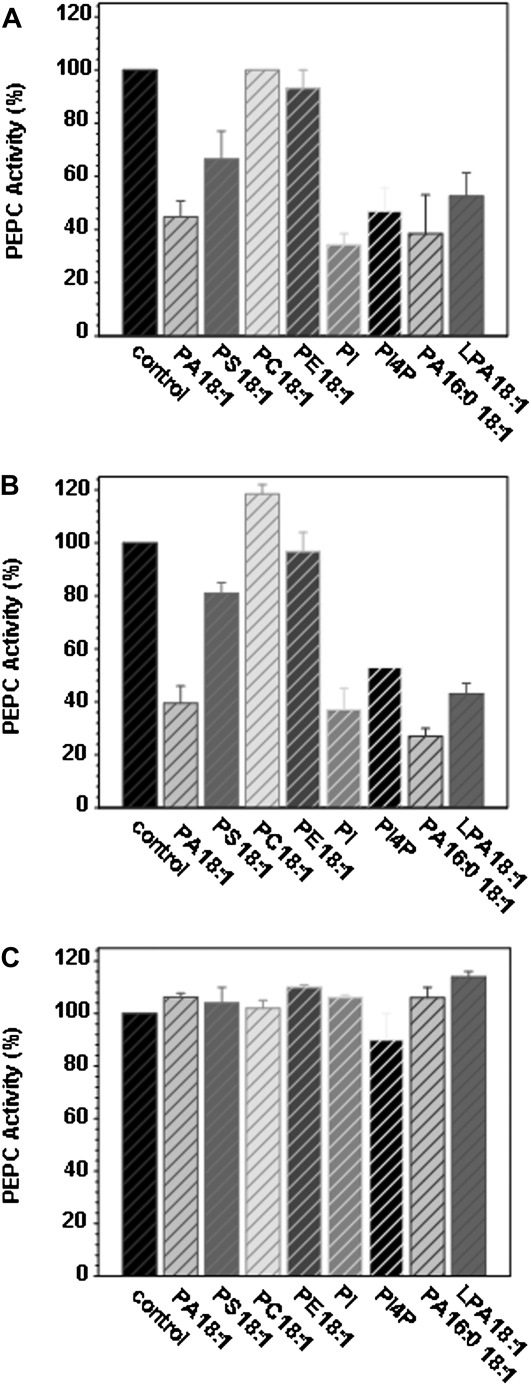
Next, we investigated whether PA, or other phospholipids, affected PEPC activity. PEPC activity was measured in protein extracts from sorghum and maize (Zea mays) leaves. Addition of 50 μm PA decreased PEPC activity to approximately 40% of the control activity for both plants (Fig. 2, A and B). A similar degree of inhibition was observed for other anionic phospholipids, including phosphatidylinositol, phosphatidylinositol-4-P, and lyso-PA. Phosphatidylserine showed a partial inhibition, while the control lipids phosphatidylcholine and phosphatidylethanolamine had no effect. Interestingly, C3-type PEPC from Arabidopsis leaves, which did not bind PA beads (Testerink et al., 2004), was not affected by PA or other anionic phospholipids (Fig. 2C).
Figure 2.
Sorghum and maize PEPC activity are inhibited by anionic phospholipids. Crude extracts were incubated at 30°C during 30 min in the presence of 50 μm of the phospholipids indicated, and PEPC activity was assayed at pH 8.0 as described in Supplemental Materials and Methods S1. A, Sorghum leaves. B, Maize leaves. C, Arabidopsis leaves. Data are means ± se of three to six different experiments. One-hundred percent activity corresponded to 1 unit mL−1 for sorghum and maize and to 0.1 unit mL−1 for Arabidopsis.
C4 PEPC INHIBITION BY PHOSPHOLIPIDS IS DIRECT AND INDEPENDENT OF PHOSPHORYLATION STATUS OR KNOWN ALLOSTERIC REGULATORS
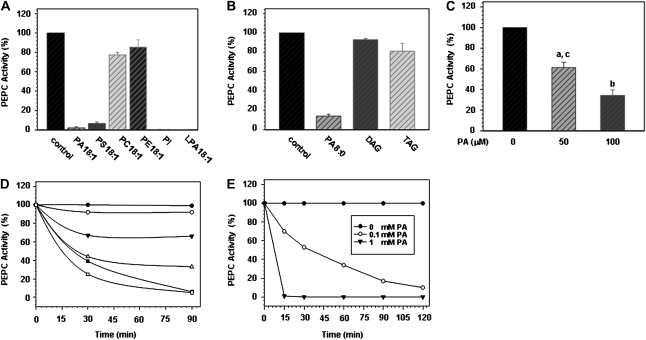
For further characterization, anionic phospholipids were tested on purified sorghum leaf PEPC. We found comparable effects for the physiological (C18:1) PA species and the water-soluble short-chain C8:0 variant of PA, but no effect of the control lipids phosphatidylcholine, phosphatidylethanolamine, or the neutral phospholipids diacylglycerol and triacylglycerol (Fig. 3, A and B). To further characterize the effect of phospholipids on purified PEPC, PA 8:0 was used. PA caused a dose- and time-dependent inactivation of purified sorghum leaf PEPC (Fig. 3, C and D). The concentration of PA necessary to inhibit PEPC activity by 50% (IC50) was calculated to be about 65 μm (Fig. 3D). This increased with increasing the amount of PEPC in the incubation mixture (Supplemental Fig. S1). At 1 mm, PA caused a rapid and complete PEPC inactivation (Fig. 3E), which could not be reversed by preincubation with bovine serum albumin or passage through a Sephadex column (data not shown). The inactivation was not due to precipitation or proteolytic cleavage of the purified PEPC. Instead, native gel electrophoresis of purified PEPC in the absence or presence of PA revealed that PA induced the formation of a higher molecular weight complex (Supplemental Fig. S2).
Figure 3.
Effect of PA on purified sorghum leaf PEPC. A and B, Anionic phospholipids inhibit purified sorghum leaf PEPC. DAG, Diacylglycerol; TAG, triacylglycerol. C, Purified PEPC was incubated with PA (8:0) in 0.1 m Tris pH 8.0 at 30°C for 30 min as described in Supplemental Materials and Methods S1. Data are means ± se of three to six different experiments. a, P < 0.05 versus 0; b, P < 0.01 versus 0; c, P < 0.05 versus 100 (t test). D and E, Time courses of PEPC inactivation at different concentrations of PA (8:0). D, Black diamonds, 0 μm; white diamonds, 20 μm; black triangles, 40 μm; white triangles, 60 μm; black squares, 80 μm; white squares, 100 μm. Data are means of three experiments. Specific activity of the purified PEPC preparation was 50 unit mg−1 protein. The results in Figure 3A corresponded to a concentration of 1 unit μm L−1 (50 μm lipid, 30 min); for Figure 3B this was 3 unit mL−1 (0.1 mm lipid, 30 min) as in Figure 3, C to E. se was always less than 12%.
Allosteric modulators (such as Glc-6-P and l-malate) are known to modify PEPC conformation. Nevertheless, inclusion of G-6-P or malic acid did not change the effect of PA on PEPC (Supplemental Fig. S4). Preincubation of the enzyme with 5 mm PEP prior to the addition of PA did not prevent inactivation either (Supplemental Fig. S4). The effect of the phosphorylation status of PEPC was investigated by specifically phosphorylating the purified enzyme's target Ser with the catalytic subunit of mammalian protein kinase A before incubation with PA. While this treatment clearly increased the IC50 of PEPC toward malate inhibition from 0.4 to 0.8 mm (data not shown), it had no effect on the PEPC-PA interaction (Supplemental Fig. S5). The incubation of PA-inactivated PEPC with protein kinase A (and the rest of the components of the phosphorylation mixture) did not restore PEPC activity either (data not shown). Varying the pH of the incubation mixture from pH 6 to pH 8 had only a slight effect, as lower pH accelerated the rate of inhibition by PA, but maximal inhibition was reached after 1 h in all cases (Supplemental Fig. S3). To monitor whether redox state could change the interaction between PA and PEPC, dithiothreitol was included in the incubation mixture. No effect was observed on the inactivation of PEPC caused by PA (data not shown).
MODIFIED PEPC IS PRESENT IN CRUDE MEMBRANE FRACTIONS OF SORGHUM LEAF EXTRACTS
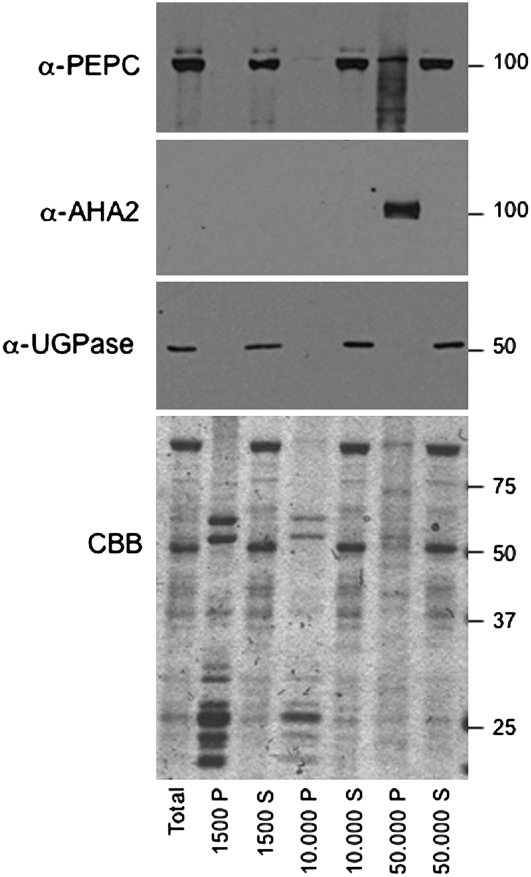
To investigate whether PEPC could interact with anionic phospholipids in vivo, its cellular location was studied using a crude biochemical fractionation approach. Sorghum leaf protein extracts were centrifuged at increasing speed to separate soluble and membrane-bound proteins (Fig. 4). As expected, PEPC protein was predominantly present in the soluble fraction, but a subpool could be detected in the 50,000g pellet fraction, which contained the plasma membrane (PM) and presumably also intracellular membranes. Judging from the absence of the cytosolic protein UGPase, there was little or no contamination with cytosolic proteins. Moreover, further treatment with Brij58 to release entrapped soluble proteins did not decrease the amount of PEPC (Supplemental Fig. S6). Interestingly, PEPC in the 50,000g pellet fraction appeared to be partially degraded, or modified, as multiple bands were detected. This was not due to overall proteolysis in this fraction since the PM marker protein AHA2 or other proteins visualized by Coomassie Brilliant Blue staining were not degraded (Fig. 4).
Figure 4.
Presence of PEPC in soluble and crude membrane fractions of sorghum leaf extracts. Western analyses of PEPC, AHA2 (PM marker), and UGPase subunits (cytosolic marker) in supernatant (S) and pellet (P) fractions after sequential centrifugation at 1,500g, 10,000g, and 50,000g (Supplemental Materials and Methods S1). Total protein for each fraction was analyzed on Coomassie Brilliant Blue (CBB)-stained gels.
ANIONIC PHOSPHOLIPIDS AS NOVEL REGULATORY FACTORS AFFECTING C4 PEPC ACTIVITY
Like the C3 PEPC proteins from Arabidopsis and tomato suspension cells, C4-type PEPC from sorghum and maize leaves is shown here to bind PA. Moreover, binding of PA and other anionic phospholipids resulted in inhibition of the enzyme in vitro. Little or no effect was found for neutral, zwitterionic, or positively charged phospholipids (Fig. 3), ruling out a nonspecific effect of the fatty acyl chains, which are known to activate E. coli PEPC (Izui et al., 2004). Thus, anionic phospholipids represent novel regulators that inhibit PEPC activity through an unknown mechanism that is independent of pH, other allosteric regulators, or substrate (PEP) binding. The effect seems specific for C4-type PEPC, as C3-type PEPC activity in Arabidopsis leaves (Fig. 2C) or Arabidopsis or sorghum roots (data not shown) were not affected.
Interestingly, low sequence homology of the Arabidopsis PEPC homolog PPC3 (aa41-100) was found with the endophilin-A1 BAR domain. This domain binds anionic phospholipids and is proposed to sense membrane curvature (Dawson et al., 2006). In the maize PEPC crystal structure, this stretch appears to be exposed (Matsumura et al., 2002) and our comparisons of BAR and PEPC structures indicate some structural resemblance (F. McLoughlin and C. Testerink, unpublished data). Whether this fragment is indeed the part that recognizes phospholipids in PEPC remains to be established.
A ROLE FOR MEMBRANE-SURFACE CHARGE IN REGULATING PEPC ACTIVITY IN VIVO?
We provide evidence that sorghum PEPC is partially associated with membranes (Fig. 4). Although in general PEPC is thought to be a cytosolic enzyme (Chollet et al., 1996), in vitro association of maize PEPC with the chloroplast membrane has been reported before (Wu and Wedding, 1992). As the protein does not have any predicted transmembrane regions, we assume that it is associated peripherally.
Several proteins involved in intracellular vesicular trafficking and signaling have been identified as anionic phospholipid-binding proteins (Testerink and Munnik, 2005; Stace and Ktistakis, 2006; Arisz et al., 2009), but less is known about the effect of phospholipids on plant metabolic enzymes. Yet, we have also identified the major glycolytic enzyme GAPDH as a PA-binding protein using PA beads (F. McLoughlin and C. Testerink, unpublished data). It was reported recently that a subpool of several glycolytic enzymes, including GAPDH, dynamically associate with mitochondria, thus allowing regulation and channeling of glycolytic intermediates at this location specifically (Graham et al., 2007). Possibly, a subpool of PEPC at the chloroplast or other cellular membranes could have a similar function.
Using crude biochemical fractionation studies, we not only detected the presence of PEPC in the membrane fraction, but strikingly also observed multiple bands. This indicates that PEPC is partially degraded or modified specifically in the membrane fraction and further confirms that it is a not a contamination from the cytosolic protein pool (Fig. 4). Recently, PEPC from castor (Ricinus communis) oil seeds was shown to be monoubiquitinated (Uhrig et al., 2008), and PEPC polyubiquitination resulting in proteolysis has also been reported (Klockenbring et al., 1998). While ubiquitination in general is known to affect PM binding (Mukhopadhyay and Riezman, 2007), it is not known whether ubiquitination of PEPC would affect its cellular localization.
We hypothesize that the negative charge of the chloroplast or other intracellular membranes will affect PEPC activity. Differences in membrane-surface charge of individual cellular membranes have been reported recently using fluorescent biosensors (Yeung et al., 2006, 2008). The action of phospholipid-metabolizing enzymes is known to affect membrane charge and thereby the localization of proteins (Roth, 2008; Yang et al., 2008). How PEPC activity and cellular location would be affected by dynamic changes in the overall amount of negatively charged membrane lipids is an interesting topic for further investigation that will require the in vivo analysis of GFP fusions of the C4 PEPC protein.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effect of PEPC concentration on the inhibition of its activity by PA (C8:0).
Supplemental Figure S2. Electrophoretic mobility shift of sorghum-purified PEPC.
Supplemental Figure S3. Effect of pH on the inhibition of PEPC activity by PA.
Supplemental Figure S4. Effect of Glc-6-P, malic acid, and PEP on the inhibition of PEPC activity by PA.
Supplemental Figure S5. No effect of phosphorylation of PEPC on its sensitivity to PA.
Supplemental Figure S6. Brij58 treatment releases entrapped cytosolic proteins from the 50,000g pellet but does not decrease PEPC in this fraction.
Supplemental Materials and Methods S1. Plant material and growth conditions, enzyme extraction and PEPC activity analysis, purification and characterization of sorghum PEPC, PA-binding assays, and subcellular fractionation of sorghum leaf.
Supplementary Material
Acknowledgments
The authors thank Maike Stam and Marieke Louwers for providing the maize plants and Ludek Tikovsky and Harold Lemereis for their excellent care of all the plants. We thank Michael Palmgren, Ming-Che Shih, and William Plaxton for providing antibodies, Teun Munnik and Frank Takken for critically reading the manuscript, and Edgar Kooijman for discussions and advice.
This work was supported by the Netherlands Organization for Scientific Research (grant nos. NWO–Veni 700.52.401 and Vidi 700.56.429 to C.T.), the Dirección General de Investigación del Ministerio de Ciencia y Tecnología (grant no. BFU2007–61431/BMC), as well as the Junta de Andalucía (BIO298). J.A.M. was financed by a Formacion del Personal Investigador fellowship from Universidad de Sevilla (Spain).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christa Testerink (c.s.testerink@uva.nl).
The online version of this article contains Web-only data.
References
- Anthony RG, Henriques R, Helfer A, Meszaros T, Rios G, Testerink C, Munnik T, Deak M, Koncz C, Bogre L (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J 23 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisz SA, Testerink C, Munnik T (2009) Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta 1791 869–875 [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47 273–298 [DOI] [PubMed] [Google Scholar]
- Cornell RB, Taneva SG (2006) Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr Protein Pept Sci 7 539–552 [DOI] [PubMed] [Google Scholar]
- Dawson JC, Legg JA, Machesky LM (2006) Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol 16 493–498 [DOI] [PubMed] [Google Scholar]
- Graham JW, Williams TC, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ (2007) Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. Plant Cell 19 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH (2006) Membrane binding domains. Biochim Biophys Acta 1761 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izui K, Matsumura H, Furumoto T, Kai Y (2004) Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol 55 69–84 [DOI] [PubMed] [Google Scholar]
- Johnson JE, Xie M, Singh LM, Edge R, Cornell RB (2003) Both acidic and basic amino acids in an amphitropic enzyme, CTP:phosphocholine cytidylyltransferase, dictate its selectivity for anionic membranes. J Biol Chem 278 514–522 [DOI] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Shaw J, Masle J (2009) Biochemical characterization of two wheat phosphoethanolamine N-methyltransferase isoforms with different sensitivities to inhibition by phosphatidic acid. J Biol Chem 284 31962–31971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockenbring T, Meinhard M, Schnabl H (1998) The stomatal phosphoenolpyruvate carboxylase—a potential target for delective proteolysis during stomatal closure? J Plant Physiol 152 222–229 [Google Scholar]
- Lemmon MA (2008) Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9 99–111 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Xie Y, Shirakata S, Inoue T, Yoshinaga T, Ueno Y, Izui K, Kai Y (2002) Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases. Structure 10 1721–1730 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315 201–205 [DOI] [PubMed] [Google Scholar]
- Munnik T, Testerink C (2009) Plant phospholipid signaling—‘in a nutshell’. J Lipid Res 50 S260–S265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Weidner A, Pech A, Stubbs MT, Tittmann K (2008) Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli. Proc Natl Acad Sci USA 105 17390–17395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo HG (2003) Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch Biochem Biophys 414 189–196 [DOI] [PubMed] [Google Scholar]
- Roth MG (2008) Molecular mechanisms of PLD function in membrane traffic. Traffic 9 1233–1239 [DOI] [PubMed] [Google Scholar]
- Stace CL, Ktistakis NT (2006) Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta 1761 913–926 [DOI] [PubMed] [Google Scholar]
- Stahelin RV (2009) Lipid binding domains: more than simple lipid effectors. J Lipid Res (Suppl) 50 S299–S304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneva S, Dennis MK, Ding Z, Smith JL, Cornell RB (2008) Contribution of each membrane binding domain of the CTP:phosphocholine cytidylyltransferase-alpha dimer to its activation, membrane binding, and membrane cross-bridging. J Biol Chem 283 28137–28148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C, Dekker HL, Lim ZY, Johns MK, Holmes AB, Koster CG, Ktistakis NT, Munnik T (2004) Isolation and identification of phosphatidic acid targets from plants. Plant J 39 527–536 [DOI] [PubMed] [Google Scholar]
- Testerink C, Larsen PB, van der Does D, van Himbergen JAJ, Munnik T (2007) Phosphatidic acid binds to and inhibits the activity of Arabidopsis CTR1. J Exp Bot 58 3905–3914 [DOI] [PubMed] [Google Scholar]
- Testerink C, Munnik T (2005) Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 10 368–375 [DOI] [PubMed] [Google Scholar]
- Uhrig RG, She YM, Leach CA, Plaxton WC (2008) Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. J Biol Chem 283 29650–29657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal J, Chollet R (1997) Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci 2 230–237 [Google Scholar]
- Wu MX, Wedding RT (1992) Inactivation of maize leaf phosphoenolpyruvate carboxylase by the binding to chloroplast membranes. Plant Physiol 100 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Gad H, Lee SY, Mironov A, Zhang L, Beznoussenko GV, Valente C, Turacchio G, Bonsra AN, Du G, et al (2008) A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat Cell Biol 10 1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S (2008) Membrane phosphatidylserine regulates surface charge and protein localization. Science 319 210–213 [DOI] [PubMed] [Google Scholar]
- Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S (2006) Receptor activation alters inner surface potential during phagocytosis. Science 313 347–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.