Abstract
Nitric oxide (NO) regulates a wide range of plant processes from development to environmental adaptation. Despite its reported regulatory functions, it remains unclear how NO is synthesized in plants. We have generated a triple nia1nia2noa1-2 mutant that is impaired in nitrate reductase (NIA/NR)- and Nitric Oxide-Associated1 (AtNOA1)-mediated NO biosynthetic pathways. NO content in roots of nia1nia2 and noa1-2 plants was lower than in wild-type plants and below the detection limit in nia1nia2noa1-2 plants. NIA/NR- and AtNOA1-mediated biosynthesis of NO were thus active and responsible for most of the NO production in Arabidopsis (Arabidopsis thaliana). The nia1nia2noa1-2 plants displayed reduced size, fertility, and seed germination potential but increased dormancy and resistance to water deficit. The increasing deficiency in NO of nia1nia2, noa1-2, and nia1nia2noa1-2 plants correlated with increased seed dormancy, hypersensitivity to abscisic acid (ABA) in seed germination and establishment, as well as dehydration resistance. In nia1nia2noa1-2 plants, enhanced drought tolerance was due to a very efficient stomata closure and inhibition of opening by ABA, thus uncoupling NO from ABA-triggered responses in NO-deficient guard cells. The NO-deficient mutants in NIA/NR- and AtNOA1-mediated pathways in combination with the triple mutant will be useful tools to functionally characterize the role of NO and the contribution of both biosynthetic pathways in regulating plant development and defense.
Nitric oxide (NO) is a small ubiquitous molecule derived from nitrogen-containing precursors that is one of the earliest and most widespread signaling molecules in living organisms from metazoans to mammals (Torreilles, 2001). The regulatory functions of NO have been extensively studied in mammals, where it is synthesized from Arg through the activity of NO synthases (Knowles and Moncada, 1994). By contrast, the biosynthesis and function of this molecule in plants are largely unknown. During the last 10 years, NO biosynthesis in plants has been one of the most controversial topics in plant biology (Durner and Klessig, 1999; Wendehenne et al., 2001; del Río et al., 2004; Zeier et al., 2004; Lamotte et al., 2005; Meyer et al., 2005; Modolo et al., 2005; Crawford, 2006; Crawford et al., 2006; Zemojtel et al., 2006a). Despite the controversy about its biosynthesis, it is now clear that NO regulates many physiological processes of plants, including seed germination, cell death, defense responses against pathogens, stomata function, senescence, and flowering (Beligni and Lamattina, 2000; Pedroso et al., 2000; Neill et al., 2002; Lamattina et al., 2003; He et al., 2004; Romero-Puertas et al., 2004; Wendehenne et al., 2004; Delledonne, 2005; Guo and Crawford, 2005; Simpson, 2005; Grün et al., 2006; Melotto et al., 2006; Planchet et al., 2006; Ali et al., 2007; Mishina et al., 2007).
The molecular mechanisms underlying the control of seed dormancy and germination are still poorly characterized. Genetic data support a central role of abscisic acid (ABA) in regulating seed dormancy, whereas gibberellins promote germination (Finkelstein et al., 2008; Holdsworth et al., 2008). In addition, NO has been lately characterized as a new component in the signaling pathway leading to dormancy breakage. NO-releasing compounds reduce dormancy in a NO-dependent manner in Arabidopsis (Arabidopsis thaliana), some warm-season grasses, and certain barley (Hordeum vulgare) cultivars (Bethke et al., 2004; Sarath et al., 2006). More recently, the aleurone layer cells have been characterized as responsive to NO, gibberellins, and ABA, thus becoming a primary determinant of seed dormancy in Arabidopsis (Bethke et al., 2007).
Two main enzyme-based pathways have been proposed to be functional for NO biosynthesis in plants. One is based on the activity of nitrate reductases (Meyer et al., 2005; Modolo et al., 2005), and another one, yet undefined, is based on the direct or indirect function of the Nitric Oxide-Associated1/Resistant to Inhibition by Fosfidomycin1 (AtNOA1/RIF1) protein. It has been also reported that NO synthesis from nitrite occurs in mitochondria associated with mitochondrial electron transport (Planchet et al., 2005) and also that this pathway is mainly functioning in roots under anoxia (Gupta et al., 2005). Moreover, the balance between mitochondrial nitrite reduction and superoxide-dependent NO degradation seems to be derived from factors controlling NO levels in Arabidopsis (Wulff et al., 2009). It has been recently reported that the synthesis of NO in floral organs requires nitrate reductase activity (Seligman et al., 2008) and also that homologues of AtNOA1 participate in NO biosynthesis in diatoms (Vardi et al., 2008), mammals (Zemojtel et al., 2006b; Parihar et al., 2008a, 2008b), and Nicotiana benthamiana (Kato et al., 2008). Recently, the identification of the rif1 mutant, carrying a null mutation in the AtNOA1 locus (At3g47450), allowed uncovering of a function for AtNOA1/RIF1 in the expression of plastome-encoded proteins (Flores-Pérez et al., 2008). Moreover, another recent report claims that AtNOA1 is not a NO synthase but a cGTPase (Moreau et al., 2008), likely playing a role in ribosome assembly and subsequent mRNA translation to proteins in the chloroplasts.
To date, it is not clear if both pathways coexist in plants and, if so, the corresponding contributions of each pathway to NO biosynthesis. In this work, we have addressed the functions of both pathways in Arabidopsis by generating a triple mutant in both nitrate reductases and AtNOA1 that is severely impaired in NO production. Further characterization of NO-deficient plants allowed us to identify a functional cross talk between NO and ABA in controlling seed germination and dormancy as well as plant resistance to water deficit.
RESULTS
Impaired NO Biosynthesis in Triple nia1nia2noa1-2 Mutant Plants
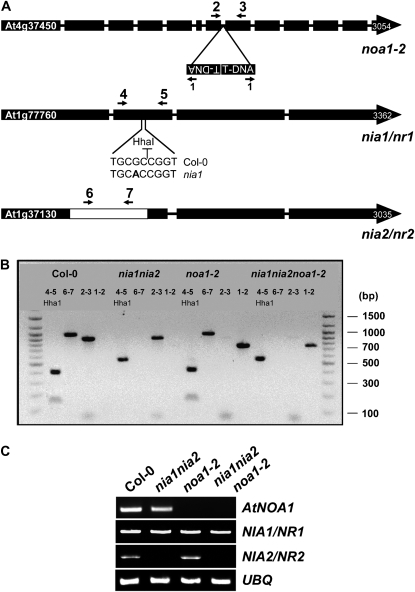
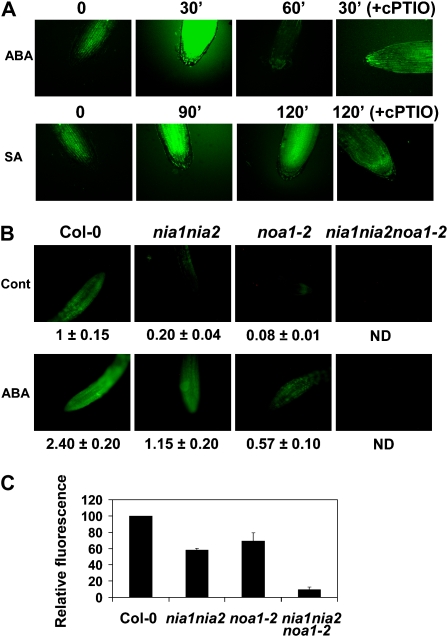
To elucidate how NO is synthesized in Arabidopsis, we generated a mutant plant simultaneously impaired in protein AtNOA1 and nitrate reductases (NR1/NIA1 and NR2/NIA2) with previously proposed NO biosynthetic activities (Yamasaki et al., 1999; Desikan et al., 2002; Rockel et al., 2002; Guo et al., 2003; Guo and Crawford, 2005; Meyer et al., 2005; Modolo et al., 2005). First, we identified and characterized a noa1 mutant allele from the Syngenta collection of T-DNA insertion lines (line SAIL_507_E11) that we called noa1-2. This mutant allele has a T-DNA insertion in the seventh intron (Fig. 1A) and resulted in a null mutant, as demonstrated by the undetectable levels of AtNOA1 transcript (Fig. 1C), and a growth phenotype undistinguishable from that of noa1-1 (Guo et al., 2003; data not shown). We then crossed nia1nia2 double mutant plants (Wilkinson and Crawford, 1993) with noa1-2 mutant plants and searched for a triple nia1nia2noa1-2 mutant plant in the F2 progeny. PCR-based genotyping of F2 plants allowed us to find plants with homozygous mutations in the three genes (Fig. 1B). Plants with nia1nia2noa1-2 mutant genotype were, like the parental noa1-2 and nia1nia2 plants, null for AtNOA1 and NIA2/NR2 transcripts, respectively (Fig. 1C). Since nia1 is a point mutation that did not truncate the open reading frame, the levels of mutant NIA1 transcript were similar to the endogenous transcript in wild-type plants. To assess whether the triple mutant in genes coding for the potential NO biosynthetic enzymes led to an actual reduced NO content, roots from different mutant and wild-type plants were analyzed. We used a cell-permeable NO-sensitive diamino fluorescein diacetate (DAF-FM DA; Invitrogen) to detect NO in roots. The basal levels of NO-associated fluorescence were low in untreated roots of wild-type plants but strongly increased upon treatment with compounds such as ABA or salicylic acid (SA; Fig. 2A). These compounds have been already characterized as inducers of NO biosynthesis (Guo et al., 2003; Zottini et al., 2007). We found that ABA was a stronger and faster inducer than SA (Fig. 2A). The detected fluorescence was specifically associated with the production of NO because of the observed reduced NO-associated fluorescence detected in SA- or ABA-treated roots in the presence of the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO; Fig. 2A). The NO-associated fluorescence was analyzed in untreated and ABA-treated roots of mutant versus wild-type plants. Figure 2B shows that the basal level of NO-associated fluorescence was strongly reduced in the roots of nia1nia2 and noa1-2 plants and undetectable in the roots of the triple mutant plants. Moreover, ABA-treated roots of the nia1nia2 and noa1-2 plants accumulated NO levels lower than those detected in the roots of ecotype Columbia (Col-0) plants, and the roots of the triple mutant plants were unable to sustain an ABA-induced accumulation of NO in the roots (Fig. 2B). nia1nia2noa1-2 plants were thus severely impaired in both basal and induced synthesis of NO as a result of the simultaneous block of AtNOA1- and NIA/NR-based biosynthetic pathways. Since NO biosynthesis may be different in different organs of the plants (Kolbert et al., 2008; Seligman et al., 2008) and to rule out a root-specific effect, we have also analyzed the accumulated production of NO in wild-type and NO-deficient whole seedlings by measuring the NO that diffused out of the cells into the medium with the non-cell-permeable 4,5-diaminofluorescein (DAF-2) and subsequently measuring the fluorescence associated with the corresponding triazole derivative. To assess whether NO production detected in nia1nia2noa1-2 seedlings was biologically or chemically synthesized, we performed measurements with seedlings from the different genotypes and the corresponding thermally inactivated seedlings as controls for nonenzymatic production of NO. We detected similar levels of basal production of NO in every sample from thermally inactivated seedlings that corresponded to nonenzymatic production. After subtraction of nonenzymatically produced NO, nia1nia2 and noa1-2 plants accumulated lower levels of NO than wild-type plants, and nia1nia2noa1-2 plants displayed an additive reduction to levels below 10% of those detected in wild-type seedlings (Fig. 2C).
Figure 1.
Generation of the nia1nia2noa1-2 triple mutant. A, Diagram showing the position of the T-DNA insertion and primers (indicated with arrows) used for genotyping (Supplemental Table S1) of noa1-2 and the nia1 and nia2 mutations in the corresponding genes. The HhaI restriction site for genotyping of nia1/nr1 is also shown. B, PCR-based genotyping of wild-type and mutant plants. Ethidium bromide-stained amplicons of different combinations of primers are shown. C, RT-PCR-based analysis of AtNOA1, NIA1/NR1, and NIA2/NR2 transcripts. Total RNAs from the indicated genotypes were extracted from 10-d-old seedlings, treated with DNase, reverse transcribed, and separated on 1% agarose gels. Ubiquitin10 (UBQ) expression was used as a loading control.
Figure 2.
NO production in Col-0, nia1nia2, noa1-2, and nia1nia2noa1-2 roots and seedlings. A, Roots from 5- to 7-d-old Col-0 seedlings treated with 50 μm ABA or 1 mm SA were loaded with 15 μm DAF-FM DA as described in “Materials and Methods.” Images were taken with a confocal microscope at the indicated times upon treatment. Roots were pretreated with 500 μm cPTIO before incubation with DAF-FM DA where indicated (+cPTIO). B, NO accumulation in Col-0, nia1nia2, noa1-2, and nia1nia2noa1-2 roots treated or not with 50 μm ABA for 30 min. Roots from 5- to 7-d-old seedlings (10 plants per experiment, three replicates) were loaded with DAF-FM DA and then treated or not with ABA. Images were captured with a Nikon fluorescence microscope 30 min after treatment with equal settings for every image. NO-derived fluorescence is shown as means ± se of three independent experiments. Cont, Control; ND, not detected. C, NO production in Col-0, nia1nia2, noa1-2, and nia1nia2noa1-2 seedlings using DAF-2 fluorescein. NO that was synthesized in the seedlings and diffused out of cells into the medium was trapped by DAF-2, and the fluorescence associated with the corresponding triazole derivative was quantified with a TECAN fluorimeter. Means ± se are shown. The experiment was repeated three times with similar results.
NO-Deficient Mutant Plants Display Alterations in Development
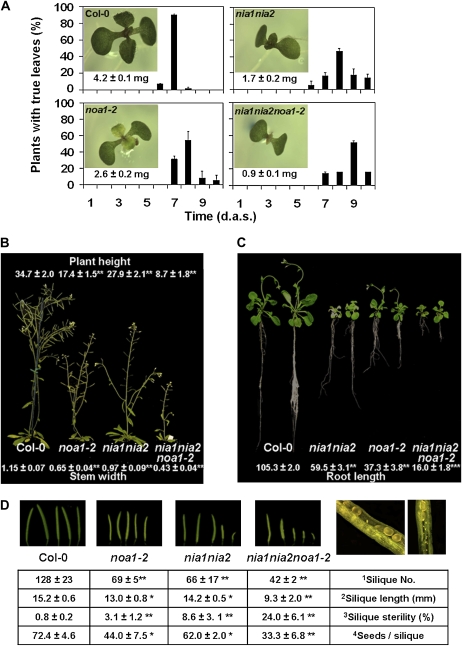
nia1nia2noa1-2 plants displayed reduced shoot and root vegetative growth as a result of additive effects from nia1nia2 and noa1-2 mutant phenotypes (Fig. 3). The delayed growth was already observed at the early seedling stage. NO-deficient seedlings showed their first pair of leaves significantly later than wild-type plants, and this phenotype was additive in the triple mutant plants (Fig. 3A). Adult triple mutant plants were semidwarf, with reduced height and stem diameter (25% and 37% of wild-type plants, respectively; Fig. 3B). The semidwarf phenotype observed in the shoots correlated also with significantly reduced root growth (56%, 35%, and 15% of wild-type roots for nia1nia2, noa1-2, and nia1nia2noa1-2 roots, respectively; Fig. 3C). This causes an altered root-to-shoot fresh weight ratio for the NO-deficient genotypes, with the ratio for nia1nia2noa1-2 being around three times lower than in the wild type (0.33 ± 0.02 and 0.12 ± 0.01, respectively). NO deficiency also correlated well with reduced size and number of siliques as well as with increased number of aborted seeds, thus leading to decreased seed yield (Fig. 3D). To assess whether the small size of the seedlings and the short roots of mutants were due to lower NO levels, we compared the length of roots and the weight of different genotype seedlings treated or not with NO. Supplemental Figure S1 shows that the fresh weight of the whole seedlings and the length of their roots were between 20% and 40% larger in NO-treated nia1nia2 and noa1-2 seedlings than in untreated seedlings. This reversion effect was more evident for NO-treated nia1nia2noa1-2 seedlings, and their roots were between 90% and 95% larger than untreated controls (Supplemental Fig. S1).
Figure 3.
Developmental phenotypes of nia1nia2noa1-2 plants. A, Fresh weight (mg) per seedling (n = 12) of the indicated genotypes grown for 7 d on MS plates. The appearance of the first pair of true leaves was scored daily from day 0 to day 10 after sowing (d.a.s.; n = 200). Histograms show the percentage of seedlings from the indicated genotypes displaying their first pair of true leaves at the indicated times. All plants were grown in long-day conditions on MS plates. Means ± sd are shown. B, Size and shape on the indicated genotypes grown in soil under long-day conditions at 22°C. Plant height (cm) was scored when plants reached its higher size (n = 10). The width (mm) of two portions from the base of the main stem (n = 5) was measured with ImageJ. Means ± sd are shown. C, Length (mm) of the main root of Col-0, nia1nia2, noa1-2, and nia1nia2noa1-2 plants grown on MS vertical plates under long-day conditions. Means ± sd from 12 plants per genotype are shown. D, Reduced size of siliques in NO-deficient plants. Details of the aborted seeds contained in nia1nia2noa1-2 siliques are shown. 1Silique number per plant (n = 10). 2A total of 10 mature fruits per plant were collected from the main inflorescence (n = 5), and their average lengths were determined using ImageJ software. 3The incidence of fruit sterility was calculated based on the number of seedless fruits versus the total number of fruits per plant (n = 10). 4The average number was determined for seeds contained in two mature fruits per plant (n = 5). Means ± sd are shown. Asterisks indicate statistical significance versus the Col-0 control in each case (* P < 0.05, ** P < 0.01, *** P < 0.001, Student's t test).
Interactions between NO and ABA in Dormancy, Seed Germination, and Establishment
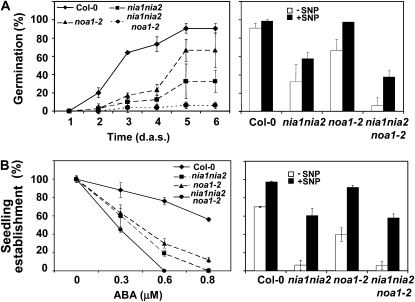
NO has been characterized as a regulator of seed dormancy and germination, dehydration responses, and oxidative damage, all ABA-related processes. Most of the work supporting this function has been conducted with biochemical/physiological approaches based on application of exogenous NO donors, many of which have uncontrolled side effects potentially interfering with the analysis. Nevertheless, genetic evidence supporting a role for NR/NIA and AtNOA1 in ABA-NO interactions has also been reported (Desikan et al., 2002; Guo et al., 2003). We thus decided to characterize NO-modulated and ABA-triggered responses in the different NO-deficient mutants, including the triple nia1nia2noa1-2 mutant plants generated in this work. Freshly harvested seeds from wild-type and NO-deficient mutant plants were sown, and the percentage of germination was calculated daily to day 6 after sowing. Figure 4A (left) shows that 2 d after sowing, when 20% of the wild-type seeds have already germinated, less than 5% of the NO-deficient mutant seeds had germinated. Moreover, the maximum germination potential for the wild-type seeds was reached by day 5 after sowing, whereas NO-deficient mutant seeds were more dormant than wild-type seeds. noa1-2, nia1nia2, and nia1nia2noa1-2 seeds reached 65%, 25%, and less than 10% germination, respectively, by day 6 after sowing (Fig. 4A, left). When freshly harvested seeds were exposed to a NO-enriched environment, the increased dormancy phenotype of the noa1-2 mutant seeds was released and the germination was similar to that observed for the wild-type seeds (Fig. 4A, right). However, the increased dormancy of either nia1nia2 or the triple mutant seeds was not fully released by sodium nitroprusside (SNP) treatment (Fig. 4A, right). Nevertheless, NO treatment led to an improved germination potential of nia1nia2noa1-2 seeds. Close to 40% of the nia1nia2noa1-2 seeds germinated in the NO-enriched environment, contrasting with around 6% that germinated in the absence of NO treatment (Fig. 4A, right).
Figure 4.
The NO-deficient mutant plants show hypersensitivity to ABA-regulated germination. A, Germination potential of freshly harvested nonstratified seeds on MS plates was scored daily until day 6 after sowing (d.a.s.; left). Assays performed in the presence (+ SNP; black bars) or absence (− SNP; white bars) of NO gas derived from a light-exposed 100 μm solution of SNP contained in separate vessels (right). Germination was scored at day 5 after sowing, and values are means ± se of three independent experiments with at least 200 seeds per experiment. B, Percentage of seeds that germinated and developed green expanded cotyledons (seedling establishment) in MS medium supplemented with the indicated ABA concentrations. At least 200 seeds were sown, stratified for 3 d at 4°C, and scored after 12 d (left). The experiment was repeated four times with similar results. Similar assays performed in MS medium plus 0.5 μm ABA in the presence (+ SNP; black bars) or absence (− SNP; white bars) of NO vapors (from a 100 μm solution of SNP) scored at day 12 after sowing (right). Values are means ± se. MS plates contained MS salts, 1% Suc, and 0.8% agar, pH 5.7.
ABA inhibits germination and is essential for the acquisition and maintenance of seed dormancy (Koornneef et al., 2002; Gubler et al., 2005). It was originally reported that the break of Arabidopsis seed dormancy by the NO released from SNP did not occur in ABA-treated seeds (Bethke et al., 2004). However, further experimental data indicated that NO gas was able to break seed dormancy also in ABA-imbibed seeds (Bethke et al., 2006). Establishment assays with NO-deficient mutant seeds in the presence of increasing concentrations of ABA indicated that all mutants were hypersensitive to ABA. Figure 4B (left) shows that at 0.6 μm ABA, around 80% of the wild-type seeds and only 35% and 20% of the noa1-2 and nia1nia2 mutant seeds established. Moreover, nia1nia2noa1-2 seeds displayed a 100% inhibition of seed establishment at 0.6 μm ABA (Fig. 4B, left), suggesting that NIA/NR- and AtNOA1-mediated pathways of NO biosynthesis are additive in terms of ABA-mediated inhibition of seedling establishment. The additive effect was also observed when germination, as endosperm rupture, was quantified with time in NO-deficient seeds in the presence of ABA (Supplemental Fig. S2). Due to the well-known effects of sugar content on seed germination and ABA signaling (for a recent review, see Graham, 2008), we also performed experiments in medium containing ABA with or without Suc. The effect of ABA on seedling establishment and germination was similar in medium containing or not Suc (Supplemental Fig. S3). The application of exogenous NO released ABA inhibition of seedling establishment of wild-type and NO-deficient mutant seeds (Fig. 4B, right). Reversion was almost complete for noa1-2 seedlings and sufficient to go from below 10% to around 60% of seedling establishment in nia1nia2 and nia1nia2noa1-2 seeds (Fig. 4B, right).
Since osmotic stress-mediated inhibition of germination is ABA dependent, we also tested whether NO deficiency may also alter the germination in medium supplemented with NaCl or mannitol. Table I shows that NO-deficient mutant seeds were more sensitive to osmotic stress-inhibited germination than wild-type seeds, with nia1nia2 and nia1nia2noa1-2 seeds showing stronger effects than noa1-2 seeds.
Table I.
Effects of mannitol or salt treatment on seed germination of wild-type and NO-deficient plants
Seeds from NO-deficient mutant plants (at least 200 per genotype) were sown on MS medium plus 1% (w/v) Suc (control) or that medium supplemented with 125 mm NaCl or 250 mm mannitol as indicated. Percentage values of seed germination are means of four replicates ± se.
| Treatment | Col-0 | nia1nia2 | noa1-2 | nia1nia2noa1-2 |
|---|---|---|---|---|
| Control | 100 | 96 ± 2 | 100 | 96 ± 3 |
| Mannitol | 77.97 ± 2.67 | 15.1 ± 0.94 | 51.98 ± 1.19 | 14.13 ± 2.05 |
| NaCl | 50.77 ± 6.88 | 1.94 ± 0.99 | 14.64 ± 4.81 | 0 |
Analysis of ABA-Responsive Gene Expression in NO-Deficient Plants
Quantitative real-time PCR was used to monitor the expression of the ABA-inducible RD29b (Yamaguchi-Shinozaki and Shinozaki, 1993) and RAB18 (Jeannette et al., 1999) genes. Table II shows that all NO-deficient mutant seedlings contained around 2-fold transcript accumulation of RD29b and RAB18 genes compared with wild-type seedlings. Upon ABA treatment, the triple mutant show more than double induction than Col-0, whereas for nia1nia2 and noa1-2, the induction rates were also higher than in Col-0 but not as high as in the triple mutant (Table II). These data suggest that the modulation exerted by NO on ABA sensitivity may not be restricted to seeds but could also be functional in seedlings and adult plants.
Table II.
Relative transcript levels of ABA-responsive genes in NO-deficient mutant seedlings
Values are means of three independent biological replicates ± se and are shown as relative values to those detected in Col-0 wild-type seedlings. Levels were normalized with the endogenous content of ACTIN2/8 transcript.
| Treatment | Gene | Genotype |
|||
|---|---|---|---|---|---|
| Col-0 | nia1nia2 | noa1-2 | nia1nia2noa1-2 | ||
| Mock | RD29b | 1 | 2.39 ± 0.44 | 2.10 ± 0.45 | 2.65 ± 0.37 |
| RAB18 | 1 | 1.74 ± 0.14 | 1.95 ± 0.42 | 2.17 ± 0.21 | |
| ABA | RD29b | 1 | 1.31 ± 0.05 | 1.25 ± 0.10 | 2.33 ± 0.15 |
| RAB18 | 1 | 1.65 ± 0.25 | 1.63 ± 0.15 | 2.05 ± 0.20 | |
NO Deficiency Confers Enhanced Resistance to Dehydration
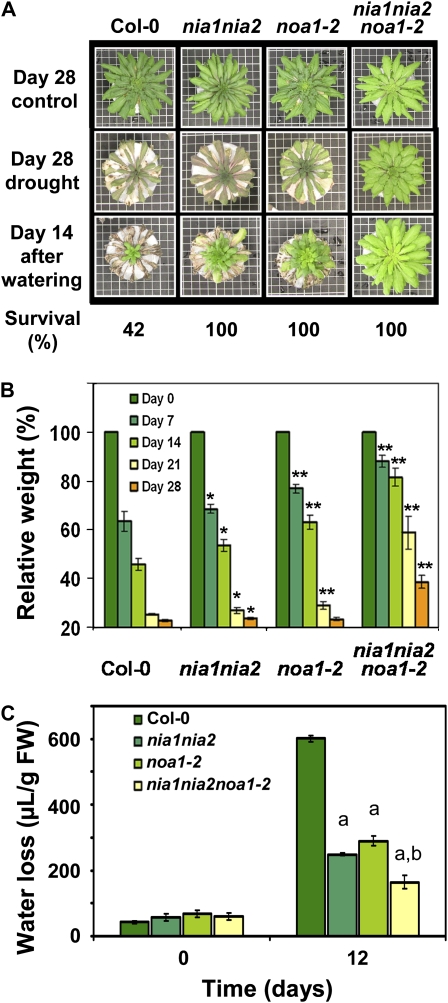
Since NO-deficient mutant plants displayed hypersensitivity to ABA, we tested whether the ABA-related phenotype of resistance to dehydration could also be observed in plants with altered levels of endogenous NO. Wild-type and NO-deficient mutant plants were subjected to water deficit. After 28 d without watering, NO-deficient plants were more resistant to dehydration than wild-type plants, with a stronger effect in nia1nia2noa1-2 plants than in nia1nia2 and noa1-2 plants (Fig. 5A). When wild-type plants showed extreme dehydration effects, the triple mutant plants kept green with no obvious dehydration symptoms (Fig. 5A). We found that the rate of weight loss due to water transpiration was significantly slower in NO-deficient than in wild-type plants, with nia1nia2noa1-2 plants showing the lowest transpiration rate and consequently being the most resistant to dehydration (Fig. 5B). Moreover, whereas 42% of the wild-type plants survived the drought treatment after restoring watering, 100% of the plants from the three different NO-deficient mutant genotypes survived (Fig. 5A). In addition, a detailed water-loss assay was performed by comparing fresh and turgid weight of rosette leaves. Under these experimental conditions, noa1-2 and nia1nia2 mutants showed a reduced water loss as compared with the wild type, retaining around two times more water than wild-type leaves (Fig. 5C). This phenotype was even stronger in nia1nia2noa1-2, which retained around four times more water than wild-type leaves under the same drought period (Fig. 5C). These results show an additive role for NIA/NR and AtNOA1 pathways on drought resistance, which is in agreement with the above described additive ABA hypersensitivity of the NO-deficient mutants.
Figure 5.
Drought resistance of NO-deficient mutant plants. A, Appearance of Col-0, nia1nia2, noa1-2, and nia1nia2noa1-2 plants 28 d after stopping watering and 14 d after restoring standard watering regimes. Plants (10 per experiment, three independent experiments) were cultivated in homogenous Jiffy 7 soil substrate under short-day conditions. Survival rate was measured as the percentage of plants developing new green rosette leaves by day 14 d after restoring watering. Means ± se are shown. B, Losses of relative weight (%) of the same individuals (plant + sealed pot) shown in A. The total weights were scored when watering was stopped and every 7 d until day 28. Values are means ± se of 10 individuals per experiment. The experiment was repeated three times with similar results. * P < 0.05, ** P < 0.005 by Student's t test. C, Quantification of transpiration-mediated water loss in plants after 12 d without watering. Data shown are average amounts of water loss measured in 10 leaves (μL g−1 fresh weight [FW]) collected from 10 different plants. aP < 0.001 by comparing NO-deficient mutants with the wild type by t test. bP < 0.005 by comparing nia1nia2noa1-2 with nia1nia2 or noa1-2 by t test.
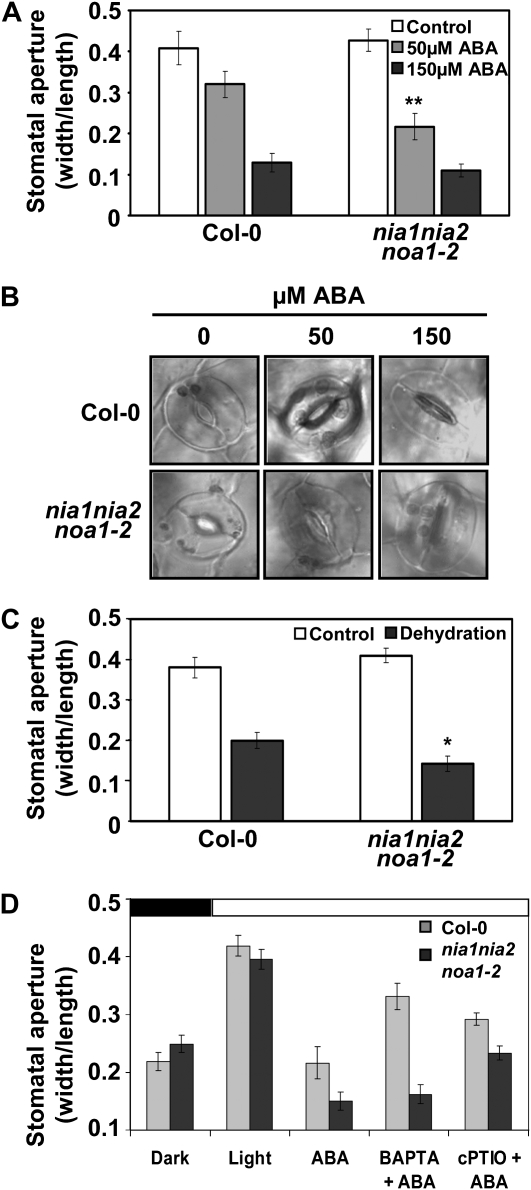
To test whether enhanced resistance to drought in NO-deficient plants was due to direct effects of ABA on stomatal closure, we measured stomatal aperture in wild-type and nia1nia2noa1-2 leaves under different conditions. First, the effects of exogenous ABA treatment and dehydration on stomatal closure of seedlings were analyzed. Figure 6, A and B, shows that stomata from nia1nia2noa1-2 leaves were significantly closer than wild-type stomata in seedlings treated with 50 μm ABA, suggesting that stomata from nia1nia2noa1-2 leaves were more efficient in ABA-induced stomatal closure. At a higher saturating concentration of 150 μm ABA, stomata from wild-type and mutant plants were both similarly closed (Fig. 6A). In addition, nia1nia2noa1-2 stomata were also significantly closer than wild-type stomata in response to dehydration of the seedlings (Fig. 6C). Since ABA is also regulating the stomata opening by light (Roelfsema and Hedrich, 2005), we analyzed this process in wild-type and nia1nia2noa1-2 seedlings. Figure 6D shows that nia1nia2noa1-2 opened stomata upon the shift to light like the wild type. ABA inhibited opening of wild-type stomata, and the effect was stronger on nia1nia2noa1-2 (Fig. 6D), thus displaying hypersensitivity to ABA in agreement with other phenotypes shown above. Moreover, the ABA-mediated inhibition of stomata opening was prevented by treatment with the cell-permeable calcium chelator 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetraacetoxymethyl ester or the NO scavenger cPTIO in wild-type plants but not in the nia1nia2noa1-2 mutant (Fig. 6D).
Figure 6.
Reduced stomatal apertures of nia1nia2noa1-2 mutant plants. A, ABA-induced stomatal closing of Col-0 and nia1nia2noa1-2 seedlings was tested in leaves with stomata preopened under light for 2.5 h and then incubated in the indicated ABA concentrations for 2.5 h under light. Data represent means ± se of 40 measured stomata per experiment. The experiment was repeated twice with similar results. B, Representative confocal microscopy images of stomata from leaves of the indicated genotype and ABA treatment used for quantification in A. C, Stomatal apertures under control (white bars) and dehydration (black bars) conditions. Dehydration conditions were set up by removing seedlings from culture medium and further incubation for 5 min in a laminar flow cabinet. D, ABA-mediated inhibition of stomata opening upon transference from darkness to light. Seedlings were incubated in the dark in opening buffer for 2.5 h. Then, they were treated for 0.5 h with 50 μm ABA, 250 μm 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetraacetoxymethyl ester (BAPTA-AM), or 250 μm cPTIO as indicated in “Materials and Methods” and shifted to light conditions. Stomatal aperture was measured just before (black horizontal bar) or 2.5 h after (white horizontal bar) the shift to light. * P < 0.05, ** P < 0.01 by Student's t test.
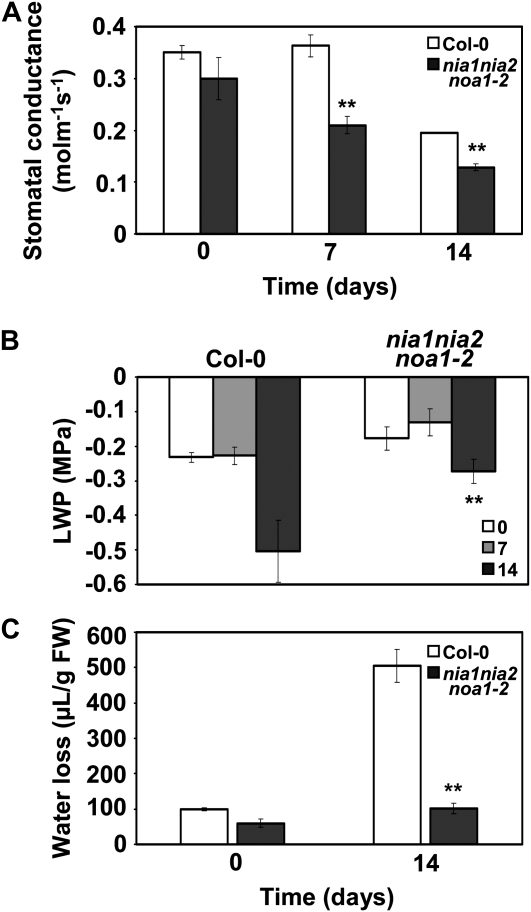
We have also confirmed altered stomatal aperture in adult nia1nia2noa1-2 plants using a noninvasive method to minimize the NO production derived from tissue damage. We found significantly lower stomatal conductance in nia1nia2noa1-2 than in wild-type plants after 7 and 14 d without watering (Fig. 7A). Besides, we also measured the leaf water potential (LWP) by psychrometry in wild-type and NO-deficient nia1nia2noa1-2 plants at those times. By 7 d, no significant changes in LWP were detected (Fig. 7B), correlating with no visual alteration in leaf turgor in any of the genotypes. By 14 d after stopping watering, when wild-type plants already showed dehydration symptoms and a decrease in LWP, only a slight decrease in LWP was observed in nia1nia2noa1-2 plants (Fig. 7B), correlating very well with the lack of a water deficit-related phenotype observed in those plants (Fig. 5).
Figure 7.
Stomatal conductance, LWP, and water losses in wild-type and nia1nia2noa1-2 plants undergoing water shortage. A, Stomatal conductance of wild-type and nia1nia2noa1-2 mutant leaves for the indicated times after stopping watering. Means of 18 replicate measurements ± se are shown. The experiment was repeated twice with similar results. B, LWP of plants undergoing dehydration after stopping watering for 0, 7, or 14 d. Values represent means of six replicate measurements ± sd. C, Water loss assay (mean ± sd) done with the same plants used in A. Water shortage conditions were as described in “Materials and Methods” and Figure 5. ** P < 0.01 by Student's t test.
DISCUSSION
The increasing evidence of NO regulating a wide array of plant physiological processes contrasts with the limited and controversial knowledge about its biosynthesis. However, most of the accumulated data on NO production and function rely on experimental approaches based on the application of NO donors and scavengers and on the analysis of targets not directly related to NO. In this paper, we have addressed the analysis of different NO-regulated processes by exploring the effects of endogenously generated NO through different pathways. First, we have confirmed that AtNOA1 participates in NO biosynthesis in Arabidopsis, since we isolated a new T-DNA mutant allele, noa1-2, that is impaired in NO production. A recent report characterizing the Nicotiana benthamiana homolog of the AtNOA1 gene is in full agreement with our data, as transgenic tobacco plants with VIGS-mediated silencing of NbNOA1 contained reduced levels of NO (Kato et al., 2008). However, the way AtNOA1 participates in NO biosynthesis is still unknown. The recent identification and characterization of the rif1 mutant point to a putative function of AtNOA1/RIF1 in the correct expression of the plastome-encoded proteins in Arabidopsis (Flores-Pérez et al., 2008). AtNOA1's role in NO biosynthesis could thus be essentially connected to chloroplast (Flores-Pérez et al., 2008) instead of mitochondrial function (Guo and Crawford, 2005). Whether the function of AtNOA1 in NO biosynthesis is related to the overall control of plastome-encoded proteins or to specific plastid targets remains unknown. The recent characterization of AtNOA1 as a GTP-binding protein with a circularly permuted GTPase domain (Moreau et al., 2008) points to a general role of this protein in ribosome function and protein translation in chloroplasts. Whether a specific still unknown protein involved in NO synthesis is not translated in noa1-2 chloroplasts, or there is an indirect effect on the chloroplastic protein synthesis, the overall function of chloroplasts remains unclear and will require more work.
On the other hand, we have also confirmed that a nitrate reductase-based mechanism represents a significant contribution in the biosynthesis of NO in Arabidopsis. In the double nia1nia2 mutant plants, with nearly null activity of the two Arabidopsis nitrate reductases NR1/NIA1 and NR2/NIA2 (Wilkinson and Crawford, 1993), we detected lower NO content than in wild-type plants. However, neither nia1nia2 nor noa1-2 mutant plants were completely impaired in basal NO production, and in fact they displayed only a partial reduction in ABA-induced NO synthesis. To obtain further insight into the function of NR/NIA- and AtNOA1-based biosynthesis of NO in Arabidopsis, we searched for a potential null NO biosynthetic mutant by crossing nia1nia2 and noa1-2 plants. After isolation of the triple homozygous nia1nia2noa1-2 mutant plants, we found an additive effect in the reduction of NO production compared with their parental plants. We did not detect basal or ABA-induced levels of NO in roots of the triple mutant plant. The analysis of NO production by whole seedlings indicated that nia1nia2noa1-2 plants are still able to produce residual (lower than 10% of wild-type plants) levels of NO. This may be explained by the existence of a still unidentified enzyme-based production of NO different from that mediated by NIA/NR and AtNOA1 pathways. That alternative pathway would be significantly less active, if active at all, in roots than in shoots. We are actively looking for such an alternative enzyme-based pathway that may be functional in Arabidopsis. We have also checked that a non-enzyme-based NO production occurred at similar levels either in NO-deficient mutant or in thermally inactivated wild-type seedlings, suggesting that there is a significant contribution of unspecific chemically produced NO in seedlings. Moreover, it is also likely that the contribution of each biosynthetic or chemical pathway may be different in different organs or developmental stages, as the expression patterns of NIA1, NIA2, and AtNOA1 genes vary along development and topology of the plant (Supplemental Fig. S4). According to data from public microarray databases (Bio-Array Resource [http://www.bar.utoronto.ca/] and Genevestigator [https://www.genevestigator.com/gv/index.jsp]), AtNOA1 transcript levels are 15- to 100-fold lower than those of NIA1 and NIA2 transcripts depending on the organ or developmental stage. Based on transcript level data, we should expect a larger contribution of the NIA/NR-mediated pathway to NO biosynthesis. However, our data suggest a larger contribution of the AtNOA1-mediated pathway in NO biosynthesis in roots, which is especially important in ABA-treated roots, and similar contributions of both pathways in basal NO production in shoots. These data suggest that there is no direct correlation between transcript levels of the potential NO biosynthetic genes and the actual levels of endogenous NO. The NO contents in the different organs of the plants are presumably due to the levels of the corresponding encoded proteins or to the activity of them. It is worthy of mention that nitrate reductase activities are more dependent on posttranslational modifications than on transcriptional regulation under certain conditions (Lea et al., 2006) and also that the activity of NIA/NR or AtNOA1 in every plant organ may be strongly modulated by the availability of the corresponding substrates.
The different NO-deficient plants we have used in this work allowed us to propose a new role for the NR/NIA- and AtNOA1-mediated biosynthesis of NO in the regulation of seed germination, dormancy, and resistance to water deficit. As expected from previously reported data on the role of NO in regulating seed dormancy and germination in connection to ABA (Bethke et al., 2004, 2006), we found that several of the NO regulatory effects were exerted through interaction with ABA. NO-deficient mutant seeds were more dormant and showed increased sensitivity to ABA-mediated inhibition of germination than wild-type seeds, which is in agreement with the proposed role for NO decreasing the sensitivity of seeds to ABA (Bethke et al., 2006). This effect was more severe in nia1nia2noa1-2 seeds than in their parental seeds, suggesting that there is a clear correlation with the endogenous NO levels. Accordingly, NO-deficient seedlings had increased basal and induced expression of ABA-responsive genes.
The altered sensitivity to ABA is likely the cause for NO-deficient mutant plants showing increased resistance to water deficit. ABA promotes closure and prevents opening of stomata (Neill et al., 2008). NO has been proposed to be a component of the signaling pathway involved in the stomata closure triggered by ABA (Desikan et al., 2002; Guo et al., 2003; García-Mata and Lamattina, 2007). Since NO-deficient plants are markedly resistant to water deficit, the reduced water losses in NO-deficient plants may be due to hypersensitivity to ABA, thus leading to NO-independent inhibition of stomata opening and enhanced closure by ABA. Actually, we observed that the triple nia1nia2noa1-2 mutant showed a hypersensitive response to dehydration or exogenous ABA treatment in stomata closure or in the inhibition of stomata opening. Moreover, leaves of nia1nia2noa1-2 plants showed a consequent decreased stomatal conductance, a drastic reduction in water loss by transpiration, and no significant alteration in LWP upon water shortage. These data suggest that ABA-mediated regulation of stomata closure may not be necessarily dependent on de novo biosynthesis of NO through any of the proposed NIA/NR- and AtNOA1-mediated pathways. These results contrast with Desikan et al. (2002) proposing that NR/NIA-dependent NO production was essential for ABA-mediated stomatal closure. However, they could not observe a wilting phenotype in nia1nia2 plants, suggesting that ABA can regulate stomatal closure through a mechanism independent of NO biosynthesis. This is also in agreement with enhanced stomata closure detected in nia1nia2 or the single nia1 and nia2 mutants subjected to dehydration (Ribeiro et al., 2009). Moreover, the ABA inhibition of stomata opening was not affected in the nia1nia2noa1-2 mutant, in agreement with previously reported data on nia1nia2 plants (Desikan et al., 2002). Therefore, our data support that NO-deficient mutants displayed an enhanced ABA-mediated stomatal closure and inhibition of opening due to ABA hypersensitivity. This process was essentially mediated by a mechanism independent of de novo NO biosynthesis, as we demonstrate that nia1nia2noa1-2 stomata closed efficiently by ABA despite levels of NO in their guard cells that were undetectable (Supplemental Fig. S5). Moreover, we have observed that the application of a NO scavenger (cPTIO) prevented the inhibition of stomata opening by ABA in wild-type plants, as reported previously (García-Mata and Lamattina, 2007; Ribeiro et al., 2009), whereas constitutive shortage of NO in NO-deficient mutants did not prevent it (Desikan et al., 2002; this work). On the other hand, although it has been reported that changes in endogenous calcium levels play an important role in ABA-mediated inhibition of stomata opening (Roelfsema and Hedrich, 2005) and our data are consistent with that in wild-type seedlings, we found that this process was calcium independent in the nia1nia2noa1-2 mutant. Among calcium-independent signaling components involved in the regulation of stomata closure by ABA, we are currently assessing whether ABA-induced calcium-independent kinases might be deregulated in NO-deficient backgrounds.
This work supports that NR/NIA- and AtNOA1-mediated production of NO are both functional and account for most of the NO biosynthesis in Arabidopsis seedlings. Several ABA-related phenotypes found in NO-deficient plants seem to be due to a function for NO as an endogenous negative regulator of the sensitivity to ABA. Under conditions where ABA-promoted synthesis of NO is impaired, plant cells respond by increasing the sensitivity to the primary stimulus; thus, NO deficiency correlates with enhanced ABA-activated responses.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and NO Treatments
Wild-type Arabidopsis (Arabidopsis thaliana Col-0) was the genetic background of every mutant plant used in this work. Seeds from the nia1nia2 mutant were obtained from the Nottingham Arabidopsis Stock Centre seed bank (N2356). Seeds from noa1-1 were a kind gift from Nigel Crawford (University of California, San Diego), and the noa1-2 allele was obtained from the SAIL collection of T-DNA insertions (SAIL_507_E11). Seeds were grown in soil mixture or Murashige and Skoog (MS) medium as described previously (Castillo and León, 2008).
NO treatments were performed by photochemically mediated release of NO gas from a solution of SNP that was always contained in internal vessels separated from MS medium, as reported previously (Bethke et al., 2006).
Generation of the Triple nia1nia2noa1-2 Mutant
Emasculated flowers from nia1nia2 plants were crossed with pollen from noa1-2 flowers. T1 plants were self-pollinated, and T2 plants were genotyped by PCR. Primers to amplify the wild-type NIA2 gene sequence that is deleted in the nia2 mutant and cleaved-amplified polymorphic sequence primers for the nia1 point mutation are described in Supplemental Table S1. A region containing the nucleotide change (G to A) of nia1 was amplified with cleaved-amplified polymorphic sequence primers and further digested with HhaI to discriminate nia1 from wild-type sequences (Fig. 1B). Plants homozygous for the T-DNA insertion in the AtNOA1 locus were genotyped by PCR with specific primers for AtNOA1 located upstream and downstream of the T-DNA insertion as well as a primer from the T-DNA left border as described in Figure 1. The transcript levels were quantified by reverse transcription (RT)-PCR with specific primers (Supplemental Table S1) for each gene.
Germination Assays
To test ABA sensitivity, seeds were sown in MS medium supplemented or not with 1% (w/v) Suc, 0.8% (w/v) agar, and increasing concentrations of (±)-cis,trans-ABA (Sigma) after stratification 3 d at 4°C. Germination was scored as endosperm rupture, and seedling establishment was assessed by quantifying seedlings with green expanded cotyledons at day 12 after sowing. To test the effect of osmotic stress, seeds were sown in medium supplemented with 125 mm NaCl or 250 mm mannitol and quantified as above. Seeds harvested at the same time were used to carry on these assays. For dormancy assays, freshly harvested seeds from yellowing siliques at stage 17B (Ferrándiz et al., 1999) of the primary bolt were used. Seeds were sown without stratification treatment on MS plates (MS salts, 1% [w/v] Suc, and 0.8% [w/v] agar, pH 5.7), and germination was quantified as endosperm rupture for 6 d.
NO Detection by Fluorescence and Confocal Microscopy
The endogenous levels of NO in roots and stomatal guard cells were determined by staining with DAF-FM DA as described (Guo et al., 2003). To analyze the kinetics of NO production, seedlings were first treated with 50 μm ABA, 1 mm SA, or not treated, as a control, for the indicated times and subsequently loaded with 15 μm DAF-FM DA. NO-associated fluorescence was detected with a fluorescence Nikon Eclipse microscope or with a TCS SL confocal laser-scanning microscope (Leica), using unchanged parameters for every measurement. Fluorescence was quantified as described previously (García-Mata and Lamattina, 2007). The specificity of NO-related fluorescence detection was assessed by treatment with 0.5 mm of the NO scavenger cPTIO (Sigma).
To quantify NO produced by seedlings, 10- to 15-d-old Col-0 and NO-deficient mutant seedlings were submerged in 650 μL of 10 μm DAF-2 in phosphate-buffered saline and kept in darkness for 1 h. The NO-derived fluorescence was then quantified in replicates on a 96-well black plate (Costar) using a TECAN fluorimeter with excitation filter of 492 nm and emission filter of 535 nm. The fluorescence values were the result of subtracting the fluorescence values of thermally inactivated seedlings (heated for 10 min at 99°C), as a control of non-enzyme-based synthesis of NO, from total fluorescence in noninactivated seedlings. Values were normalized to the fresh weight of seedlings.
RNA Isolation and Analysis
Total RNA was isolated from 10- to 12-d-old seedlings, separated, and analyzed by RT-PCR techniques as described previously (Castillo and León, 2008). RAB18 and RD29B transcript levels were quantified by quantitative RT-PCR using specific primers (Supplemental Table S1).
Drought Assays
Plants with similar rosette sizes were selected to minimize biomass-related factors in further drought experiments. NO-deficient seeds were sown before wild-type seeds in Jiffy 7 substrate (Clause), grown under short-day conditions, and exposed to drought as follows. Three independent replicates with 10 plants per experiment grown under standard watering conditions were not watered for 28 d. Evaporation from the soil was minimized by covering pots with plastic wrap film. Pots were weighed and photographed before stopping watering and every 7 d until day 28. To quantify drought resistance, plants were then watered again with the same demineralized water quantity for an additional 14 d, and the percentage of plants surviving drought treatment was calculated for every genotype.
Assays to estimate the transpiration-mediated water loss were performed as follows. Plants (10 individuals per experiment, three independent experiments) were grown under normal watering conditions and subjected to drought stress by completely terminating irrigation and minimizing soil evaporation by covering pots with plastic wrap film. Ten leaves were removed at the indicated time points, weighed, incubated in demineralized water for 3 h, and weighed again. The difference in weight was considered as water loss.
Stomatal Aperture, Conductance, and Psychrometric Measurements
For stomata aperture measurements, the same adaxial regions of the first true leaves of four different plants were captured with a TCS SL confocal laser-scanning microscope (Leica), and width and length of the aperture were measured with ImageJ software (National Institutes of Health). Prior to measurements, plants grown for 10 d under long-day conditions were incubated in stomata-opening buffer (30 mm KCl and 10 mm MES-KOH, pH 6.1) on 24-well multiwell plates (Costar) for 2.5 h under cool-white light (150–200 μE m−2 s−1) at 22°C. To induce stomata closure, plants were incubated in the appropriate ABA concentrations for 2.5 h under light. For inhibition of stomata opening, experiments were done by applying chemicals carefully under green light to avoid light-induced stomata opening as reported previously (García-Mata and Lamattina, 2007).
Stomatal conductances were measured on short-day cultivated plants with the Steady State Difusión Porometer SC-1 (Decagon Devices) as indicated by the manufacturer at 0, 7, and 14 d after stopping watering on the adaxial side of three leaves of six different plants at each time point. LWP was determined in 5-mm leaf discs of short-day cultivated plants using the dew-point method with a C-52 sample chamber (Wescor) connected to a psychrometer switchbox (Ps-10) and to a dew-point microvoltimeter (model HT-33T; Wescor). Measurements were performed according to the manufacturer's instructions at 0, 7, and 14 d after stopping watering. Potential values were determined in three leaf discs obtained from three leaves belonging to six different plants for each time point and genotype.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Rescue of vegetative developmental phenotypes of NO-deficient mutants by NO.
Supplemental Figure S2. The NO-deficient mutant plants show hypersensitivity to ABA in germination assays.
Supplemental Figure S3. Suc does not affect ABA sensitivity in NO-deficient mutant plant germination and establishment.
Supplemental Figure S4. NIA1, NIA2, and AtNOA1 expression levels in different tissues and developmental stages.
Supplemental Figure S5. NO in guard cells of wild-type and nia1nia2noa1-2 stomata.
Supplemental Table S1. Oligonucleotides used for genotyping and RT-PCR.
Supplementary Material
Acknowledgments
We gratefully acknowledged the assistance of M.D. Gómez (Instituto de Biología Molecular y Celular de Plantas [IBMCP]) in microscopy techniques. We thank Silvia Rubio (IBMCP) for help in ABA sensitivity tests of seed germination and Pedro Rodriguez (IBMCP) for critical reading of the manuscript. We also thank Manuel Agustí and Carlos Mesejo (IAM) for assistance and discussion on psychrometric measurements.
This work was supported by Ministerio de Educación y Ciencia and Fondo Europeo de Desarrollo Regional funds (grant nos. GEN2003–20477–C02–02, BIO2005–00222, and BIO2008–00839 to J.L.) and by the Consejo Superior de Investigaciones Científicas (Bancaja Program fellowship to J.L.-J.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: José León (jleon@ibmcp.upv.es).
The online version of this article contains Web-only data.
References
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA (2007) Death don't have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L (2000) Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210 215–221 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Gubler F, Jacobsen JV, Jones RL (2004) Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta 219 847–855 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Aoyama N, Chung YY, Still DW, Jones RL (2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol 143 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Libourel IGL, Jones RL (2006) Nitric oxide reduces seed dormancy in Arabidopsis. J Exp Bot 57 517–526 [DOI] [PubMed] [Google Scholar]
- Castillo MC, León J (2008) Expression of the beta-oxidation gene 3-ketoacyl-CoA thiolase 2 (KAT2) is required for the timely onset of natural and dark-induced leaf senescence in Arabidopsis. J Exp Bot 59 2171–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM (2006) Mechanisms for nitric oxide synthesis in plants. J Exp Bot 57 471–478 [DOI] [PubMed] [Google Scholar]
- Crawford NM, Galli M, Tischner R, Heimer YM, Okamoto M, Mack A (2006) Response to Zemojtel et al: Plant nitric oxide synthase: back to square one. Trends Plant Sci 11 526–527 [Google Scholar]
- Delledonne M (2005) NO news is good news for plants. Curr Opin Plant Biol 8 390–396 [DOI] [PubMed] [Google Scholar]
- del Río LA, Corpas FJ, Barroso JB (2004) Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry 65 783–792 [DOI] [PubMed] [Google Scholar]
- Desikan R, Graffiths R, Hancock J, Neill S (2002) A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc Natl Acad Sci USA 99 16314–16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J, Klessig DF (1999) Nitric oxide as a signal in plants. Curr Opin Plant Biol 2 369–374 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Pelaz S, Yanofsky MF (1999) Control of carpel and fruit development in Arabidopsis. Annu Rev Biochem 68 321–354 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Arizumi T, Steber C (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59 387–415 [DOI] [PubMed] [Google Scholar]
- Flores-Pérez U, Sauret-Güeto S, Gas E, Jarvis P, Rodríguez-Concepción M (2008) A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. Plant Cell 20 1303–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2007) Abscisic acid (ABA) inhibits light-induced stomatal opening through calcium- and nitric oxide-mediated signaling pathways. Nitric Oxide 17 143–151 [DOI] [PubMed] [Google Scholar]
- Graham IA (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59 115–142 [DOI] [PubMed] [Google Scholar]
- Grün S, Lindermayr C, Sell S, Durner J (2006) Nitric oxide and gene regulation in plants. J Exp Bot 57 507–516 [DOI] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8 183–187 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Crawford NM (2005) Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell 17 3436–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Okamoto M, Crawford NM (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302 100–103 [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Stoimenova M, Kaiser WM (2005) In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J Exp Bot 56 2601–2609 [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305 1968–1971 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D (2008) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13 7–13 [DOI] [PubMed] [Google Scholar]
- Jeannette E, Rona JP, Bardat F, Cornel D, Sotta B, Miginiac E (1999) Induction of RAB18 gene expression and activation of K+ outward rectifying channels depend on an extracellular perception of ABA in Arabidopsis thaliana suspension cells. Plant J 18 13–22 [DOI] [PubMed] [Google Scholar]
- Kato H, Asai S, Yamamoto-Katou A, Yoshioka H, Doke N, Kawakita K (2008) Involvement of NbNOA1 in NO production and defense responses in INF1-treated Nicotiana benthamiana. J Gen Plant Pathol 74 15–23 [Google Scholar]
- Knowles RG, Moncada S (1994) Nitric oxide synthases in mammals. Biochem J 298 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Z, Bartha B, Erdei L (2008) Exogenous auxin-induced NO synthesis is nitrate reductase-associated in Arabidopsis thaliana root primordia. J Plant Physiol 165 967–975 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5 33–36 [DOI] [PubMed] [Google Scholar]
- Lamattina L, García-Mata C, Graziano M, Pagnussat G (2003) Nitric oxide: the versatility of an extensive signal molecule. Annu Rev Plant Biol 54 109–136 [DOI] [PubMed] [Google Scholar]
- Lamotte O, Courtois C, Barnavon L, Pugin A, Wendehenne D (2005) Nitric oxide in plants: the biosynthesis and cell signalling properties of a fascinating molecule. Planta 221 1–4 [DOI] [PubMed] [Google Scholar]
- Lea US, Leydecker MT, Quilleré I, Meyer C, Lillo C (2006) Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol 140 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126 969–980 [DOI] [PubMed] [Google Scholar]
- Meyer C, Lea US, Provan F, Kaiser WM, Lillo C (2005) Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynth Res 83 181–189 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Lamb C, Zeier J (2007) Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ 30 39–52 [DOI] [PubMed] [Google Scholar]
- Modolo LV, Augusto O, Almeida IM, Magalhaes JR, Salgado I (2005) Nitrite as the major source of nitric oxide production by Arabidopsis thaliana in response to Pseudomonas syringae. FEBS Lett 579 3814–3820 [DOI] [PubMed] [Google Scholar]
- Moreau M, Lee GI, Wang Y, Crane BR, Klessig DF (2008) AtNOS/A1 is a functional Arabidopsis thaliana cGTPase and not a nitric oxide synthase. J Biol Chem 283 32957–32967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill S, Barros R, Bright J, Desikan R, Hancock J, Harrison J, Morris P, Ribeiro D, Wilson I (2008) Nitric oxide, stomatal closure, and abiotic stress. J Exp Bot 59 165–176 [DOI] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT (2002) Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol 128 13–16 [PMC free article] [PubMed] [Google Scholar]
- Parihar A, Parihar MS, Chen Z, Ghafourifar P (2008. a) mAtNOS1 induces apoptosis of human mammary adenocarcinoma cells. Life Sci 82 1077–1082 [DOI] [PubMed] [Google Scholar]
- Parihar MS, Parihar A, Chen Z, Nazarewicz R, Ghafourifar P (2008. b) mAtNOS1 regulates mitochondrial functions and apoptosis of human neuroblastoma cells. Biochim Biophys Acta 1780 921–926 [DOI] [PubMed] [Google Scholar]
- Pedroso MC, Magalhaes JR, Durzan D (2000) A nitric oxide burst precedes apoptosis in angiosperm and gymnosperm callus cells and foliar tissues. J Exp Bot 51 1027–1036 [DOI] [PubMed] [Google Scholar]
- Planchet E, Gupta KJ, Sonoda M, Kaiser WM (2005) Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J 41 732–743 [DOI] [PubMed] [Google Scholar]
- Planchet E, Sonoda M, Zeier J, Kaiser WM (2006) Nitric oxide (NO) as an intermediate in the cryptogein-induced hypersensitive response: a critical re-evaluation. Plant Cell Environ 29 59–69 [DOI] [PubMed] [Google Scholar]
- Ribeiro DM, Desikan R, Bright J, Confraria A, Harrison J, Hancock JT, Barros RS, Neill SJ, Wilson ID (2009) Differential requirement for NO during ABA-induced stomatal closure in turgid and wilted leaves. Plant Cell Environ 32 46–57 [DOI] [PubMed] [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53 103–110 [PubMed] [Google Scholar]
- Roelfsema MR, Hedrich R (2005) In the light of stomatal opening: new insights into ‘the Watergate’. New Phytol 167 665–691 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Perazzolli M, Zago ED, Delledonne M (2004) Nitric oxide signaling functions in plant-pathogen interactions. Cell Microbiol 6 795–803 [DOI] [PubMed] [Google Scholar]
- Sarath G, Bethke PC, Jones R, Baird LM, Hou G, Mitchell RB (2006) Nitric oxide accelerates seed germination in warm-season grasses. Planta 223 1154–1164 [DOI] [PubMed] [Google Scholar]
- Seligman K, Saviani EE, Oliveira HC, Pinto-Maglio CA, Salgado I (2008) Floral transition and nitric oxide emission during flower development in Arabidopsis thaliana is affected in nitrate reductase-deficient plants. Plant Cell Physiol 49 1112–1121 [DOI] [PubMed] [Google Scholar]
- Simpson GG (2005) NO flowering. Bioessays 27 239–241 [DOI] [PubMed] [Google Scholar]
- Torreilles J (2001) Nitric oxide: one of the more conserved and widespread signaling molecules. Front Biosci 6 D1161–D1172 [DOI] [PubMed] [Google Scholar]
- Vardi A, Bidle KD, Kwityn C, Hirsh DJ, Thompson SM, Callow JA, Falkowski P, Bowler C (2008) A diatom gene regulating nitric-oxide signaling and susceptibility to diatom-derived aldehydes. Curr Biol 18 895–899 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Durner J, Klessig DF (2004) Nitric oxide: a new player in plant signalling and defense responses. Curr Opin Plant Biol 7 449–455 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J (2001) Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci 6 177–183 [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Crawford NM (1993) Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol Gen Genet 239 289–297 [DOI] [PubMed] [Google Scholar]
- Wulff A, Oliveira HC, Saviani EE, Salgado I (2009) Nitrite reduction and superoxide-dependent nitric oxide degradation by Arabidopsis mitochondria: influence of external NAD(P)H dehydrogenases and alternative oxidase in the control of nitric oxide levels. Nitric Oxide 21 132–139 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236 331–340 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y, Takahashi S (1999) An alternative pathway for nitric oxide production in plants: new features of an old enzyme. Trends Plant Sci 4 128–129 [DOI] [PubMed] [Google Scholar]
- Zeier J, Delledonne M, Mishina T, Severi E, Sonoda M, Lamb C (2004) Genetic elucidation of nitric oxide signaling in incompatible plant-pathogen interactions. Plant Physiol 136 2875–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemojtel T, Fröhlich A, Palmieri MC, Kolanczyk M, Mikula I, Wyrwicz LS, Wanker EE, Mundlos S, Vingron M, Martasek P, et al (2006. a) Plant nitric oxide synthase: a never-ending story? Trends Plant Sci 11 524–525; author reply 526–527 [DOI] [PubMed] [Google Scholar]
- Zemojtel T, Kolanczyk M, Kossler N, Stricker S, Lurz R, Mikula I, Duchniewicz M, Schuelke M, Ghafourifar P, Martasek P, et al (2006. b) Mammalian mitochondrial nitric oxide synthase: characterization of a novel candidate. FEBS Lett 580 455–462 [DOI] [PubMed] [Google Scholar]
- Zottini M, Costa A, De Michele R, Ruzzene M, Carimi F, Lo Schiavo F (2007) Salicylic acid activates nitric oxide synthesis in Arabidopsis. J Exp Bot 58 1397–1405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.